Abstract
Background:
Bacteria are increasingly resistant to antibiotics used to treat life-threatening infections in critically ill patients. The carbapenems represent the last line of defense against Gram-negative rods that are increasingly resistant to all other classes of β-lactam antibiotics used to treat life-threatening infections in critically ill patients. Carbapenem resistance in Gram-negative rods is most commonly caused by expression of carbapenemases, enzymes that hydrolyze the β-lactam ring of carbapenem antibiotics rendering them inactive. All of the available diagnostic tests rely on bacterial growth rendering them time consuming; therefore, rapid diagnostic tests are needed to identify multidrug (including carbapenem)-resistant bacteria.
Results:
We report the development of a novel LC–MS/MS method that detects carbapenemase activity from bacterial isolates. Incubation of a bacterial suspension with physiological levels of ertapenem leads to carbapenemase-mediated drug hydrolysis that produces a specific metabolite with an 18 Da increase in m/z within 1 h. Using the ratio of metabolite:parent, detected by LC–MS/MS from the culture, the sensitivity, specificity and a threshold cutoff for carbapenemase production (interpretive criteria) have been determined.
Conclusion:
A 100% correlation of our LC–MS/MS assay with the modified Hodge test (functional test for carbapenemase production) and PCR emphasizes the robust nature of this assay. The assay requires minimal hands-on time and a straightforward protocol allowing convenient implementation into clinical laboratories. Inclusion of stable isotope-labeled standard will further increase the robustness of the assay. This assay offers several advantages over other similar assays that use MALDI-TOF MS analysis.
Infections caused by antibiotic-resistant bacteria are becoming increasingly common worldwide, and patients infected with multidrug-resistant (MDR) bacteria have longer hospital stays and increased morbidity and mortality compared with patients infected with susceptible bacteria [1–5]. An important risk factor for these poor outcomes is the delayed institution of appropriate antibiotic therapy and there is great interest in the development of rapid assays to identify resistant bacteria [6]. Nucleic acid amplified tests (NAATs) targeting genes that confer resistance and immunoassays targeting gene products are available for the detection of nearly all MDR Gram-positive bacteria, such as methicillin-resistant Staphylococcus aureus, and their use can decrease the time needed to identify methicillin-resistant S. aureus infections and colonization leading to improved patient outcomes and infection control measures, respectively [7–9].
Resistance in Gram-negative rods (GNRs) is more complex. β-lactam antibiotics (e.g., penicillins, cephalosporins, carbapenems and monobactams) are the mainstay of empiric therapy for infections caused by Gram-negative pathogens, and resistance to β-lactams most commonly arises from the expression of β-lactamase enzymes capable of hydrolyzing and inactivating β-lactam antibiotics [10,11]. Among β-lactams, carbapenems are most resistant to β-lactamases hydrolysis, but carbapenemases capable of degrading all β-lactam antibiotics are endemic in regions of the USA and throughout the world [12,13]. The great genetic and enzymatic diversity among GNR-expressed β-lactamases has limited the development of rapid tests capable of identifying carbapenemase-producing organisms to better direct therapeutic and infection-control interventions. Some clinical microbiology laboratories use NAATs for the detection of Klebsiella pneumoniae carbapenemase (KPC), but this approach fails to detect other classes of clinically significant carbapenemases, for example, NDM, IMP, VIM and so on [14–17]. Additionally, the modified Hodge test (MHT) may detect expression of functional carbapenemases and is used by some laboratories. However, it adds significant time (24–48 h) to detection and is recommended only for the detection of KPC by the CDC. For these reasons, phenotypic methods that require monitoring bacterial growth in the presence of an antibiotic remain the standard of care, but they can take up to 18 h after bacterial isolation, during which time patients may be on inappropriate cephalosporin or carbapenem therapy.
Carbapenemases.
Diverse class of β-lactamase enzymes that confer resistance to cephalosporins, cephamycins and carbapenem classes of antibiotics.
Modified Hodge test.
For isolates of the Enterobacteriaceae family, carbapenemase production by the test isolate allows growth of a carbapenem-susceptible strain towards a carbapenem disc and results in characteristic cloverleaf-like growth pattern.
The ideal assay for identifying carbapenemase-producing MDR GNRs would be both rapid and able to detect carbapenem hydrolysis mediated by multiple enzyme classes. All β-lactamases hydrolyze the β-lactam ring leading to the addition of H2O, adding 18 Da to the molecular weight of the compound. MALDI-TOF-MS has recently been shown to detect β-lactamase and carbapenemase activity from bacteria suspended in saline and one LC–MS/MS assay for β-lactamase activity has been described for ampicillin [18–22]. We hypothesized that LC–MS/MS, with its excellent analytical sensitivity and proven utility for small-molecule detection, could reliably detect carbapenemase activity by monitoring for the appearance of the hydrolyzed metabolite of a carbapenem antibiotic from a complex biological matrix. Hydrolysis of each carbapenem depends on the relative concentrations of enzyme, drug and the corresponding Michaelis–Menten constant, Km. Although the use of a cocktail of multiple carbapenem antibiotics as a substrate in the assay is attractive, the presence of multiple substrates for the carbapenemases in the assay will lead to competition for enzyme binding. This will cause incomplete and/or inconsistent hydrolysis and complications in data interpretation. Therefore, we focused on optimizing the LC and MS/MS conditions, and used ertapenem to illustrate detection of carbapenemase activity from bacterial isolates suspended in growth media.
Experimental
▪ Materials & reagents
Columbia blood agar, MacConkey agar, Mueller-Hinton agar and tryptic soy broth (TSB) were obtained from Remel (KS, USA). Disks were also from Remel. Formic acid and acetonitrile were purchased from Sigma-Aldrich (MO, USA). A collection of clinical Enterobacteriaceae isolates (Yale-New Haven Hospital, CT, USA) and type strains from the American Type Culture Collection (ATCC; VA, USA) were acquired through appropriate material transfer agreements. The clinical strains were selected based on their resistance to carbapenem and/or extended spectrum cephalosporin antibiotics.
Ertapenem was obtained from the Yale-New Haven Hospital pharmacy, reconstituted as directed, frozen in aliquots at -70°C, individually thawed as needed, and diluted to the indicated concentration in TSB. Hydrolyzed ertapenem was generated by incubating ertapenem (500 µg/ml) with KPC-positive Klebsiella pneumoniae isolate ATCC-1705 overnight at 37°C. Complete hydrolysis was confirmed by LC–MS/MS. Pure antibiotic solutions were used to set-up and optimize LC–MS/MS parameters, and proof-of-principle experiments were conducted with KPC-producing (ATCC 1705) and KPC-negative (ATCC 1706) K. pneumoniae reference strains. Ion-suppression effects by the matrix (TSB) were performed by directly infusing ertapenem and hydrolyzed ertapenem at a flow rate of 20 µl/min and 5 µl of blank TSB sample injected through the UHPLC. No significant ion suppression occurred during the retention time window for each analyte (Supplementary Figure 1).
▪ Antimicrobial susceptibility testing
Phenotypic antimicrobial testing was performed as described [23]. Briefly, a 0.5 McFarland (McF) bacterial suspension in 0.45% NaCl was prepared from isolated bacterial colonies and spread uniformly over Mueller-Hinton agar plates (Thermo Scientific, KS, USA). Antibiotic discs were dispensed, and plates were incubated for 16–18 h at 37°C. The sizes of zones of growth inhibition were measured and recorded, and susceptibility was determined according to Clinical and Laboratory Standards Institute criteria [23].
The MHT to assess for carbapenemase production was performed as described [23]. Briefly, the ertapenem-susceptible Escherichia coli strain ATCC 25922 was streaked as a lawn on Mueller-Hinton agar. A 10 µg ertapenem disk was placed on the inoculated plate, and screen-positive isolates were streaked from the disk outward. The MHT was interpreted as positive when in-growth of the susceptible E. coli strain was noted along the path of the experimental isolate. The K. pneumoniae ATCC strains 1705 and 1706 were used as positive and negative controls, respectively. MHT results were independently read by two experienced observers.
▪ Genotypic characterization of bacterial isolates
Genomic DNA was prepared from each isolate using 1 µl loopful of bacteria suspended in 100 µl PrepMan® Ultra (Life Technologies Ltd, Paisley, UK) according to manufacturer’s instructions, and 5 µl of extracted genomic DNA was added to 25 µl of GeneAmp® Fast PCR master mix (Life Technologies Ltd) along with primers specific for the amplification of KPC, NDM-1, IMP, VIM or OXA (Supplementary Table 1). The presence of gene-specific amplicons was detected by performing gel electrophoresis (Supplementary Figure 2). PCR reactions mixed with the TackIt cyan/orange loading buffer were run on a 2% agarose gel containing SYBR Safe DNA gel stain and visualized using Safe Imager 2.0. All the chemicals, reagents and the Safe Imager were from Life Technologies (NY, USA).
▪ Detection of carbapenemase activity using MS
Isolated bacterial colonies were selected from freshly grown overnight cultures streaked on blood agar and suspended to 0.5 McF in 2.0 ml of TSB containing 4 µg/ml ertapenem. The resulting mixtures were capped and incubated at 37°C for the indicated periods of time, after which 0.3 ml aliquots were removed and mixed with 0.6 ml of acetonitrile in microcentrifuge tubes. After 30 s of vigorous mixing, the solutions were centrifuged at 16,000 × g for 2 min to pellet bacteria and precipitate proteins. An aliquot of 100 µl of the supernatant was then mixed with 500 µl of deionized water for LC–MS/MS analysis.
Sample analysis was performed using a Prominence UFLCXR LC system (Shimadzu, MD, USA) coupled to an AB Triple Quad™ 5500 MS/MS (AB Sciex, MA, USA) detector. 5 µl of the above prepared solutions were injected onto a Kinetex pentafluorophenyl column (2.6 µm, 100 Å, 50 × 3 mm; Phenomenex, CA, USA) equilibrated with 10% acetonitrile plus 0.1% formic acid at 40°C and separated using a 0.6-min gradient from 10 to 25% acetonitrile plus 0.1% formic acid. The mobile phases A and B were deionized water and acetonitrile, respectively, both supplemented with 0.1% formic acid. The binary flow gradient profile is shown (Figure 1C). The 5500 Triple Quad mass spectrometer was used in positive ion mode. MS voltages and energies were manually tuned by infusion of 0.1 µg/ml solution of each analyte in 10% methanol with 0.1% of formic acid, with an integrated syringe pump using Analyst 1.6 software (AB Sciex, MA, USA). The IonSpray Voltage was maintained at 5000 V and the temperature of the turbo-V source was set at 600°C. Air was used as the nebulizer gas (GS1) at a pressure of 50 psi and the heater gas (GS2) was set to 50 psi. Nitrogen was used as the curtain gas at a pressure of 30 units and as the collision gas at 7 arbitrary units. The MS/MS analysis was performed using the SRM mode of operation, and by monitoring the protonated molecular ions of ertapenem and hydrolyzed ertapenem. The dwell time for each transition was set at 30 ms. SRM transitions for each analyte and their respective settings were as follows:
Ertapenem – Q1 mass: 476 m/z; Q3 mass: 432 m/z; declustering potential: 70 V; entrance potential: 10 V; collision energy: 12 V; and collision cell exit potential: 18 V;
Hydrolyzed ertapenem – Q1 mass: 494 m/z; Q3 mass: 450 m/z; declustering potential: 70 V; entrance potential: 10 V; collision energy: 13 V; and collision cell exit potential: 17 V.
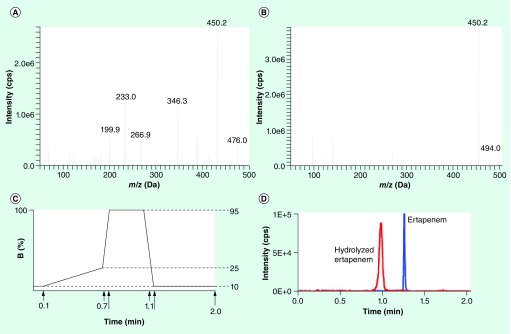
Figure 1. MS and LC parameters for ertapenem and hydrolyzed ertapenem.
Product ion scans obtained by direct infusion of 0.1 µg/ml solution of (A) ertapenem, Q1 m/z = 476 Da and (B) hydrolyzed ertapenem, Q1 m/z = 494 Da. The predominant products (fragments) for ertapenem and hydrolyzed ertapenem were 432 and 450 m/z, respectively. (C) 2-min LC program using deionized H2O + 0.1% formic acid as solvent A and acetonitrile + 0.1% formic acid as solvent B. (D) Detection of ertapenem (blue trace) and hydrolyzed ertapenem (red trace) by LC–MS/MS from a test mixture of ‘intensity-matched’ samples of ertapenem and hydrolyzed ertapenem.
Results
▪ Identification of ertapenem & hydrolyzed ertapenem SRMs
We first determined the optimal tuning parameters for the detection of intact ertapenem and its hydrolyzed form on the AB Triple Quad™ 5500 MS/MS system (AB Sciex). Using the analyst software version 1.6, we manually tuned source and MS/MS parameters by infusion of 0.1 µg/ml solution of ertapenem in 10% methanol with 0.1% of formic acid, with an integrated syringe pump. The published SRM used for ertapenem in positive ESI mode was 476→432 m/z, and we also identified this precursor/product ion pair as the optimal SRM (Figure 1A) [24].
While no purified hydrolyzed ertapenem is commercially available, intact ertapenem spontaneously hydrolyzes when incubated overnight at 37°C as reconstituted according to the Invanz® product insert in H2O. This solution was used for identification and tuning MS parameters for the hydrolyzed metabolite of ertapenem. The major compound found in this solution was the hydrolyzed (+18 Da) form of ertapenem; no other major reaction products were found. No SRMs for hydrolyzed ertapenem are published, but the primary product ion detected for hydrolyzed ertapenem also demonstrated the loss of 44 Da. Thus, the final SRM used for the detection of hydrolyzed ertapenem was 494→450 m/z (Figure 1B).
▪ Development of the LC method
A 0.6-min gradient from 10 to 25% acetonitrile plus 0.1% formic acid using a Kinetex pentafluorophenyl (2.6 µm, 100 Å, 50 × 3 mm) column at 40°C, with 10% acetonitrile plus 0.1% formic acid as the mobile phase, was chosen for optimal retention and separation of intact (retention time = 1.23 min) and hydrolyzed ertapenem (retention time = 0.95 min; Figure 1C & D).
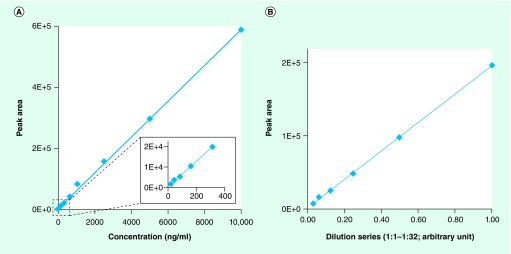
LC–MS/MS demonstrates excellent sensitivity and linearity for the detection of small molecules, and we observed a linear range of detection of 500-fold for unhydrolyzed ertapenem ranging from 20 to 10 µg/ml (Figure 2A). Because hydrolyzed ertapenem of known concentration is not commercially available, we had to perform serial dilutions of hydrolyzed ertapenem in TSB to determine its linearity of detection. Using these LC–MS/MS conditions, hydrolyzed ertapenem was detected with excellent linearity across a 32-fold dilution series (Figure 2B).
Figure 2. LOD and linear dynamic range for detection of ertapenem and hydrolyzed ertapenem.
(A) 10 µg/ml solution of ertapenem, used for a twofold dilution series up to 512-fold in tryptic soy broth, was detected by LC–MS/MS. (B) Hydrolyzed ertapenem, prepared by incubation of ertapenem with Klebsiella pneumoniae carbapenemase-postive bacteria for 24 h, was used for a twofold dilution series in tryptic soy broth and detected by LC–MS/MS.
▪ Detection of ertapenem hydrolysis within 20 min
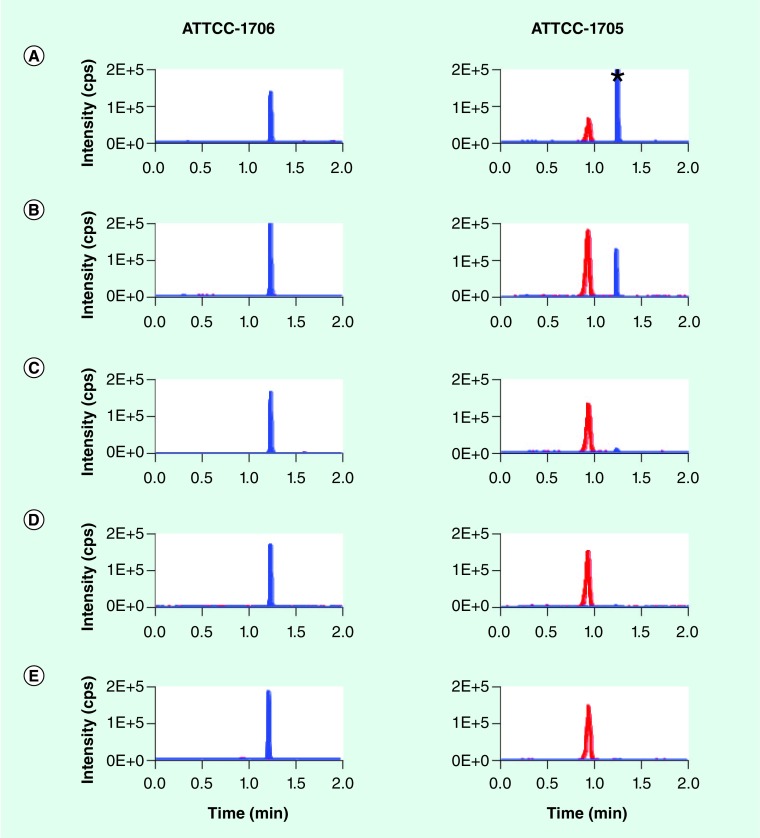
We performed a time-course study to compare the appearance of hydrolyzed ertapenem in standard 0.5 McF suspensions of ertapenem-resistant, non-carbapenemase-producing K. pneumoniae strain ATCC-1706, and ertapenem-resistant, KPC-2-producing K. pneumoniae strain ATCC-1705 (Figure 3). Even in the short period of time required to inoculate and sample the inoculated tubes, hydrolyzed ertapenem was consistently detected in KPC-expressing bacteria at the ‘t = 0’ time point. Hydrolyzed ertapenem levels were increased at 20 min in the presence of KPC-2 activity, and intact ertapenem was completely absent after 40 min of incubation. Additionally, nonspecific hydrolysis was insignificant for the carbapenemase-negative bacteria even after extended 18 h incubation.
Figure 3. LC–MS/MS analysis to detect hydrolysis of ertapenem by Klebsiella pneumoniae carbapenemase-positive bacteria.
A total of 4 µg/ml ertapenem was added to a 0.5 McFarland suspension (in tryptic soy broth) of Klebsiella pneumoniae carbapenemase-negative ATCC1706 and Klebsiella pneumoniae carbapenemase-positive ATCC1705 bacterial strains grown overnight on blood agar. Cultures were incubated at 37°C for 18 h. At indicated times, samples were harvested and subjected to LC–MS/MS for detection of ertapenem (blue trace) and hydrolyzed ertapenem (red trace). (A) 0 min, (B) 20 min, (C) 40 min, (D) 60 min and (E) 18 h. The ratio of the total integrated peak area of hydrolyzed ertapenem and ertapenem was used as an indicator for positive carbapenem hydrolysis activity. The ratios for the ATCC1706 strain were: 0 min: 0.01; 20 min: 0.01; 40 min: 0.01; 60 min: 0.01; and 18 h: 0.11; while those for ATCC1705 were: 0 min: 0.33; 20 min: 4.81; 40 min: 29.83; 60 min: 77.55; and 18 h: 1218.99.
*Indicates maximum intensity 4.74E5.
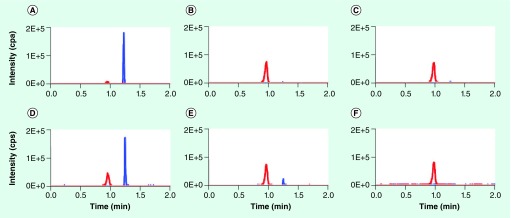
Carbapenemase activity and expression could vary substantially by enzyme subtype, host species and individual clinical isolate, among other factors. To determine specificity of this assay, each isolate was subjected to PCR testing for carbapenemase and ESBL genes, conventional antibiotic-susceptibility testing by disk diffusion testing, the MHT for phenotypic carbapenemase expression, and our MS-based rapid assay for carbapenemase activity. Representative chromatograms are shown in Figure 4. Visual inspection of all chromatograms revealed excellent assay specificity; hydrolyzed ertapenem was detected only in isolates positive for carbapenemase gene expression (Table 1). To define objective and quantitative interpretive criteria for carbapenemase-positive isolates, we calculated the ratio of hydrolyzed ertapenem peak area and intact ertapenem peak area after 1 h of incubation. All of the isolates that tested negative by the MHT and lacked carbapenemase genes by PCR had ratios ≤0.03, while all of the isolates that were phenotypically or genotypically positive for carbapenemase expression had ratios ≥0.10. Receiver–operator analysis of the internal intensity ratio, using the MHT as the gold standard, revealed an optimum ratio threshold of 0.05, which provided a sensitivity and specificity of 100% for the sample set and estimated 95% confidence intervals for the true sensitivity and specificity of the assay of 81–100% and 84–100%, respectively. Interestingly, one E. coli strain that was negative by PCR for all carbapenemase genes tested was carbapenem-resistant phenotypically, and tested positive by both the MHT and MS assay. Additionally, ATCC strain BAA2146, a K. pneumoniae isolate that is positive for NDM-1, was also positive by the MS assay.
Figure 4. Representative chromatograms obtained by LC–MS/MS analysis of ertapenem hydrolysis by various bacterial strains.
Each panel shows chromatograms for ertapenem (blue trace) and hydrolyzed ertapenem (red trace) obtained by performing the ertapenem hydrolysis assay for 1 h using the indicated bacterial strains. (A) ATCC-700603 (ESBL-positive, KPC-negative control Klebsiella pneumoniae), (B) clinical KPC-positive Klebsiella oxytoca isolate, (C) clinical KPC-positive Serratia marcescens isolate, (D) ATCC-BAA2146 (NDM-1-positive K. pneumoniae), (E) clinical KPC-positive K. pneumoniae isolate, and (F) clinical KPC-positve Escherichia coli isolate.
Table 1. Sensitivity and specificity of the LC–MS/MS ertapenem hydrolysis assay.
| Species | Antibiotic resistance | Presence of genes that encode enzymes | Modified Hodge test result | Metabolite:parent ratio at 1 h | |||||
|---|---|---|---|---|---|---|---|---|---|
| Ertapenem | Meropenem | Ceftazidime | Cefotaxime | β-lactamase | Carbapenemase | ||||
| K. pneumoniae (ATCC-1705) | R | R | R | R | KPC | + | 6.65 | ||
| K. pneumoniae (ATCC-BAA2146) | R | R | R | R | NDM-1 | + | 0.57 | ||
| K. pneumoniae | R | R | R | R | SHV | KPC | + | 9.95 | |
| S. marcescens | R | R | S | S | TEM | KPC | + | 80.25 | |
| S. marcescens | R | R | R | R | TEM | KPC | + | 2180.00 | |
| K. oxytoca | R | R | R | R | TEM | KPC | + | 79.92 | |
| K. pneumoniae | R | R | R | R | SHV, TEM | KPC | + | 0.36 | |
| E. coli | R | R | S | R | TEM | KPC | + | 2350.00 | |
| E. coli | R | R | R | R | TEM | KPC | + | 2220.00 | |
| K. pneumoniae | R | R | R | R | SHV, TEM | KPC | + | 43.12 | |
| E. coli | R | R | S | S | TEM | + | 0.10 | ||
| K. pneumoniae (ATCC-1706) | R | R | S | S | - | 0.03 | |||
| K. pneumoniae | S | S | R | S | SHV, TEM | - | 0.03 | ||
| K. pneumoniae | S | S | S | S | SHV | 0.02 | |||
| K. pneumoniae | S | S | R | R | SHV | - | 0.03 | ||
| K. pneumoniae | S | S | S | R | SHV, CTX-M, TEM | - | 0.03 | ||
| E. coli | S | S | R | R | TEM, CTX-M | - | 0.02 | ||
| E. cloacae | R | S | R | R | SHV | - | 0.02 | ||
| S. marcescens | S | S | S | R | SHV | - | 0.02 | ||
| E. coli | S | S | S | R | CTX-M | - | 0.03 | ||
| K. pneumoniae | S | S | S | R | SHV, TEM, CTX-M | - | 0.02 | ||
| E. cloacae | R | S | R | R | SHV, TEM | - | 0.02 | ||
| E. cloacae | S | S | R | R | CTX-M | - | 0.03 | ||
| E. cloacae | R | S | R | R | TEM | - | 0.03 | ||
E. cloacae: Enterobacter cloacae; E. coli: Escherichia coli; K. oxytoca: Klebsiella oxytoca; K. pneumoniae: Klebsiella pneumoniae; R: Resistant; S: Sensitive; S. marcescens: Serratia marcescens.
Discussion
We have provided proof-of-concept experiments for an LC–MS/MS method that rapidly detects carbapenemase activity among a set of highly resistant Enterobacteriaceae. The results shown clearly demonstrate:
The predicted 18 Da change in drug molecular weight associated with the addition of H2O is readily detected at physiologic levels in bacterial growth broths;
KPC-expressing bacteria rapidly hydrolyze ertapenem within 20 min and statistically significant hydrolysis is detectable in all isolates within 60 min;
There is no significant hydrolysis of ertapenem detectable after 18 h incubation in TSB or after 60 min in the presence of ESBL-producing bacteria;
Results can be obtained with a straightforward protocol easily implemented in clinical laboratories.
Our sample set contains a clinical E. coli isolate that is carbapenem-resistant and MHT-positive; yet, the responsible enzyme remains unidentified despite extensive genetic testing for the five predominant carbapenemase gene families. This finding highlights the value of phenotypic resistance testing over genetic approaches, as a rapid NAAT for common carbapenemase genes would not have identified this organism as carbapenem-resistant. Phenotypic growth in the presence of drug detects resistance regardless of mechanism and remains the gold standard for susceptibility testing, but, despite the development of rapid reading protocols on some automated antibiotic-susceptibility testing platforms, it typically requires 8–12 h of growth after the isolation of a pure organism. Our assay has the potential to detect all carbapenemase-mediated resistance within 1–2 h of isolation or, potentially, even sooner.
For our study set, the degree of ertapenem hydrolysis during the incubation period varied for individual isolates, as indicated by a range of internal intensity ratios from 0.1 to >2000 for the positive isolates. This variation in enzyme activity could arise from expression of different enzyme isoforms, differing enzyme expression levels, and/or the presence of other species- or isolate-specific factors. Calculation of a peak area ratio for hydrolyzed ertapenem to intact ertapenem at 1 h (Table 1) was found to be an effective surrogate for complete hydrolysis seen at 4 h (data not shown) for all training-set isolates. As more diverse organisms and enzymes are tested, the calculation of a quantitative peak area ratio will be critical to maintain statistically valid interpretive criteria. A future clinical validation study will provide additional information that is not possible with the training set alone. Enzymes expressed by different bacterial species may hydrolyze carbapenems at a faster or slower rate than those in the training set. Different bacterial species may have metabolites that interfere with the detection of specific precursor and product ions through ion suppression, similar mass and so on. Other unanticipated interferences must also be excluded.
In this initial application of LC–MS/MS to the detection of carbapenemase activity, we have restricted our analysis to Enterobacteriaceae. In our institution, KPC is currently the dominant carbapenemase isoform, similar to the rest of the USA, and KPC is predominantly isolated from Enterobacteriaceae. Ertapenem was chosen for its high sensitivity to KPC-mediated hydrolysis and its use in the MHT, which served as our standard for comparison. However, MDR GNRs also include important non-Enterobacteriaceae such as Pseudomonas aeruginosa and Acinetobacter baumannii. These organisms are intrinsically resistant to ertapenem owing to non-carbapenemase mechanisms and, thus, ertapenem is not US FDA approved for the treatment of infections caused by non-Enterobacteriaceae. Additionally, different families of carbapenemases are expressed more frequently by non-Enterobacteriaceae. Therefore, ertapenem is likely a suboptimal choice for an MS-based carbapenemase assay targeting MDR non-Enterobacteriaceae, justifying their exclusion from the current study.
Several papers have described therapeutic drug-monitoring assays for ertapenem [25–28] and a similar approach for detection of enzyme-mediated hydrolysis of β-lactam antibiotics using MALDI-TOF instruments, which are currently being used in clinical microbiology laboratories for bacterial identification [18–21]. We propose that the use of LC–MS/MS described here offers multiple advantages over MALDI-TOF for the detection of carbapenemase activity. Publications describing MALDI-TOF for the detection of carbapenemase activity report using unusually high drug concentrations, which is presumably required for adequate detection. By contrast, LC–MS/MS allows analysis of drugs at levels similar to therapeutic concentrations and those currently used in microbiology laboratories for FDA-approved antibiotic-susceptibility testing methods [23]. Additionally, published MALDI-TOF methods utilize aqueous buffer or normal saline to suspend and incubate bacterial colonies instead of standard growth media. Again, this is presumably required to reduce background MS signals owing to the abundance of small molecules found in bacterial growth broths, which is not a problem with the method presented here due to a combination of chromatographic separation and SRM detection. Finally, although not a concern during analysis of abundant bacterial colonies, future testing of primary specimens would likely require a degree of growth enrichment, which would be difficult to accomplish in the absence of growth media.
LC–MS/MS is widely utilized for the quantitative analysis of small molecules in biologic specimens. Specific identification of molecules of interest is assured by a combination of chromatographic retention time and SRM detection. For toxicology testing and therapeutic drug monitoring, a minimum of three identifiers or their equivalent are required by multiple regulatory agencies and represent standard practice in the field [29]. Additionally, effective retention of ertapenem and its hydrolyzed metabolite on a chromatographic column, in order to separate them from salts, other polar molecules and amphiphilic lipids, reduces the risk of ionization interferences affecting ion quantitation. Accurate quantitation will be further assured by the future incorporation of isotope-labeled compounds as internal standards. The ability to reliably and reproducibly quantitate the intensity ratio for hydrolyzed ertapenem relative to the parent drug has allowed us to statistically validate a defined threshold for distinguishing positive from negative results. Simplistic, straightforward and objective criteria for judging assay results will be essential for effective utilization of this method in a routine clinical microbiology laboratory. By contrast, mass spectra generated with MALDI-TOF carbapenemase assays are more complex, often requiring multifactorial interpretation criteria that may not be sufficiently robust for routine clinical use.
We recognize that the assay presented here will not detect carbapenem resistance arising from nonenzymatic mechanisms such as efflux pumps and porin mutations. Nevertheless, we expect that the introduction of a rapid assay to detect carbapenemase activity in bacterial isolates will have significant, beneficial clinical impact. The negative consequences of hospital-acquired infections with MDR bacteria on patient outcomes, length of stay and hospital costs are all well-established [1–3]. Furthermore, each day that appropriate antibiotic therapy is delayed due to unrecognized resistance results in increased morbidity and mortality for affected patients, and increased institutional costs [30]. Carbapenemases are frequently plasmid-encoded and easily transferable between GNRs, even across species, and this contributes to their well-described ability to cause outbreaks within healthcare facilities [31]. Although the current MS-based assay has the potential to significantly aid the identification of such outbreaks, we anticipate that the ability of LC-coupled triple-quadrupole MS to effectively analyze specific compounds in complex biological samples will allow extension of the current work to primary patient specimens and epidemiologic monitoring, which is where the greatest clinical impact will be realized.
Future perspective
With increased attention being given to the ever increasing threat of antibiotic-resistant bacterial (‘superbug’) infections and a lack of effective treatment, we believe that development of rapid tests that can be easily performed in the clinical laboratory will become an integral part of clinical management of patients with hospital-acquired infections. Additionally, the availability of diagnostic tests developed using the LC–MS/MS and MALDI-TOF platforms combined with the ease of use and application support provided for the instruments will allow efficient adoption of these platforms in the clinical laboratory. Meaningful progress will be made in the development of MS-based diagnostic tests, and novel, companion or replacement assays that increase sensitivity, specificity and/or shorten the time of other existing diagnostic tests.
Executive summary.
Background
Currently, the diagnosis of carbapenem-resistant infections relies on measuring bacterial growth in the presence of drug and requires 48–96 h, during which patients are treated empirically.
Rapid diagnostic tests, including nucleic acid amplified tests (NAATs) and antigen-based measurement of expressed carbapenems, suffer from low sensitivity and specificity.
Experimental
A novel LC–MS/MS assay that detects parent and hydrolyzed metabolites of ertapenem by carbapenemases expressed in clinical isolates is introduced.
Results
Analysis of ertapenem hydrolysis by known American Type Culture Collection strains that express Klebsiella pneumoniae carbapenemase and NDM-1, along with ESBL-expressing strains as controls, were used to design and test assay parameters.
Sensitivity and specificity of the assay were determined by testing characterized clinical isolates and comparing results with those from novel NAAT, modified Hodge test and the gold-standard Kirby–Bauer test.
Using receiver-operator curve analysis of the metabolite:parent ratio, objective diagnostic interpretive criteria for carbapenemase-positive isolates were established.
Discussion
The LC–MS/MS assay has several highlights, including high sensitivity and specificity, easy sample preparation, 15-min hands-on time, 2-min MS run time, detection of carbapenemase activity by novel carbapenemases, and 100% correlation with the modified Hodge test and NAATs.
This method offers rapid detection of carbapenemase-resistant infections, but will require adoption of the LC–MS/MS technology in the clinical microbiology laboratory.
The MALDI-TOF platform, which is traditionally not suited for the detection of small molecules and metabolites, has been used for development of a similar application, but suffers from a lack of sensitivity, specificity and defined objective diagnostic criteria.
Conclusion
Further development of this novel LC–MS/MS assay will require evaluation and inclusion of an internal standard that will aid in establishing diagnostic criteria. The finalized assay can then be submitted for US FDA approval.
Supplementary Material
Footnotes
Financial & competing interests disclosure
M Hodsdon, T Murray and D Peaper are co-founders, and A Tichy and M Kulkarni are employees, of M/Z Diagnostics, Inc., a commercial entity that has licensed from Yale University the intellectual property associated with this manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1 . Evans HL, Lefrak SN, Lyman J et al. Cost of Gram-negative resistance. Crit. Care Med. 35(1) 89–95 (2007). [DOI] [PubMed] [Google Scholar]
- 2 . Giske CG, Monnet DL, Cars O, Carmeli Y. Clinical and economic impact of common multidrug-resistant Gram-negative bacilli. Antimicrob. Agents Chemother. 52(3) 813–821 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3 . Mauldin PD, Salgado CD, Hansen IS, Durup DT, Bosso JA. Attributable hospital cost and length of stay associated with health care-associated infections caused by antibiotic-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 54(1) 109–115 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4 . Marchaim D, Gottesman T, Schwartz O et al. National multicenter study of predictors and outcomes of bacteremia upon hospital admission caused by Enterobacteriaceae producing extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 54(12) 5099–5104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5 . Roberts RR, Hota B, Ahmad I et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin. Infect. Dis. 49(8) 1175–1184 (2009). [DOI] [PubMed] [Google Scholar]
- 6 . Bilavsky E, Schwaber MJ, Carmeli Y. How to stem the tide of carbapenemase-producing Enterobacteriaceae? Proactive versus reactive strategies. Curr. Opin. Infect. Dis. 23(4) 327–331 (2010). [DOI] [PubMed] [Google Scholar]
- 7 . Arcenas RC, Spadoni S, Mohammad A et al. Multicenter evaluation of the LightCycler MRSA advanced test, the Xpert MRSA Assay, and MRSASelect directly plated culture with simulated workflow comparison for the detection of methicillin-resistant Staphylococcus aureus in nasal swabs. J. Mol. Diagn. 14(4) 367–375 (2012). [DOI] [PubMed] [Google Scholar]
- 8 . Verhoeven PO, Grattard F, Carricajo A et al. Quantification by real-time PCR assay of Staphylococcus aureus load: a useful tool for rapidly identifying persistent nasal carriers. J. Clin. Microbiol. 50(6) 2063–2065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9 . Calfee DP, Salgado CD, Classen D et al. Strategies to prevent transmission of methicillin-resistant Staphylococcus aureus in acute care hospitals. Infect. Control Hosp. Epidemiol. 29(Suppl. 1) S62–S80 (2008).18840090 [Google Scholar]
- 10 . Sundsfjord A, Simonsen GS, Haldorsen B, Lundblad EW, Samuelsen O. Broad-spectrum β-lactamases in Gram-negative bacteria. Tidsskr. Nor. Lægeforen. 128(23) 2741–2745 (2008). [PubMed] [Google Scholar]
- 11 . Malloy AM, Campos JM. Extended-spectrum β-lactamases: a brief clinical update. Pediatr. Infect. Dis. J. 30(12) 1092–1093 (2011). [DOI] [PubMed] [Google Scholar]
- 12 . Ho J, Tambyah PA, Paterson DL. Multiresistant Gram-negative infections: a global perspective. Curr. Opin. Infect. Dis. 23(6) 546–553 (2010). [DOI] [PubMed] [Google Scholar]
- 13 . Kumarasamy KK, Toleman MA, Walsh TR et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10(9) 597–602 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14 . Francis RO, Wu F, Della-Latta P, Shi J, Whittier S. Rapid detection of Klebsiella pneumoniae carbapenemase genes in Enterobacteriaceae directly from blood culture bottles by real-time PCR. Am. J. Clin. Pathol. 137(4) 627–632 (2012). [DOI] [PubMed] [Google Scholar]
- 15 . Hindiyeh M, Smollan G, Grossman Z et al. Rapid detection of blaKPC carbapenemase genes by internally controlled real-time PCR assay using bactec blood culture bottles. J. Clin. Microbiol. 49(7) 2480–2484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16 . Hindiyeh M, Smollen G, Grossman Z et al. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46(9) 2879–2883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17 . Schechner V, Straus-Robinson K, Schwartz D et al. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47(10) 3261–3265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18 . Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49(9) 3321–3324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19 . Hrabak J, Walkova R, Studentova V, Chudackova E, Bergerova T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49(9) 3222–3227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20 . Kempf M, Bakour S, Flaudrops C et al. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS ONE 7(2) e31676 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21 . Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J. Clin. Microbiol. 50(3) 927–937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22 . Grundt A, Findeisen P, Miethke T, Jager E, Ahmad-Nejad P, Neumaier M. Rapid detection of ampicillin resistance in Escherichia coli by quantitative mass spectrometry. J. Clin. Microbiol. 50(5) 1727–1729 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23 . Cockerill F. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement Clinical and Laboratory Standards Institute, Wayne, PA, USA (2011). [Google Scholar]
- 24 . Lefeuvre S, Venisse N, Marchand S, Bachelet M, Couet W. A simple and sensitive liquid chromatography-tandem mass spectrometry assay for the quantification of ertapenem in microdialysate. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 862(1–2) 242–245 (2008). [DOI] [PubMed] [Google Scholar]
- 25 . D’Avolio A, Baietto L, De Rosa FG et al. A simple and fast method for quantification of ertapenem using meropenem as internal standard in human plasma in a clinical setting. Ther. Drug Monit. 30(1) 90–94 (2008). [DOI] [PubMed] [Google Scholar]
- 26 . Legrand T, Chhun S, Rey E et al. Simultaneous determination of three carbapenem antibiotics in plasma by HPLC with ultraviolet detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 875(2) 551–556 (2008). [DOI] [PubMed] [Google Scholar]
- 27 . McWhinney BC, Wallis SC, Hillister T, Roberts JA, Lipman J, Ungerer JPJ. Analysis of 12 β-lactam antibiotics in human plasma by HPLC with ultraviolet detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878(22) 2039–2043 (2010). [DOI] [PubMed] [Google Scholar]
- 28 . la Marca G, Giocaliere E, Villanelli F et al. Development of an UPLC–MS/MS method for the determination of antibiotic ertapenem on dried blood spots. J. Pharm. Biomed. Anal. 61(0) 108–113 (2012). [DOI] [PubMed] [Google Scholar]
- 29 .College of American Pathologists. Chemistry and Toxicology Checklist College of American Pathologists, Northfield, IL, USA (2012). [Google Scholar]
- 30 . Slama TG. Gram-negative antibiotic resistance: there is a price to pay. Crit. Care 12(Suppl. 4) S4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31 . Mathers AJ, Cox HL, Kitchel B et al. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. MBio 2(6) e00204–e00211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.