SUMMARY
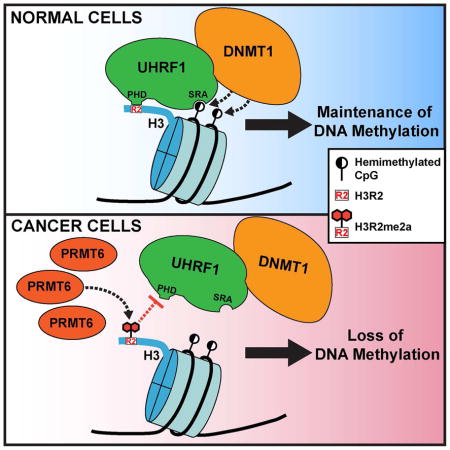
DNA methylation plays crucial roles in chromatin structure and gene expression. Aberrant DNA methylation patterns, including global hypomethylation and regional hypermethylation, are associated with cancer and implicated in oncogenic events. How DNA methylation is regulated in developmental and cellular processes and dysregulated in cancer is poorly understood. Here, we show that PRMT6, a protein arginine methyltransferase responsible for asymmetric dimethylation of histone H3 arginine 2 (H3R2me2a), negatively regulates DNA methylation and that PRMT6 upregulation contributes to global DNA hypomethylation in cancer. Mechanistically, PRMT6 overexpression impairs chromatin association of UHRF1, an accessory factor of DNMT1, resulting in passive DNA demethylation. The effect is likely due to elevated H3R2me2a, which inhibits the interaction between UHRF1 and histone H3. Our work identifies a mechanistic link between protein arginine methylation and DNA methylation, which is disrupted in cancer.
Keywords: PRMT6, arginine methylation, UHRF1, DNMT1, DNA methylation, cancer
In Brief
Veland et al. find that PRMT6, an arginine methyltransferase responsible for histone H3 arginine 2 (H3R2) methylation, negatively regulates maintenance DNA methylation by impairing UHRF1 recruitment to chromatin. The authors also find that PRMT6 upregulation contributes to global DNA hypomethylation in cancer cells.
INTRODUCTION
In mammals, DNA methylation (5-methylcytosine, 5mC) is mostly restricted to CpG dinucleotides and plays crucial roles in many biological processes. Aberrant DNA methylation is associated with cancer. Specifically, cancer cells generally exhibit global hypomethylation and loci-specific hypermethylation, which are implicated in genomic instability and tumor suppressor silencing, respectively (Baylin and Jones, 2016). However, the mechanisms underlying these alterations remain largely unclear.
DNA methylation patterns are established by the de novo DNA methyltransferases DNMT3A and DNMT3B and maintained primarily by the maintenance enzyme DNMT1. DNMT1 recruitment to hemi-methylated CpG sites during DNA replication depends on UHRF1, a multi-domain protein (Bostick et al., 2007, Sharif et al., 2007). The SRA (SET- and RING-associated) domain of UHRF1 preferentially binds hemi-methylated DNA and plays an important role in loading DNMT1 onto newly synthesized DNA (Bostick et al., 2007, Sharif et al., 2007, Liu et al., 2013). The RING domain-mediated ubiquitination of lysine residues in the N-terminal tail of histone H3 promotes DNMT1 association with H3 (Nishiyama et al., 2013, Qin et al., 2015, Harrison et al., 2016). Moreover, the TTD (tandem Tudor domain) and PHD (plant homeodomain) cooperatively interact with the N-terminal tail of H3 by recognizing a specific histone modification signature. Specifically, the TTD exhibits affinity for di- and tri-methylated lysine 9 (H3K9me2/me3) (Rothbart et al., 2012, Rothbart et al., 2013, Liu et al., 2013), whereas PHD-mediated binding to H3 is disrupted by arginine 2 (H3R2) methylation (Rajakumara et al., 2011, Hu et al., 2011, Wang et al., 2011, Lallous et al., 2011). Recent studies show that the SRA domain interaction with DNA stimulates TTD-PHD-mediated H3 binding and that hemi-methylated DNA recognition allosterically activates RING domain-mediated H3 ubiquitination (Harrison et al., 2016, Fang et al., 2016). These data suggest that UHRF1 targets DNMT1 to newly replicated DNA through complex interactions with chromatin.
The observation that the UHRF1 PHD specifically binds unmodified, but not H3R2-methylated, N-terminal tail of H3 suggests that DNA methylation may be modulated by H3R2 methylation. Arginine methylation is carried out by the PRMT family, consisting of nine members (Bedford and Richard, 2005). PRMT6 is the primary enzyme responsible for asymmetric dimethylation of H3R2 (H3R2me2a) (Guccione et al., 2007, Hyllus et al., 2007, Iberg et al., 2008). Notably, PRMT6 is frequently overexpressed in cancer cells and implicated in tumorigenic functions (Yang and Bedford, 2013). In this study, we show that PRMT6 negatively regulates DNA methylation by impairing UHRF1 association with chromatin and that its overexpression contributes to global DNA hypomethylation in cancer.
RESULTS
PRMT6 overexpression induces global DNA hypomethylation in mESCs
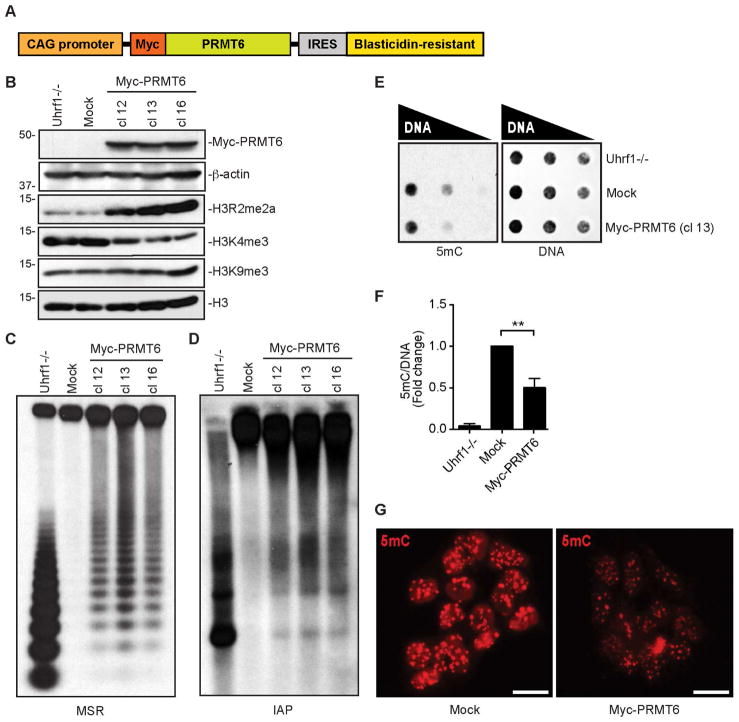
Given that H3R2 methylation disrupts UHRF1-H3 interaction (Rajakumara et al., 2011, Hu et al., 2011, Wang et al., 2011, Lallous et al., 2011), we hypothesized that PRMT6, the primary enzyme responsible for H3R2me2a, regulates DNA methylation. To test the idea, we generated mouse embryonic stem cell (mESC) clones overexpressing human PRMT6 by stable transfection of a bicistronic vector expressing Myc-tagged PRMT6 and the blasticidin-resistant gene (Figure 1A). mESCs offer an ideal experimental system for studying DNA methylation regulators, as their survival and proliferation are not affected by DNA methylation loss (Tsumura et al., 2006).
Figure 1. Overexpression of PRMT6 in mESCs induces global DNA hypomethylation.
(A) Diagram of Myc-PRMT6 plasmid.
(B) Western blots showing the levels of Myc-PRMT6 and histone marks in stable mESC clones. Mock, mESCs transfected with empty vector. Uhrf−/−, Uhrf1 knockout mESCs.
(C and D) Southern blots showing DNA methylation at MSR (C) and IAP (D) after digestion of genomic DNA with the methylation-sensitive restriction enzyme HpaII.
(E) Dot blot analysis of genomic DNA with 5mC antibody (left). The same membrane was stained with SYTOX Green to verify equal DNA loading (right).
(F) Quantification of data in (E) by densitometry using Image J. Shown are relative 5mC levels (mean + SD from three independent experiments). Paired t test was used to determine statistical significance. **P < 0.01.
(G) IF analysis with 5mC antibody. Scale bars, 15 μm.
As expected, PRMT6 overexpression resulted in elevated H3R2me2a levels, which correlated with reduced H3K4me3 levels, consistent with previous reports that H3R2 methylation antagonizes H3K4me3 (Guccione et al., 2007, Kirmizis et al., 2007, Hyllus et al., 2007). As controls, H3K9me3 and total H3 showed no alterations (Figure 1B). Notably, PRMT6 overexpression also led to increases in arginine methylation of non-histone proteins (Figure S1A). The stable clones maintained the mESC state, as judged by colony morphology, growth rates, and expression of the pluripotency factors Nanog, Sox2 and Oct4 (Figures S1B–S1D).
To assess the impact of PRMT6 overexpression on DNA methylation, we first analyzed the minor satellite repeats (MSR) and intracisternal A-particle (IAP) retrotransposons. Southern blot analysis of genomic DNA digested with the methylation-sensitive restriction enzyme HpaII revealed that cells expressing Myc-PRMT6 exhibited marked DNA hypomethylation compared to cells transfected with the empty vector (mock) (Figures 1C and 1D). We then confirmed global DNA hypomethylation in PRMT6-overexpressing mESCs with dot blot and immunofluorescence (IF) analyses using a 5mC antibody (Figures 1E–1G). The effect of PRMT6 on DNA methylation depends on its catalytic activity, as an inactive PRMT6 mutant (E155Q) failed to induce DNA hypomethylation in mESCs (Figures S2A–S2C). Together, these results demonstrate that PRMT6 and its methyltransferase activity negatively regulate global DNA methylation.
PRMT6 overexpression impairs Uhrf1 association with chromatin
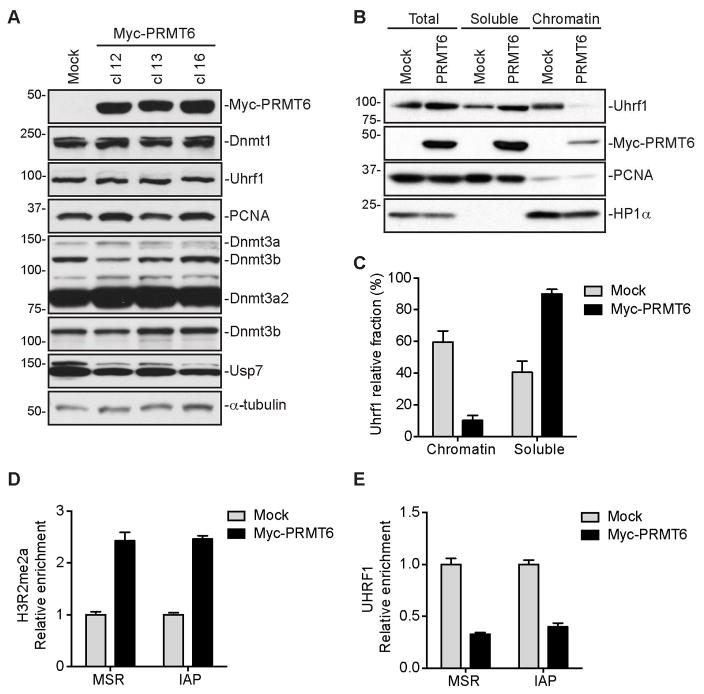
To determine the mechanism by which PRMT6 induces DNA hypomethylation, we first examined the expression of Dnmts, as well as key regulators of DNA methylation, including Uhrf1 (murine Uhrf1 is also known as Np95), PCNA and Usp7. None of the proteins showed changes in PRMT6-overexpressing cells (Figure 2A). We then tested the hypothesis that increased H3R2me2a negatively affects Uhrf1 binding to chromatin. Nuclear fractionation experiments revealed that Uhrf1 chromatin association reduced dramatically in PRMT6-overexpressing mESCs compared to mock mESCs (~10% vs. ~60%) (Figures 2B and 2C). Chromatin immunoprecipitation (ChIP) analysis confirmed that increased H3R2me2a levels correlated with decreased Uhrf1 enrichment at MSR and IAP regions (Figures 2D and 2E). These results support our hypothesis that higher H3R2me2a levels induced by PRMT6 overexpression impair Uhrf1 association with chromatin, resulting in a failure in maintaining DNA methylation.
Figure 2. Uhrf1 chromatin association is impaired in mESCs overexpressing PRMT6.
(A) Western blots showing the levels of DNA methylation enzymes and regulators. Note that mESCs express two major Dnmt3a isoforms (Dnmt3a and Dnmt3a2) and that the Dnmt3a antibody cross-reacts with Dnmt3b.
(B) Nuclear fractionation assay showing Uhrf1 chromatin association. PCNA and HP1α were used as controls for soluble and chromatin-associated proteins, respectively.
(C) Quantification of data in (B) by densitometry using Image J. Shown are percentages of soluble and chromatin-associated Uhrf1 in each sample (mean + SD from three independent experiments).
(D and E) ChIP assays showing relative enrichment of H3R2me2a (D) and Uhrf1 (E) at MSR and IAP regions (mean + SD from three independent experiments).
PRMT6 upregulation correlates with DNA hypomethylation in human cancers
Global DNA hypomethylation is a hallmark of cancer cells (Baylin and Jones, 2016), but the underlying mechanisms are poorly understood. Given that overexpression of PRMT6 is reported in multiple types of cancer (Yang and Bedford, 2013), we postulated that PRMT6 upregulation might contribute to global DNA hypomethylation.
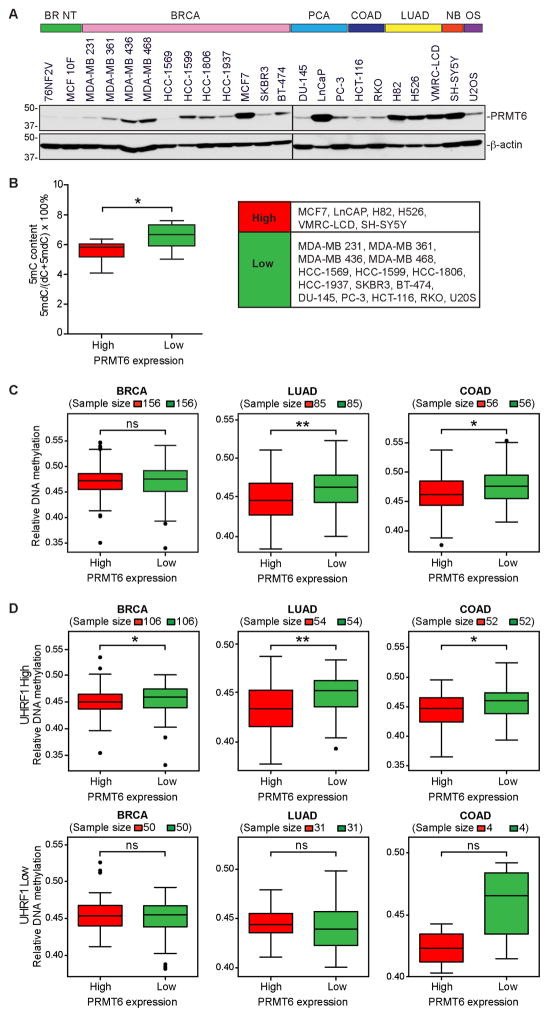
We first assessed the correlation between PRMT6 expression and DNA methylation in a panel of human cell lines. Compared to non-tumorigenic breast cell lines 76NF2V and MCF 10F, most cancer cell lines examined exhibited upregulation of PRMT6, although the levels varied greatly (Figure 3A). The 5mC content in the cancer cell lines was measured by liquid chromatography and tandem mass spectrometry (LC-MS/MS) (Table S1). Based on the relative levels of PRMT6 (Figure 3A), we divided the cancer cell lines into two groups: the PRMT6-high group had significantly lower levels of 5mC than the PRMT6-low group (Figure 3B).
Figure 3. PRMT6 expression inversely correlates with DNA methylation in human cancer cells.
(A) Western blots showing PRMT6 levels in human cancer cell lines. BR NT, breast non-tumorigenic; BRCA, breast cancer; PCA, prostate cancer; COAD, colorectal adenocarcinoma; LUAD, lung adenocarcinoma; NB, neuroblastoma; OS, osteosarcoma.
(B) Comparison between PRMT6-high and PRMT6-low cell lines for 5mC levels (determined by LC-MS/MS, see Table S1).
(C and D) Correlation of PRMT6 expression and DNA methylation data from the TCGA database. The mean DNA methylation levels between cancer samples with the highest (top 20%) and lowest (bottom 20%) PRMT6 expression in each cancer type were compared, either without considering UHRF1 expression (C) or by dividing all samples into UHRF1-high (upper 70%) and UHRF1-low (lower 30%) groups before analysis (D). Note that the 30% cutoff for UHRF1 expression was based on merged datasets of the three cancer types and, therefore, the sample numbers of UHRF1-high and UHRF1-low groups in each cancer type do not make up precisely 70% and 30%, respectively.
Wilcoxon rank sum non-parametric test with two-tailed P value was used to determine the significance of differences in (B–D). *P < 0.05; **P < 0.01; ns, not significant.
We next asked whether PRMT6 upregulation correlates with DNA hypomethylation in primary tumor samples by employing The Cancer Genome Atlas (TCGA) database. Data downloaded from the cBioPortal for Cancer Genomics showed wide variations in PRMT6 expression in all cancer types (Figure S3). We selected three common cancer types, i.e. breast cancer, lung cancer and colorectal cancer, because a large amount of DNA methylation data is available in the TCGA database. When all samples of each cancer type were included in the analyses, no clear correlation was observed between PRMT6 expression and DNA methylation levels, which is not surprising because both PRMT6 and DNA methylation levels are highly variable. However, comparisons of the samples with the highest 20% and lowest 20% of PRMT6 expression revealed a significant inverse correlation between PRMT6 expression and DNA methylation in lung cancer and colorectal cancer, but not in breast cancer (Figure 3C). Based on our hypothesis, DNA methylation may not be affected by PRMT6 if UHRF1 expression is low. Therefore, we first divided the samples into UHRF1-high (upper 70%) and UHRF1-low (lower 30%) groups and then compared DNA methylation levels in samples with high (top 20%) and low (bottom 20%) PRMT6 expression. Consistent with our hypothesis, we observed a significant inverse correlation between PRMT6 expression and DNA methylation in the UHRF1-high groups of all the three cancer types (Figure 3D upper panel) and no correlations in the UHRF1-low groups (Figure 3D lower panel). Together, these data suggest that PRMT6 upregulation contributes to global DNA hypomethylation in cancer.
PRMT6 depletion or inhibition restores global DNA methylation in MCF7 cells
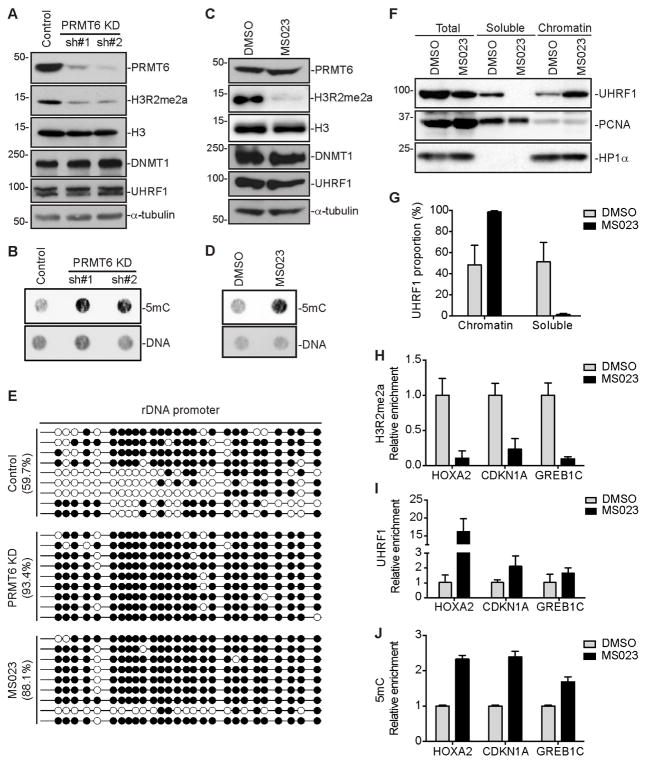
To validate the significance of PRMT6 upregulation in DNA hypomethylation, we assessed the impact of PRMT6 knockdown (KD) and inhibition in MCF7 cells, a breast cancer cell line with high PRMT6 expression (Figure 3A). Stable expression of PRMT6 shRNAs, which efficiently depleted PRMT6 (Figure 4A), or treatment with MS023, a PRMT inhibitor that is potent for PRMT6 (Eram et al., 2016), resulted in substantial decreases in H3R2me2a, but no changes in DNMT1 and UHRF1 levels (Figures 4A and 4C). Consistent with previous reports (Neault et al., 2012, Stein et al., 2012, Phalke et al., 2012, Kleinschmidt et al., 2012, Eram et al., 2016), PRMT6 KD or MS023 treatment resulted in defects in MCF7 proliferation (Figures S4A–S4D). Dot blot analysis showed that PRMT6 depletion or inhibition led to increases in global DNA methylation (Figures 4B and 4D). We verified the results by bisulfite sequencing analysis of a region in the 45S ribosomal DNA (rDNA) promoter, which is partially methylated in MCF7 cells (Karahan et al., 2015). Indeed, PRMT6 KD or MS023-treated cells had markedly higher levels of DNA methylation (~90%) than control cells (~60%) (Figure 4E). Both de novo and maintenance methylation probably contributed to the restoration of DNA methylation levels.
Figure 4. PRMT6 depletion or inhibition restores DNA methylation in MCF7 cells.
(A and C) Western blot analysis of MCF7 cells stably transfected with PRMT6 shRNAs (A) or treated with MS023 (C).
(B and D) 5mC dot blot analysis of samples in (A) and (C), respectively. DNA loading was verified by SYTOX Green staining.
(E) Bisulfite sequencing analysis of a 45S rDNA promoter region containing 27 CpG sites. Open circles, unmethylated CpGs; Filled circles, methylated CpGs.
(F) Nuclear fractionation of MCF7 cells treated with MS023 for UHRF1 chromatin association. PCNA and HP1α were used as controls for soluble and chromatin-associated proteins, respectively.
(G) Quantification of data in (F) by densitometry using Image J. Shown are percentages of soluble and chromatin-associated UHRF1 in each sample (mean + SD from three independent experiments).
(H–J) ChIP or MeDIP assays showing relative enrichment of H3R2me2a (H) and UHRF1 (I) or relative DNA methylation levels (J) at HOXA2, CDKN1A and GREB1C promoter regions (mean + SD from three independent experiments).
MS023 treatment was performed at 10 μM for 4 days for all experiments.
See also Figure S4.
In agreement with our hypothesis, UHRF1 chromatin association was substantially enhanced in cells treated with MS023 (Figures 4F and 4G). In addition, we analyzed UHRF1 occupancy at the promoter regions of three PRMT6 target genes, HOXA2, CDKN1A and GREB1C (Hyllus et al., 2007, Kleinschmidt et al., 2012, Phalke et al., 2012, Mann et al., 2014). ChIP and methylated-DNA immunoprecipitation (MeDIP) analyses confirmed that H3R2me2a reduction induced by MS023 correlated with increases in UHRF1 enrichment and DNA methylation levels at these loci (Figures 4H–4J). Collectively, these results suggest that, in MCF7 cells, PRMT6 upregulation plays a critical role in inducing global DNA hypomethylation by impairing UHRF1 chromatin binding.
DISCUSSION
Various epigenetic mechanisms act cooperatively in regulating chromatin structure and gene activity. While it has been well established that DNA methylation and several histone modifications, notably lysine methylation, are functionally linked (Rothbart et al., 2012, Du et al., 2015), much less clear is the crosstalk between DNA methylation and arginine methylation. In this study, we demonstrate that PRMT6 is a negative regulator of DNA methylation. We show that overexpression of PRMT6 in mESCs compromises Uhrf1 association with chromatin. Consistent with previous evidence that H3R2 methylation inhibits Uhrf1-H3 interaction (Rajakumara et al., 2011, Hu et al., 2011, Wang et al., 2011, Lallous et al., 2011, Rothbart et al., 2013), our results indicate that the catalytic activity of PRMT6 is required for its effect on DNA methylation. Thus, we propose that PRMT6, by generating H3R2me2a, impairs recruitment of the Dnmt1-Uhrf1 complex to newly replicated DNA, resulting in passive DNA demethylation. However, we cannot rule out the possibility that PRMT6-mediated methylation of other arginine residues on histone or non-histone proteins also contributes to DNA methylation changes.
While the relevance of PRMT6 in regulating DNA methylation in normal developmental and cellular processes remains to be determined, we provide evidence that PRMT6 overexpression contributes to global DNA hypomethylation in cancer cells. We show that PRMT6 expression levels inversely correlate with DNA methylation levels in both cancer cell lines and primary cancer tissues. Moreover, PRMT6 depletion or inhibition leads to restoration of DNA methylation levels in MCF7 cells, suggesting a causal link between PRMT6 overexpression and DNA hypomethylation. While most PRMT6 high-expressing cell lines have relatively low levels of DNA methylation, some cell lines that are severely hypomethylated (e.g., MDA-MB 231, SKBR3) show no obvious PRMT6 upregulation (Figure 3A and Table S1), suggesting that multiple mechanisms are involved in DNA hypomethylation in cancer. Some of the mechanisms likely affect the functionality of the DNMT1-UHRF1 complex. UHRF1 could positively or negatively impact DNA methylation. On one hand, UHRF1 is essential for DNMT1 recruitment to newly replicated DNA to maintain DNA methylation (Bostick et al., 2007, Sharif et al., 2007). On the other hand, UHRF1, an E3 ubiquitin ligase, can ubiquitinate UHRF1 itself, DNMT1 and DNMT3A, leading to their degradation (Jenkins et al., 2005, Du et al., 2010, Qin et al., 2011, Jia et al., 2016). UHRF1 is highly expressed in many cancers (Yang and Bedford, 2013), which likely contributes to DNA methylation changes. Indeed, overexpression of human UHRF1 in zebrafish hepatocytes leads to Dnmt1 mislocalization and degradation, DNA hypomethylation, and hepatocellular carcinoma (Mudbhary et al., 2014).
PRMT6 is overexpressed in multiple types of cancer. In breast cancer, PRMT6 levels positively correlate with tumor stages, suggesting that PRMT6 may contribute to tumor progression (Phalke et al., 2012). Nevertheless, we observed that the benign breast cancer cell line MCF7 has higher levels of PRMT6 than the aggressive cell line MDA-MB 231 (Figure 3A), indicating that the relationship between PRMT6 expression and cancer invasiveness is complex. The differences could be attributed to the different cancer subtypes that the cell lines represent. How PRMT6 overexpression contributes to tumorigenesis remains to be determined. PRMT6 generally functions as a transcriptional repressor, although it has also been shown to act as a co-activator of nuclear receptors such as estrogen receptor (Yang and Bedford, 2013). In this study, we demonstrate that PRMT6 overexpression contributes to DNA hypomethylation, which could lead to genomic instability and expression of cancer-promoting genes. Importantly, the effect of PRMT6 on DNA methylation is reversible, as PRMT6 depletion or inhibition restores DNA methylation levels in MCF7 cells. This raises the possibility of targeting PRMT6 for cancer therapy. However, it remains to be determined to what extent PRMT6-dependent DNA hypomethylation contributes to tumorigenesis and maintenance of the tumor phenotype.
EXPERIMENTAL PROCEDURES
Cell culture and manipulations
mESCs culture and generation of stable clones were described previously (Dan et al., 2017). Human cancer cell lines were cultured according to instructions of American Type Culture Collection. For PRMT6 KD, MCF7 cells transfected with shRNA plasmids were selected and maintained in medium containing 1 μg/ml of puromycin. To inhibit PRMT6 activity, cells were treated with 10 μM of MS023 (Eram et al., 2016).
Nuclear fractionation
Nuclear fractionation in mESCs was carried out as described previously (Mendez and Stillman, 2000) with the following modification: buffer B was replaced by buffer N [15 mM Tris-HCl (pH 7.5), 200 mM NaCl, 60 mM KCl, 5 mM MgCl2, 1 mM CaCl2, 0.3% NP-40, and 1X protease inhibitor cocktail (Thermo Scientific)]. For experiments in MCF7 cells, NaCl in buffer N was adjusted to 100 mM.
DNA methylation analyses
Southern blot, dot blot and IF analyses of DNA methylation were performed as described previously (Dan et al., 2017). For bisulfite sequencing analysis, bisulfite conversion was performed using EZ DNA Methylation Kit (Zymo), PCR products cloned using NEB PCR cloning kit, and individual clones sequenced. Quantification of 5mC content was done by LC-MS/MS.
ChIP and MeDIP
ChIP assays were performed as described previously (Dan et al., 2017). For MeDIP, RNA-free genomic DNA was sonicated and incubated with 2 μg of 5mC antibody (Millipore) in MeDIP buffer [10 mM Na-Phosphate (pH 7.0), 140 mM NaCl, and 0.05% Triton X-100] for 3 hours at 4°C. Dynabeads M-280 sheep anti-mouse IgG (Thermo Scientific) were used to precipitate DNA complexes.
Bioinformatics and statistical analysis
PRMT6 expression data and DNA methylation data for breast cancer, lung adenocarcinoma and colorectal adenocarcinoma samples in TCGA database were analyzed. The mean DNA methylation levels of samples with the highest (top 20%) and lowest (bottom 20%) PRMT6 expression were compared either without taking UHRF1 expression into consideration or by first dividing the samples into UHRF1-high (upper 70%) and UHRF1-low (lower 30%) groups. Wilcoxon rank sum non-parametric test with two-tailed P value was used to determine the significance of differences in Figures 3B–3D. Paired t test with two-tailed P value was used for Figure 1F.
Supplementary Material
Highlights.
PRMT6 overexpression induces histone H3R2me2a and DNA hypomethylation in mESCs
Uhrf1 association with chromatin is impaired in mESCs overexpressing PRMT6
PRMT6 upregulation correlates with DNA hypomethylation in cancer cells
PRMT6 depletion or inhibition restores DNA methylation in MCF7 cells
Acknowledgments
We thank S. Richard for PRMT6 cDNA, K. Keyomarsi for 76NF2V cell line, the shRNA and ORFeome Core at MD Anderson Cancer Center (MDACC) for the shRNA plasmids, and the Institute for Applied Cancer Science at MDACC for the PRMT inhibitor MS023. This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT, R1108 to T.C. and RP170002 to NGS Core at MDACC) and the National Institutes of Health (1R01AI12140301A1 to T.C., R01GM110058 to B.D.S., R00CA181343 and R35GM124736 to S.B.R., DK062248 to M.T.B., and CA16672 to CCSG Cores at MDACC). N.V., S.G. and J.D. were supported by scholarships from the Center for Cancer Epigenetics at MDACC. N.V. was also supported by a CPRIT Research Training Award (RP140106) and was awarded the Andrew Sowell-Wade Huggins Scholarship Fund. T.C. is a CPRIT Scholar in Cancer Research.
Footnotes
AUTHOR CONTRIBUTIONS
N.V., B.D.S., S.B.R., M.T.B. and T.C. designed the experiments and analyzed the data. N.V., S.H., S.G., J.D. and S.B.R. performed the experiments. Y.Z. did bioinformatics and statistical analyses. N.V. and T.C. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760– 1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Dan J, Rousseau P, Hardikar S, Veland N, Wong J, Autexier C, Chen T. Zscan4 Inhibits Maintenance DNA Methylation to Facilitate Telomere Elongation in Mouse Embryonic Stem Cells. Cell Rep. 2017;20:1936–1949. doi: 10.1016/j.celrep.2017.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eram MS, Shen Y, Szewczyk MM, Wu H, Senisterra G, Li F, Butler KV, Kaniskan HU, Speed BA, Dela Sena C, et al. A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases. ACS Chem Biol. 2016;11:772–781. doi: 10.1021/acschembio.5b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Cheng J, Wang J, Zhang Q, Liu M, Gong R, Wang P, Zhang X, Feng Y, Lan W, et al. Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat Commun. 2016;7:11197. doi: 10.1038/ncomms11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Harrison JS, Cornett EM, Goldfarb D, Darosa PA, Li ZM, Yan F, Dickson BM, Guo AH, Cantu DV, Kaustov L, et al. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. Elife. 2016;5:e17101. doi: 10.7554/eLife.17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Wang P, Lin Y, Xu Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 2011;21:1374–1378. doi: 10.1038/cr.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell. 2005;16:5621– 5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Li P, Fang L, Zhu H, Xu L, Cheng H, Zhang J, Li F, Feng Y, Li Y, et al. Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer. Cell Discov. 2016;2:16007. doi: 10.1038/celldisc.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahan G, Sayar N, Gozum G, Bozkurt B, Konu O, Yulug IG. Relative expression of rRNA transcripts and 45S rDNA promoter methylation status are dysregulated in tumors in comparison with matched-normal tissues in breast cancer. Oncol Rep. 2015;33:3131–3145. doi: 10.3892/or.2015.3940. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt MA, De Graaf P, Van Teeffelen HA, Timmers HT. Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin-dependent kinase inhibitors. PLoS One. 2012;7:e41446. doi: 10.1371/journal.pone.0041446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallous N, Legrand P, Mcewen AG, Ramon-Maiques S, Samama JP, Birck C. The PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readers. PLoS One. 2011;6:e27599. doi: 10.1371/journal.pone.0027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- Mann M, Zou Y, Chen Y, Brann D, Vadlamudi R. PELP1 oncogenic functions involve alternative splicing via PRMT6. Mol Oncol. 2014;8:389–400. doi: 10.1016/j.molonc.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neault M, Mallette FA, Vogel G, Michaud-Levesque J, Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012;40:9513–9521. doi: 10.1093/nar/gks764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502:249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, Thike AA, Kuznetsov VA, Tan PH, Voorhoeve PM, et al. p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 2012;40:9534– 9542. doi: 10.1093/nar/gks858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112:439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forne I, Pichler G, Horl D, Fellinger K, et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015;25:911–929. doi: 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908– 912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Stein C, Riedl S, Ruthnick D, Notzold RR, Bauer UM. The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res. 2012;40:9522– 9533. doi: 10.1093/nar/gks767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Shen J, Yang Z, Chen P, Zhao B, Hu W, Lan W, Tong X, Wu H, Li G, et al. Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger. Cell Res. 2011;21:1379–1382. doi: 10.1038/cr.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.