Abstract
Objective
To profile the amine/phenol submetabolome to determine potential metabolite biomarkers associated with Parkinson’s disease (PD) and PD with incipient dementia.
Methods
At baseline of a 3-wave (18-month intervals) longitudinal study, serum samples were collected from 42 healthy controls and 43 PD patients. By wave 3 (year 3) 16 PD patients were diagnosed with dementia and were classified as PD with incipient dementia at baseline. Metabolomic profiling using dansylation isotope labeling liquid chromatography mass spectrometry was conducted to compare controls with full PD group, PD with no dementia and PD with incipient dementia.
Results
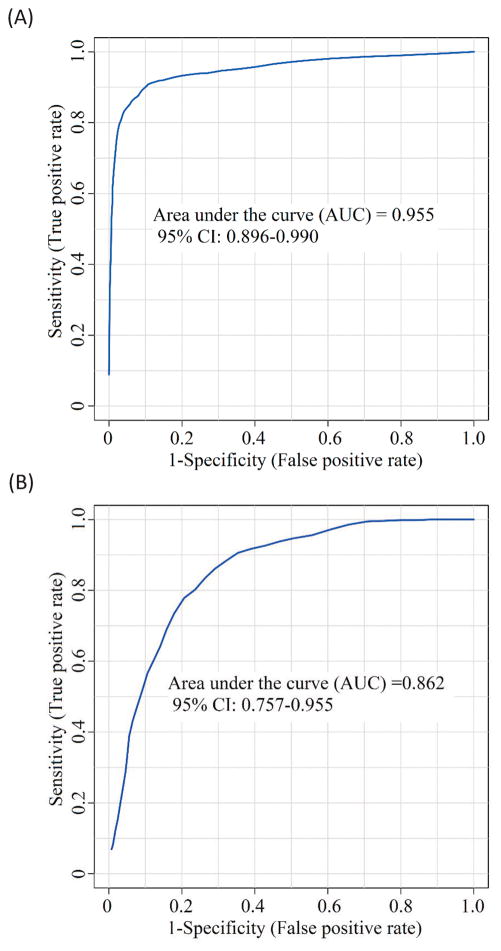
Metabolomic analyses detected 719 common metabolites in 80% of the samples. Some were significantly altered in pairwise comparison of different groups (fold-change of >1.2 or <0.83 with q<0.05). We discriminated PD and controls by using a 5-metabolite panel, vanillic acid, 3-hydroxykynurenine, isoleucyl-alanine, 5-acetylamino-6-amino-3-methyluracil and theophylline. The Receiver Operating Characteristic curve produced an Area-Under-the-Curve value of 0.955 with 87.5% sensitivity and 93.0% specificity. In comparing PD with no dementia with PD with incipient dementia we used an 8-metabolite panel, His-Asn-Asp-Ser, 3, 4-dihydroxyphenylacetone, desaminotyrosine, hydroxy-isoleucine, alanyl-alanine, putrescine [-2H], purine [+O] and its riboside. This produced an Area-Under-the-Curve value of 0.862 with 80.0% sensitivity and 77.0% specificity.
Conclusions
The significantly altered metabolites can be used to differentiate (1) PD patients from healthy controls with high accuracy and (2) the stable PD with no dementia group from those with incipient dementia. Following further validation in larger cohorts, these metabolites could be used for both discrimination and establishing prognosis in PD.
Keywords: Parkinson’s disease, metabolomics, biomarkers, metabolic pathways, dementia
Introduction
Parkinson’s disease (PD) is a common progressive neurodegenerative disorder associated with the loss of dopaminergic neurons in the substantia nigra and production of Lewy bodies composed of α-synuclein proteins.[1] Clinical PD diagnosis is based, in part, on impaired motor abilities such as bradykinesia, rigidity, tremor and postural instability. To date, no definitive single or set of biomarkers for PD have been discovered.[2] PD misdiagnosis rates were about 10% in 2001[3] and 6% in 2009[4], rates which may depend on duration and stage of disease.[5] Therefore, the first goal of this research was to use systematic and unbiased metabolomics technology to detect a set of biomarkers that reflect disease pathways and might contribute to accurate discrimination. In addition to motoric impairment, up to 80% of PD patients eventually show cognitive impairment, including dementia, over the course of the disease, compromising quality of life and raising economic costs.[6] Therefore, the second goal of this research was to discover biomarkers that can be useful in discriminating PD patients who may remain dementia free (PDND) from those at risk for developing dementia. A better understanding of the metabolic pathways of PD patients with incipient dementia (PDID) can lead to improved disease monitoring and interventions targeted to patients according to dementia risk (see Supplemental Note S1.1 for the definition of dementia).
Metabolomics is an emerging field for biomarker discovery in human aging and neurodegenerative diseases.[7] A previous metabolomics study on cerebrospinal fluid (CSF) detected an increase in concentration of 3-hydroxykynurenine and decrease in concentration of glutathione in PD patients, suggesting the involvement of neurotoxicity and oxidative stress in PD pathogenesis. [8] Several potential biomarkers have also been identified in serum samples, which is less invasive to collect, including those involved in oxidative stress, [9] purine metabolism, [10] caffeine and xanthine metabolism. [11] However, because of the complexity of the metabolome, new technologies providing larger coverage and better quantitative capability will enable the discovery of more specific metabolic biomarkers, for both PD discrimination and early PDID sub-classification. Chemical isotope labeling liquid chromatography mass spectrometry (CIL LC-MS) that uses different labeling reagents to target chemical-group-based submetabolomes is a relatively new analytical platform for generating comprehensive and quantitative metabolomic profiles for biomarker research.[12] In the present study, we applied dansylation CIL LC-MS targeting the amine/phenol submetabolome to find the key metabolic differences in serum between two sets of groups. The overall study used a longitudinal design with baseline serum collection and two 18-month follow-ups. Using the baseline serum we performed two comparisons of metabolomics profiles: (1) PD patients and healthy controls and (2) PD patients who remained dementia-free for three years and PDID patients who developed dementia within this interval.
Methods
Participants
Clinically established PD patients (n = 52) and age-and-sex matched healthy controls (n = 50) between 64–84 years old volunteered for a 3-wave (18-month intervals) longitudinal study in Edmonton, Canada as previously detailed.[13, 14] At baseline PD patients (1) met standard criteria for PD, (2) did not meet criteria for atypical parkinsonism, and (3) did not have unstable health conditions compromising survival. Patients who developed abnormal imaging such as a stroke or atypical features with follow up were also excluded. They were recruited from movement disorder clinics, the Parkinson’s Society of Alberta, and from community neurologists. The control group was recruited by advertisement in seniors’ centers and magazines, control and patient contacts, and general medicine clinics. The University of Alberta health ethics review board approved this study and all participants provided informed consent. For both groups, we excluded participants with baseline dementia, stroke, atypical parkinsonism, or attrition. In addition, one control participant was excluded as an outlier in the metabolomics analysis. Thus, there were 43 patients and 42 controls in the final groups. Participants performed three waves of standardized assessments, including assessment for cognitive function and dementia (see Supplemental Note S1.2 for more detail). Of the 43 baseline PD patients 16 were diagnosed with dementia at wave 3. No further blood was taken and therefore the analyses (group comparison and prediction of dementia group at waver 3) were based on baseline blood work. Note that the exclusion described above was done before the metabolomic analysis. One outlier in the control group was excluded after the analysis, as its metabolomic data were too different from those of other participants. A possible reason of having this outlier could be contamination during sample collection.
We performed two pairwise metabolomics analyses. The first comparison evaluated the metabolomic profiles of the full available baseline groups, including 43 PD (no dementia at baseline) patients (M age = 70.71 years; sex = 44% female) and 42 controls (M age = 71.49 years; sex = 45% female) (see Table 1). The second comparison evaluated the profiles for two PD subgroups, those who remained dementia-free at wave 3 (n = 27; M age = 69.58 years) and those who were diagnosed with dementia at wave 3. The latter were classified post hoc as PDID at baseline (n = 16; M age = 72.62 years). Duration of disease did not differ significantly between PDID (9.59±5.1 years) and those remaining cognitively intact (7.75±4.1, p=0.19).
Table 1.
Baseline demographic and clinical characteristics.
| Control | PD | p-value | PDND | PDID | p-value | |
|---|---|---|---|---|---|---|
| N | 42 | 43 | -- | 27 | 16 | -- |
| Age (years) | 71.49 (5.01) | 70.71 (4.14) | .434 | 69.58 (3.55) | 72.62 (4.46) | .018 |
| Education (years) | 15.00 (3.42) | 14.28 (2.98) | .303 | 14.74 (3.36) | 13.50 (2.07) | .190 |
| Sex (F/M) | 19/23 | 19/24 | .923 | 12/15 | 7/9 | .966 |
| MMSE | 28.56 (1.48) | 28.33 (1.67) | .503 | 28.85 (1.29) | 27.36 (1.91) | .006 |
| Folate (nmol/L) | 879.81 (236.25) | 842.56 (207.45) | .442 | 799.00 (185.45) | 916.06 (227.41) | .073 |
| Vitamin B12 (pmol/L) | 393.79 (198.05) | 293.26 (112.82) | .005 | 295.70 (92.16) | 289.13 (144.52) | .856 |
| Levodopa equivalents (mg) | N/A | 644.00 (360.06) | -- | 611.83 (392.94) | 703.76 (293.41) | .448 |
| UPDRS part 3 | N/A | 16.12 (7.90) | -- | 16.67 (8.08) | 15.19 (7.77) | .559 |
Note. PD, Parkinson’s disease; PDND, Parkinson’s disease no dementia; PDID, Parkinson’s disease incipient dementia; MMSE, Mini Mental State Exam; UPDRS, Unified Parkinson’s Disease Rating Scale
Baseline comparisons are shown in Table 1. PD patients did not differ from controls in age, education, sex distribution or cognitive status. The PDND and PDID subgroups were similar, differing slightly only on age and initial cognitive status (both means above impairment cut-offs). While they did not differ statistically on the key comparisons of levodopa equivalents and the Unified Parkinson’s Disease Rating Scale, the levodopa equivalent dose was higher in the PDID, despite slightly lower UPDRS part 3.
Serum samples and dansylation LC-MS metabolomic profiling
Serum was collected at baseline only from all participants and stored at −80 °C. To reveal small concentration variations of metabolites in comparative samples, we applied the CIL LC-MS technique to overcome potential inaccuracy due to matrix effects, ion suppression, or instrumental drift in MS detection. In our workflow, individual samples were labeled using 12C-dansyl chloride and a pooled sample generated by mixing small aliquots of samples was labeled by 13C-dansyl chloride (Supplemental Note S2). Each 12C-labeled sample was mixed with an aliquot of 13C-pooled sample, followed by LC-MS analysis of the mixture. All the labeled metabolites were detected as 13C- and 12C-peak pairs and the peak ratios were determined and used for quantitative metabolomic analysis of the individual samples.
We minimized variations in total sample amount in different samples in order to detect the individual metabolite concentration differences in comparative samples more accurately by performing sample normalization. Specifically, we applied an LC-UV method to determine the total concentration of dansyl-labeled metabolites based on the UV absorption of the dansyl group[15] (Supplemental Note S2). Before the LC-MS analysis, we mixed the 12C-labeled individual sample with the same total amount of 13C-labeled pool, according to the total concentrations of labeled metabolites, for sample amount normalization.
For LC-MS analysis, a Bruker maXis impact high-resolution quadrupole time-of-flight (Q-TOF) mass spectrometer with electrospray ionization (Bruker, Billerica, MA) combined with an Agilent 1100 HPLC was used (Supplemental Note S2).
Data processing and statistical analysis
The 12C-/13C-peak pairs from each LC-MS run were extracted by the IsoMS software.[16] IsoMS-Align was used to align the peak pair data from different samples by retention time and accurate mass. The missing ratio values were filled back by using the Zero-fill program.[17] IsoMS-Quant[18] was used to generate the final metabolite-intensity table, which was exported to SIMCA-P+ 12 (Umetrics AB, Umeå, Sweden) for analysis. We followed the method described in the work of LeWitt et al.[8] for statistical analysis (Supplemental Note S1.3). To avoid over-fitting, we calculated the q-value for each p-value using QVALUE.[19] Supplemental Table T1 lists the p-values and q-values for Control vs. PD and PDND vs. PDID. Metabolite identification was done using a DnsID standards library[20] for positive identification as well as the HMDB library and EML database for putative identification (see Supplemental Note S1.4 for the definition of putative identification).[21]
Results
Submetabolome and metabolite identification
Dansylation labeling LC-MS targets the analysis of the amine/phenol submetabolome; many metabolomic pathways contain the amine- and phenol-containing metabolites. A total of 719 metabolites were commonly detected in 80% of the 85 samples. Among them, 66 metabolites were positively identified using an in-house developed dansyl standards library consisting of 273 compounds (Supplemental Table T2). For the remaining peak pairs, accurate mass search with a mass accuracy tolerance of 0.005 Da putatively identified 333 metabolites using the HMDB database and 282 metabolites in the EML database using MyCompoundID[21] (Supplemental Table T3). In total, 681 of the 719 metabolites (95%) were either definitely or putatively identified.
Comparative metabolome analysis for PD biomarker discovery
The Partial Least Squares-Discriminant Analysis (PLS-DA) and Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA) score plots are shown in Supplemental Figure S1. Figure S1A and Figure S1B show that there was a significant difference between the healthy controls (in green) and the PD patients (in red). This group separation was validated in a permutation test (Figure S1C). Note that in our data analysis the PLS and OPLS methods were used as multivariate calibration processes, and the resulting score plots helped with visualizing the inter-sample and inter-group variances. For the multivariate classifications (Control vs. PD, or PDND vs. PDID), we used random forest analyses. The model performance indicators (the R2 and Q2 values) are provided in the corresponding score plots. Supplemental Note S1.5 describes the PLS-DA model.
The volcano plot shown in Figure S1D displays 28 metabolites with Fold Change (FC) > 1.2, q < 0.1 (in red) and 48 metabolites with FC < 0.83, q < 0.1 (in blue). Supplemental Table T4 lists the significant metabolites identified. Four significant metabolites, identified as 4-hydroxy-benzenepropanedioate, vanillylmandelic acid-isomer, alpha-methyldopa, and methylguanine, were observed only in the PD group. The dansyl library identified citrulline, methionine sulfoxide, pantothenic acid, glycyl-valine, pipecolic acid, serotonin, vanillic acid, vanillylmandelic acid, theophylline, and hydroxykynurenine.
Among significant metabolites, there were several catecholamine metabolites. Some metabolites from the tryptophan pathway were also detected and quantified. Biopterin had an increased concentration in the PD group. Some significant metabolites in Supplemental Table T4 were related to oxidative stress, such as citrulline and methionine sulfoxide. Finally, caffeine metabolites were detected as altered in PD.
Comparative metabolome analysis of PD with and without incipient dementia
Supplemental Figures S2A and S2B show the PLS-DA and OPLS-DA score plots, respectively, for the comparison between PDND and PDID. The permutation test result is shown in Figure S2C. These analyses indicate that the two groups were separated based on the metabolomic data set.
The volcano plot (Figure S2D) shows 21 metabolites with FC > 1.2, q < 0.1 (in red) and 15 metabolites with FC < 0.83, q < 0.1 (in blue). Among the 36 significant metabolites, 16 were identified (see Supplemental Table T5 for the list). Among these 16 metabolites, two metabolites were definitely identified by the library and the others were putatively identified based on a database search. The two definitely identified metabolites are desaminotyrosine and 5-hydroxylysine.
Common discriminating metabolites
Comparison of the metabolites listed in Supplemental Tables T4 and T5 indicate that there are five common significant metabolites. For example, 3, 4dihydroxyphenylacetone, a significant metabolite in the PDND-PDID comparison (Supplemental Table T5), also significantly differentiated PD patients from the healthy controls (Supplemental Table T4). For this metabolite, the averaged peak pair ratio in the control group was 0.006 ± 0.012. In the PDND subgroup and the PDID subgroup, the averages were 2.16 ± 1.63 and 3.34 ± 1.95, respectively.
ROC curves
Metaboanalyst 3.0 was used to generate a receiver operating characteristic (ROC) curve for differentiating PD (baseline) from healthy controls. The classification model was built by the random forest method based on five metabolites: vanillic acid, 3-hydroxykynurenine, isoleucyl-alanine, 5-acetylamino-6-amino-3-methyluracil and theophylline. The procedure for selecting the five metabolites as the panel is given in Supplemental Note S1.6. The AUC value for each of the metabolites separately was found to be 0.939, 0.781, 0.794, 0.730 and 0.714, respectively. Of interest, using vanillic acid alone, we could achieve both sensitivity and specificity at 90.0%. The discriminating power was improved by combining these five metabolites into a biomarker panel. The corresponding ROC curve shown in Figure 1A produced an AUC value of 0.955, which is within the range of 0.896–0.990 at the 95% confidence interval. Discrimination of PD from controls can be achieved at 87.5% sensitivity and 92.0% specificity. Using a permutation test, we did not find any over-fitting of the ROC results (Supplemental Note S1.7).
Figure 1.
For differentiating the PDND and PDID subgroups at baseline, we found that the discrimination power of a panel based on the two definitely identified metabolites was not strong (AUC=0.673). The univariate AUC of 5-hydroxylysine is 0.659 and that of desaminotyrosine is 0.674. Thus, we selected the following eight putatively identified biomarker candidates with highly ranked independent AUCs to form a panel: His-Asn-Asp-Ser (AUC=0.597), 3, 4-dihydroxyphenylacetone (0.677), desaminotyrosine (0.640), hydroxy-isoleucine (0.610), alanyl-alanine (0.737), putrescine [-2H] (0.736), purine [+O] (0.627) and its riboside (0.597). As can be seen in Figure 1B, the ROC curve for this biomarker panel produced an AUC value of 0.862, which is within the range of 0.757–0.955 at the 95% confidence interval. This panel can provide discrimination with sensitivity of 80.0% and specificity of 77.0%. The procedure for selecting the eight metabolites as the panel is also given in Supplemental Note S1.6. Using a permutation test, we did not find any over-fitting of the ROC results (Supplemental Note S1.7).
We have examined whether the use of any unidentified significant metabolites (there were 38 in total; see Supplemental Table T6) could increase the differentiation power. We found that there was no significant increase in performance when one or more of these were used to replace the panels described above (Supplemental Note S1.8). We also compared the significant metabolites found using PLS-DA and the random forest method; the top 10 metabolites were the same (Supplemental Note S1.9).
Discussion
We performed two two-group metabolomic comparisons for identifying novel biomarkers of PD. First, using baseline serum samples and clinical characterizations and diagnoses, we compared a PD (no dementia) group with a comparable older adult control group (no PD, dementia, or impairment). Our results showed clear differentiation between groups. A panel of 5 metabolites in the ROC analysis gave an AUC of 0.955 with sensitivity of 87.5% and specificity of 93.0%. Second, we compared two subgroups of the initial PD group, again using baseline serum samples. PD patients who have yet to meet the criteria for a dementia diagnosis represent a detectably early and more advanced transitional phase of PD. Chia and colleagues observed metabolomic changes in the cerebrospinal fluid of PD patients already diagnosed with dementia and depression.[22] Notably, these subgroups were determined three years after the baseline clinical evaluation did not detect incipient dementia. The present task met the challenge of detecting biomarkers of PD dementia prior to clinical diagnosis. Specifically, using a panel of 8 metabolites in ROC analysis, we obtained an AUC of 0.862 with sensitivity of 80.0% and specificity of 77.0%. These results hold promise for PD discrimination and prognosis, as well as identification of pathways leading to dementia within PD patient groups that may assist in identifying targets for intervention. While the markers we have identified are not in presymptomatic or early patients, patients with established disease are at risk for cognitive decline and dementia, features which are critically relevant to clinical decisions regarding future planning and treatment options (such as having deep brain stimulation).
As Gerlach and colleagues suggested, biomarkers for PD should not only be linked to fundamental features of PD neuropathology, but also be correlated to the disease progression assessed by clinical rating scales.[23] Our approach targeted the amine/phenol-containing metabolites; other metabolites would require other labeling approaches. For the first set of results, the pathway analysis showed that catecholamine metabolism, tryptophan metabolism and caffeine metabolism were the most relevant metabolomics pathways found to be affected by PD. In addition, metabolites related to oxidative stress were also identified.
Catecholamine metabolism
The metabolites in the catecholamine pathway were excluded in order to avoid the interference of levodopa medication. However, vanillylmandelic acid (VMA), which is the end product of the catecholamine pathway, was retained as a significantly changed metabolite, because some studies showed that urinary excretions of VMA and its up-stream metabolites (epinephrine and normetanehrine) were not greatly affected by levodopa.[24, 25] The fold change of VMA between the PD group and the control group was as large as 3.80, suggesting a significant metabolic change caused by PD rather than medication. In addition, the isomer of VMA was detected only in the PD group. Moreover, the vanillylmandelic acid-isomer showed very similar fold changes, although its origin and biological function is unclear.
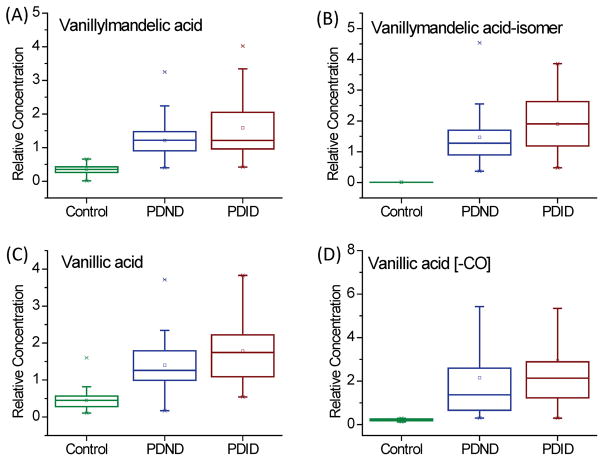
This relationship between VMA and PD has not been previously reported; this change is likely related to a disorder of the catecholamine pathway, in which dopamine is produced. Vanillic acid is a food metabolite, which is often found in the urine of humans who have consumed coffee, tea and vanilla-flavored food.[26] It may have an increased concentration in PD via conversion from VMA or homovanillic acid. The derivative of vanillic acid, vanillic acid [-CO], was also a unique metabolite in the PD group. The relative concentrations of these metabolites in the control group, the PDND subgroup and the PDID subgroup are shown as box plots in Figure 2. Despite the fact that all four have an increased concentration in the serum of PD patients, the relative concentrations in the PDID subgroup were higher than those in the PDND subgroup. These results support our previous discussion that a more advanced transitional phase of PD can be detected in PD patients biologically, even if not yet clinically.
Figure 2.
Both methyldopa and 3, 4-dihydroxyphenylacetone were increased in the PD samples. Methyldopa is known to be an aromatic-amino-acid decarboxylase (AADC) inhibitor in animals and in humans.[27] Declining levels of AADC may contribute to decreasing effectiveness of L-dopa medication over time.[28] A metabolite of methyldopa via AADC, [29] 3, 4-dihydroxyphenylacetone, was greatly increased in concentration in the PD group, but its role is unknown.
Tryptophan metabolism
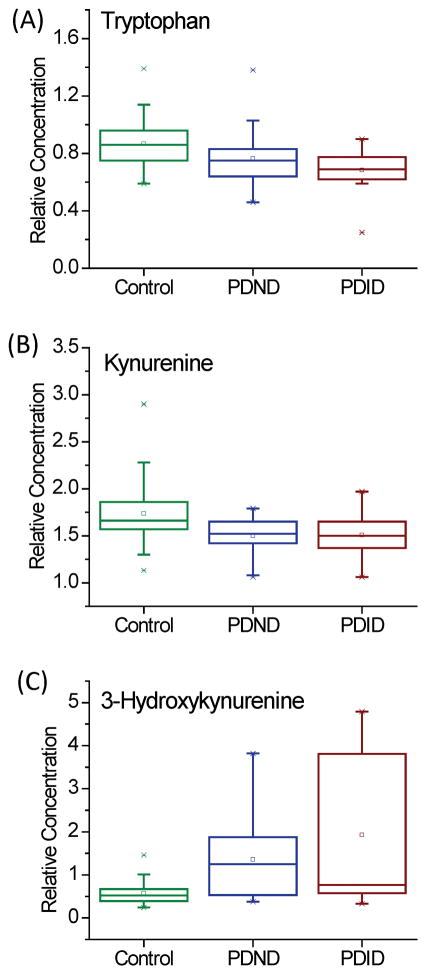
Although the concentrations of tryptophan and kynurenine did not change greatly, increased relative concentration of 3-hydroxykynurenine in the PD group was observed. Figure 3 shows the relative concentrations of tryptophan, kynureinine and 3-hydroxykynurenine in different groups. Metabolites of the kynurenine pathway are believed to play crucial roles in maintaining normal brain function.[30] Kynurenine is the down-stream metabolite of tryptophan that can be further converted to 3-hydroxykynurenine or kynurenic acid. 3-hydroxykynurenine is a neurotoxic metabolite which causes neuronal death.[31] However, kynurenic acid behaves as an endogenous neuroprotective agent.[32] Consistent with our results, LeWitt and colleagues reported that the CSF concentration of 3-hydroxykynurenine was increased in PD patients.[8]
Figure 3.
Caffeine metabolism
Although caffeine cannot be labeled by the dansylation reagent, some of its metabolites are labeled and were detected as significantly changed metabolites (Supplemental Figure S3A). Theophylline can be labeled by the dansylation reagent. Paraxanthine cannot be labeled, but its down-stream metabolite, 5-acetylamino-6-amino-3-methyluracil, was detected as a significant metabolite. Although methylxanthine was detected and was in the VIP list (Supplemental Table T4), we could not differentiate its three isomers without standards. Xanthine is the end product of caffeine metabolism. It was detected and identified by the dansyl standards library. Figures S3B–D show the relative concentrations of theophylline, 5-acetylamino-6-amino-3-methyluracil and xanthine in different groups. The concentrations of caffeine metabolites were lower in the PD group. Xanthine can also be converted from hypoxanthine and guanine in the purine pathway, and its concentration changed marginally.
It has been widely reported that coffee and tea consumption could protect against the risk of PD.[33] A possible reason for this phenomenon is that caffeine is an antagonist of the adenosine A2A receptor, which has a role in the regulation of glutamate and dopamine release.[34] Adenosine A2A antagonists can modify motor function and are being tested for the treatment of PD.[35] Caffeine metabolites, such as theophylline, may act as adenosine A2A receptor antagonists.[36] We did not determine caffeine intake in our participants; however, in recent work by Hatano and colleagues, serum levels of caffeine and caffeine metabolites were lower in PD patients than controls, even though there was no difference in caffeine consumption.[11]
Oxidative stress
Supplemental Figure S4 shows the relative concentrations of methionine, an isomer of methionine and citrulline. According to our results, methionine sulfoxide and its isomer had increased concentrations in the PD group. Methionine sulfoxide is an oxidation product of methionine with reactive oxygen species so it is considered as a biomarker of oxidative stress.[37] It has also been reported that the oxidation of methionine residues could play an important role in the aggregation of normally soluble α-synuclein in PD.[38] The average concentration of methionine sulfoxide in the two subgroups was similar, implying that the extent of peripheral oxidative stress does not increase dementia risk. On the other hand, citrulline, an oxidization product of arginine, had a decreased concentration. It is reported to be an efficient hydroxyl radical scavenger.[39] Increased oxidative damage occurs in all human neurodegenerative diseases, including PD,[40] and some studies suggest that oxidative stress may be a factor in the loss of dopaminergic neurons.[41]
Additional subgroup comparisons
Post-hoc comparisons showed the metabolic data discriminated subgroups of dementia severity (Supplemental Note S1.10) but not age (Supplemental Note S1.11).
Limitations
First, our study examined a single cohort of subjects who were on treatment for PD at one center (Supplemental Note S1.12). Future studies should replicate and extend our results. One specific direction would be to examine untreated patients in order to estimate the potential confounding of medications. In this work, although not statistically significant, the PDID group did differ from the cognitively stable PD group in some characteristics, including baseline disease duration, UPDRS part 3, and LED. We note, however, the important point that the groups did differ at baseline on global cognitive performance (MMSE), but no subject had functionally significant impairment (dementia). This highlights the importance of replicating and extending the present study with careful attention to matching on baseline characteristics. Second, diet may affect the metabolic profiles (Supplemental Note S1.12). Third, we collected blood only at baseline and thus intra-patient comparisons of metabolomic changes (Supplemental Note 1.12) or correlations of changes with PD progress (Supplemental Note 1.13) are not possible. Fourth, the dansylation labeling mainly enhances the detection of the amine/phenol-containing metabolites, but it cannot detect other important metabolites in related pathways. Applying other labeling techniques to target different groups of submetabolomes will expand the overall metabolome coverage in the future. Fifth, in this work, there was no external dataset for validation and the sample size was relatively small for validation work. In the future, more participants will be enrolled for the validation of the novel biomarker candidates comprising the panels observed in this study.
Conclusions
Metabolomics analyses of serum are useful tools for identifying novel biomarker panels for both PD discrimination and the early detection of established PD patients who are at risk for transitioning to dementia. Follow-up studies can provide insights into potential pathways of PD neuropathology, including those associated with early discrimination and identification of risk for dementia.
Supplementary Material
Footnotes
Authors’ Roles
1) Research project: A. Conception, B. Organization, C. Execution; 2) Statistical Analysis: A. Design, B. Execution, C. Review and Critique; 3) Manuscript: A. Writing of the first draft, B. Review and Critique.
W.H.: 1B, 1C, 2A, 2B, 3A
S.S.: 1B, 1C, 3B
R.A.D.: 1A, 1B, 2A, 2C, 3B
R.C: 1A, 1B, 2A, 2C, 3B
L.L.: 1A, 1B, 2A, 2C, 3B
Financial disclosures of all authors for the preceding 12 months
None.
Financial Disclosure/Conflict of Interest: None
References
- 1.Berg D, Postuma RB, Bloem B, et al. Annals of Neurology. Mov Disord. 2014;29(4):454–462. doi: 10.1002/mds.25844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller DB, O’Callaghan JP. Biomarkers of Parkinson’s disease: present and future. Metabolism. 2015;64(3):S40–S46. doi: 10.1016/j.metabol.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes AJ, Daniel SE, Lees AJ. Improved accuracy of clinical diagnosis of Lewy body Parkinson’s disease. Neurology. 2001;57(8):1497–1499. doi: 10.1212/wnl.57.8.1497. [DOI] [PubMed] [Google Scholar]
- 4.Newman EJ, Breen K, Patterson J, et al. Accuracy of Parkinson’s disease diagnosis in 610 general practice patients in the West of Scotland. Mov Disord. 2009;24(16):2379–2385. doi: 10.1002/mds.22829. [DOI] [PubMed] [Google Scholar]
- 5.Adler CH, Beach TG, Hentz JG, et al. Low clinical diagnostic accuracy of early vs advanced Parkinson disease Clinicopathologic study. Neurology. 2014;83(5):406–412. doi: 10.1212/WNL.0000000000000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svenningsson P, Westman E, Ballard C, et al. Cognitive impairment in patients with Parkinson’s disease: diagnosis, biomarkers, and treatment. The Lancet Neurology. 2012;11(8):697–707. doi: 10.1016/S1474-4422(12)70152-7. [DOI] [PubMed] [Google Scholar]
- 7.Xia J, Broadhurst DI, Wilson M, et al. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9(2):280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeWitt PA, Li J, Lu M, et al. 3-hydroxykynurenine and other Parkinson’s disease biomarkers discovered by metabolomic analysis. Mov Disord. 2013;28(12):1653–1660. doi: 10.1002/mds.25555. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanov M, Matson WR, Wang L, et al. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain. 2008;131(2):389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- 10.Johansen KK, Wang L, Aasly JO, et al. Metabolomic profiling in LRRK2-related Parkinson’s disease. PLoS One. 2009;4(10):e7551. doi: 10.1371/journal.pone.0007551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatano T, Saiki S, Okuzumi A, et al. Identification of novel biomarkers for Parkinson’s disease by metabolomic technologies. J Neurol Neurosurg Psychiatry. 2015 doi: 10.1136/jnnp-2014-309676. jnnp-2014-309676. [DOI] [PubMed] [Google Scholar]
- 12.Guo K, Li L. Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal Chem. 2009;81(10):3919–3932. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- 13.Camicioli R, Sabino J, Gee M, et al. Ventricular dilatation and brain atrophy in patients with Parkinson’s disease with incipient dementia. Mov Disord. 2011;26(8):1443–1450. doi: 10.1002/mds.23700. [DOI] [PubMed] [Google Scholar]
- 14.Sapkota S, Gee M, Sabino J, et al. Association of homocysteine with ventricular dilatation and brain atrophy in Parkinson’s disease. Mov Disord. 2014;29(3):368–374. doi: 10.1002/mds.25798. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Li L. Determination of total concentration of chemically labeled metabolites as a means of metabolome sample normalization and sample loading optimization in mass spectrometry-based metabolomics. Anal Chem. 2012;84(24):10723–10731. doi: 10.1021/ac3025625. [DOI] [PubMed] [Google Scholar]
- 16.Zhou R, Tseng C-L, Huan T, et al. IsoMS: automated processing of LC-MS data generated by a chemical isotope labeling metabolomics platform. Anal Chem. 2014;86(10):4675–4679. doi: 10.1021/ac5009089. [DOI] [PubMed] [Google Scholar]
- 17.Huan T, Li L. Counting Missing Values in a Metabolite-Intensity Data Set for Measuring the Analytical Performance of a Metabolomics Platform. Anal Chem. 2014;87(2):1306–1313. doi: 10.1021/ac5039994. [DOI] [PubMed] [Google Scholar]
- 18.Huan T, Li L. Quantitative metabolome analysis based on chromatographic peak reconstruction in chemical isotope labeling liquid chromatography mass spectrometry. Anal Chem. 2015;87(14):7011–7016. doi: 10.1021/acs.analchem.5b01434. [DOI] [PubMed] [Google Scholar]
- 19.Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J R Stat Soc Ser B-Stat Methodol. 2004;66:187–205. [Google Scholar]
- 20.Huan T, Wu Y, Tang C, et al. DnsID in MyCompoundID for Rapid Identification of Dansylated Amine-and Phenol-Containing Metabolites in LC–MS-Based Metabolomics. Anal Chem. 2015;87(19):9838–9845. doi: 10.1021/acs.analchem.5b02282. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Li R, Zhou J, et al. MyCompoundID: using an evidence-based metabolome library for metabolite identification. Anal Chem. 2013;85(6):3401–3408. doi: 10.1021/ac400099b. [DOI] [PubMed] [Google Scholar]
- 22.Chia L-G, Cheng L-J, Chuo L-J, et al. Studies of dementia, depression, electrophysiology and cerebrospinal fluid monoamine metabolites in patients with Parkinson’s disease. J Neurol Sci. 1995;133(1):73–78. doi: 10.1016/0022-510x(95)00146-s. [DOI] [PubMed] [Google Scholar]
- 23.Gerlach M, Maetzler W, Broich K, et al. Biomarker candidates of neurodegeneration in Parkinson’s disease for the evaluation of disease-modifying therapeutics. J Neural Transm. 2012;119(1):39–52. doi: 10.1007/s00702-011-0682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinterberger H, Andrews CJ. Catecholamine metabolism during oral administration of Levodopa: effects of the medication in Parkinson’s disease. Arch Neurol. 1972;26(3):245–252. doi: 10.1001/archneur.1972.00490090071005. [DOI] [PubMed] [Google Scholar]
- 25.Factor SA, Schneider AS. Peripheral catecholamine output in Parkinson’s disease: effects of drug treatment. Exp Neurol. 1995;131(1):64–68. doi: 10.1016/0014-4886(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 26.Wishart DS, Tzur D, Knox C, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35(suppl 1):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Culvenor AJ, Jarrott B. Reduction of Aromatic L-Amino Acid Decarboxylase Protein in Rats after Chronic Administration of Alpha-Methyldopa. Mol Pharmacol. 1979;15(1):86–98. [PubMed] [Google Scholar]
- 28.Coune PG, Schneider BL, Aebischer P. Parkinson’s disease: gene therapies. Cold Spring Harbor perspectives in medicine. 2012;2(4):a009431. doi: 10.1101/cshperspect.a009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertoldi M, Dominici P, Moore PS, et al. Reaction of dopa decarboxylase with α-methyldopa leads to an oxidative deamination producing 3, 4-dihydroxyphenylacetone, an active site directed affinity label. Biochemistry. 1998;37(18):6552–6561. doi: 10.1021/bi9718898. [DOI] [PubMed] [Google Scholar]
- 30.Vamos E, Pardutz A, Klivenyi P, et al. The role of kynurenines in disorders of the central nervous system: possibilities for neuroprotection. J Neurol Sci. 2009;283(1):21–27. doi: 10.1016/j.jns.2009.02.326. [DOI] [PubMed] [Google Scholar]
- 31.Chiarugi A, Meli E, Moroni F. Similarities and differences in the neuronal death processes activated by 3OH-kynurenine and quinolinic acid. J Neurochem. 2001;77(5):1310–1318. doi: 10.1046/j.1471-4159.2001.00335.x. [DOI] [PubMed] [Google Scholar]
- 32.Hartai Z, Klivenyi P, Janaky T, et al. Kynurenine metabolism in plasma and in red blood cells in Parkinson’s disease. J Neurol Sci. 2005;239(1):31–35. doi: 10.1016/j.jns.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Hu G, Bidel S, Jousilahti P, et al. Coffee and tea consumption and the risk of Parkinson’s disease. Mov Disord. 2007;22(15):2242–2248. doi: 10.1002/mds.21706. [DOI] [PubMed] [Google Scholar]
- 34.Schwarzschild MA, Xu K, Oztas E, et al. Neuroprotection by caffeine and more specific A2A receptor antagonists in animal models of Parkinson’s disease. Neurology. 2003;61(11 suppl 6):S55–S61. doi: 10.1212/01.wnl.0000095214.53646.72. [DOI] [PubMed] [Google Scholar]
- 35.Morelli M, Di Paolo T, Wardas J, et al. Role of adenosine A 2A receptors in parkinsonian motor impairment and l-DOPA-induced motor complications. Prog Neurobiol. 2007;83(5):293–309. doi: 10.1016/j.pneurobio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Yasui K, Agematsu K, Shinozaki K, et al. Theophylline induces neutrophil apoptosis through adenosine A2A receptor antagonism. J Leukocyte Biol. 2000;67(4):529–535. doi: 10.1002/jlb.67.4.529. [DOI] [PubMed] [Google Scholar]
- 37.Mashima R, Nakanishi-Ueda T, Yamamoto Y. Simultaneous determination of methionine sulfoxide and methionine in blood plasma using gas chromatography-mass spectrometry. Anal Biochem. 2003;313(1):28–33. doi: 10.1016/s0003-2697(02)00537-7. [DOI] [PubMed] [Google Scholar]
- 38.Glaser CB, Yamin G, Uversky VN, et al. Methionine oxidation, α-synuclein and Parkinson’s disease. Biochim Biophys Acta, Proteins Proteomics. 2005;1703(2):157–169. doi: 10.1016/j.bbapap.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 39.Akashi K, Miyake C, Yokota A. Citrulline, a novel compatible solute in drought-tolerant wild watermelon leaves, is an efficient hydroxyl radical scavenger. FEBS Lett. 2001;508(3):438–442. doi: 10.1016/s0014-5793(01)03123-4. [DOI] [PubMed] [Google Scholar]
- 40.Halliwell B. Oxidative stress and neurodegeneration: where are we now? J Neurochem. 2006;97(6):1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhou C, Huang Y, Przedborski S. Oxidative stress in Parkinson’s disease. Ann N Y Acad Sci. 2008;1147(1):93–104. doi: 10.1196/annals.1427.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.