Abstract
SAGA (Spt-Ada-Gcn5-Acetyltransferase) and TFIID (transcription factor IID) have been previously shown to facilitate the formation of the PIC (pre-initiation complex) at the promoters of two distinct sets of genes. Here, we demonstrate that TFIID and SAGA differentially participate in the stimulation of PIC formation (and hence transcriptional initiation) at the promoter of PHO84, a gene for the high-affinity inorganic phosphate (Pi) transporter for crucial cellular functions, in response to nutrient signaling. We show that transcriptional initiation of PHO84 occurs predominantly in a TFIID-dependent manner in the absence of Pi in the growth medium. Such TFIID dependency is mediated via the NuA4 (nucleosome acetyltransferase of H4) histone acetyltransferase (HAT). Intriguingly, transcriptional initiation of PHO84 also occurs in the presence of Pi in the growth medium, predominantly via the SAGA complex, but independently of NuA4 HAT. Thus, Pi in the growth medium switches transcriptional initiation of PHO84 from NuA4-TFIID to SAGA dependency. Further, we find that both NuA4-TFIID- and SAGA-dependent transcriptional initiations of PHO84 are facilitated by the 19S proteasome subcomplex or regulatory particle (RP) via enhanced recruitment of the coactivators SAGA and NuA4 HAT, which promote TFIID-independent and -dependent PIC formation for transcriptional initiation, respectively. NuA4 HAT does not regulate activator binding to PHO84, but rather facilitates PIC formation for transcriptional initiation in the absence of Pi in the growth medium. On the other hand, SAGA promotes activator recruitment to PHO84 for transcriptional initiation in the growth medium containing Pi. Collectively, our results demonstrate two distinct stimulatory pathways for PIC formation (and hence transcriptional initiation) at PHO84 by TFIID, SAGA, NuA4, and 19S RP in the presence and absence of an essential nutrient, Pi, in the growth media, thus providing new regulatory mechanisms of transcriptional initiation in response to nutrient signaling.
Keywords: transcription, TFIID, SAGA, NuA4, 19S RP
IN eukaryotes, transcription is initiated by the formation of the preinitiation complex (PIC), an assembly of general transcription factors and RNA polymerase II, at the core promoter (Bhaumik 2011). Activator protein (or activator) facilitates the formation of the PIC at the promoter, and hence transcriptional initiation (Bhaumik 2011). An activator contains at least a DNA-binding domain that targets activator to the UAS (upstream activating sequence) of the promoter, and an activation domain that interacts with a transcription factor to enhance PIC formation at the core promoter (Bhaumik 2011). Based on the target specificities of the activators, two distinct stimulatory pathways of transcriptional initiation have been deciphered (Bhaumik 2011). These are SAGA (Spt-Ada-Gcn5-Acetyltransferase)- and TFIID (transcription factor IID)-regulated transcriptional initiations. SAGA has histone acetyltransferase (HAT) activity for histone H3 and histone deubiquitinase activity for ubiquitylated histone H2B (Grant et al. 1997; Bhaumik 2011). SAGA consists of 15 nonessential and six essential components (Grant et al. 1997; Bhaumik 2011), while TFIID is composed of TBP (TATA-box binding protein) and a set of TAFs (TBP associated factors) (Kuras et al. 2000; Bhaumik 2011). At the SAGA-regulated genes, activator targets SAGA, which enhances PIC formation for transcriptional initiation (Bhaumik 2011). On the other hand, activator targets TFIID for PIC formation during TFIID-dependent transcriptional initiation (Bhaumik 2011). Further, NuA4 (nucleosome acetyltransferase of H4) has been shown to facilitate the recruitment of TFIID at TFIID-regulated genes (Uprety et al. 2012, 2015).
Approximately 90% of RNA polymerase II genes in yeast are regulated by TFIID, and transcription of ∼10% RNA polymerase II genes occurs via SAGA (Lee et al. 2000; Bhaumik 2011). However, both TFIID and SAGA are required for the transcription of the hexose transporter genes HXT2 and HAT4 (van Oevelen et al. 2005), and the inorganic phosphate (Pi) transporter gene PHO84 (Huisinga and Pugh 2004). Likewise, induction of RNR3 (ribonucleotide reductase 3) transcription by methyl methanesulfonate is also dependent on both TFIID and SAGA (Zhang et al. 2008). Consistently, a recent study also demonstrated the recruitment of both SAGA and TFIID in a genomic response to DNA damage or acute heat shock (Ghosh and Pugh 2011). This study further suggested that genes that are controlled by a more sustained PIC formation may recruit both TFIID and SAGA, while a long-lived presence of TFIID might lead to a greater dependence on it. However, the molecular basis for such a distinction remains elusive. Further, it is not clear whether a gene can be distinctly and predominantly regulated by either TFIID or SAGA for PIC formation (and hence transcriptional initiation) under different environmental cues. To address this, we analyzed the mechanisms of transcriptional initiation of a Pi transporter gene, PHO84, in the absence and presence of Pi (an essential nutrient to living organisms) in the growth media. Transport of Pi across the plasma membrane is the first step in its utilization in the biosynthesis of important cellular components such as nucleic acids, nucleoproteins, and phospholipids, and hence metabolic and signaling pathways (Bevington et al. 1992). Altered phosphate homeostasis is associated with serious human disorders such as hemolysis, skeletal muscle myopathy, cardiomyopathy, neuropathy, osteomalacia, tissue calcifications, and metabolic changes (Bringhurst and Leder 2006; Bergwitz and Juppner 2009). Phosphate homeostasis is maintained via the membrane Pi transporter. Much of our knowledge of the membrane transport of Pi has been learned from yeast. Pho84 is a high-affinity Pi transporter in yeast, and is conserved in metazoans (Toh-e et al. 1988; Werner and Kinne 2001; Hubbard et al. 2007; Bergwitz et al. 2012). PHO84 belongs to the family of genes involved in the phosphate-responsive signaling (PHO) pathway, whose transcriptional initiation is regulated by cellular levels of Pi via an intracellular signaling cascade from a complex of Pho80 (a cyclin) and Pho85 (a member of the p34cdc2/CDC28-related kinase family) (Toh-e et al. 1988; Kaffman et al. 1994). The Pho80-Pho85 complex phosphorylates five serine–proline (SP) dipeptides referred to as SP1–SP4 and SP6 on Pho4, the basic helix-loop-helix domain-containing transcriptional activator of PHO84 and other PHO genes (O’Neill et al. 1996; Komeili and O’Shea 1999). During phosphate starvation, downregulation of the kinase activity of the Pho80-Pho85 complex occurs, which results in a hypophosphorylated/unphosphorylated form of Pho4 (Kaffman et al. 1994; Schneider et al. 1994). This form of Pho4 associates with the UAS of PHO84 and other PHO genes, along with Pho2, to initiate transcription (Vogel et al. 1989; Bun-Ya et al. 1991; Hirst et al. 1994; Magbanua et al. 1997a; Oshima 1997; Komeili and O’Shea 2000). Under high concentration of Pi, transcription of the PHO genes is turned off because the Pho80-Pho85 complex phosphorylates Pho4 (Kaffman et al. 1994), and hyperphosphorylated Pho4 does not stay in the nucleus, but rather is localized in the cytoplasm (Komeili and O’Shea 1999).
Even though PHO84 belongs to the family of the PHO genes, it is transcribed in the YPD (yeast extract and peptone, plus 2% dextrose) growth medium that contains Pi (Bhaumik and Green 2002; Springer et al. 2003; Shukla et al. 2006a,b; Durairaj et al. 2014a). Further, we show here that transcriptional initiation at PHO84 follows two predominantly distinct pathways in the presence and absence of Pi in the growth media. In the absence of Pi in the growth medium, TFIID is primarily involved in the stimulation of PIC formation at the PHO84 core promoter (and hence transcriptional initiation). Such a function of TFIID is further mediated via NuA4 HAT. On the other hand, SAGA facilitates PIC formation at the PHO84 core promoter in the presence of Pi in the growth medium. SAGA regulation of PIC formation (and hence transcriptional initiation) of PHO84 is not dependent on NuA4 HAT. Thus, our results reveal here that transcription of the same gene can be regulated by both SAGA and TFIID under different concentrations of Pi in the growth media. Intriguingly, both TFIID and SAGA-dependent transcriptional initiations of PHO84 are facilitated by the 19S proteasome subcomplex or regulatory particle (RP). Therefore, transcriptional initiation of PHO84 is commonly promoted by the 19S RP, but differentially stimulated by NuA4-TFIID and SAGA, under different concentrations of Pi in the growth media. These results provide new regulatory mechanisms of gene activation by the 19S RP, TFIID, NuA4, and SAGA in response to an essential nutrient, Pi, for crucial cellular functions, as presented below.
Materials and Methods
Plasmids
The plasmid pFA6a-13Myc-KanMX6 was used for genomic Myc epitope tagging of the proteins of interest. The plasmids pRS416 and pRS406 were used in the PCR-based gene disruption.
Yeast strains
The taf13-ts (temperature-sensitive) mutant (WCS179) and its isogenic wild-type equivalent were obtained from the Green laboratory (Michael R. Green, University of Massachusetts Medical School) (Li et al. 2000; Shen et al. 2003). The Δspt20 (FY1097) and wild-type (FY67) strains were obtained from the Winston laboratory (Fred Winston, Harvard Medical School) (Roberts and Winston 1996). The esa1-ts mutant (LPY3291) and wild-type (LPY3498) strains were obtained from the Pillus laboratory (Lorraine Pillus, University of California, San Diego) (Clarke et al. 1999). The strain bearing the ts (temperature-sensitive) mutation in Rpt4 (rpt4-ts or sug2-13, Sc677) and its isogenic wild-type equivalent (Sc599) were obtained from the Kodadek and Johnston laboratories (Tom Kodadek and Stephen A. Johnston; UT Southwestern Medical Center) (Russell and Johnston 2001). The strain, PSY17 (Rpt2-Myc), was generated by adding multiple Myc epitope tags at the C-terminal of Rpt2 in its chromosomal locus in Sc599 (obtained from the laboratory of Stephen A. Johnston; UT Southwestern Medical Center; Russell and Johnston 2001). Multiple Myc epitope tags were added at the original chromosomal locus of ESA1 in the rpt4-ts (Sc677) and wild-type (Sc599) strains to generate BUY13 (Esa1-Myc in rpt4-ts) and BUY12 (Esa1-Myc), respectively (Uprety et al. 2012). The SPT20 gene was tagged with multiple Myc epitopes at the C-terminus in its chromosomal locus in the wild-type strain (W303a) to generate the ASY10 strain (Uprety et al. 2015). Yeast strains ARY1a and ARY2a were generated by adding multiple Myc epitope tags at the C-terminal of Pho2 in its original chromosomal locus in the Sc599 and Sc677 strains, respectively. Likewise, the VBY1a and VBY2 strains were generated by adding multiple Myc epitope tags at the C-terminal of Pho4 in its original chromosomal locus in the Sc599 and Sc677 strains, respectively. The JPY9a and JPY10 strains were generated by adding Myc epitope tags at the C-terminal of Pho2 in the wild-type (FY67) and Δspt20 (FY1097) strains, respectively. Likewise, the JPY11a and JPY12a strains were generated by adding Myc epitope tags at the C-terminal of Pho4 in the wild-type (FY67) and Δspt20 (FY1097) strains, respectively. Myc epitope tags were added at the C-terminus of Pho2 in the wild-type and esa1-ts mutant strains to generate the JPY6 and JPY7 strains, respectively.
Growth media
For studies of PHO84 in the absence of Pi, yeast cells were grown in YPD up to an OD600 of 0.5 at 30°, followed by transferring the cells to the YPD medium without Pi (YPD-Pi) for 3 hr. For experiments in the presence of Pi, growth of the cells was continued in YPD medium, after cells reached an OD600 of 0.5, for 3 hr. Subsequently, cells were cross-linked by formaldehyde for chromatin immunoprecipitation (ChIP) assay or harvested for mRNA analysis. The YPD-Pi medium was prepared by precipitating phosphate from YPD using MgSO4 and NH4OH, and adjusting the final pH to 5.8 (Wykoff and O’Shea 2001). For experiments in the taf13-ts, rpt4-ts, and esa1-ts mutants and their wild-type equivalents in YPD, yeast cells were grown in YPD at 23° up to an OD600 of 0.85, and then switched to 37° for 1 hr prior to harvesting for mRNA analysis or cross-linking for ChIP assay. For studies using the ts mutants and their wild-type equivalents in the YPD-Pi medium, cells were initially grown in YPD at 23° up to an OD600 of 0.5, followed by switching the cells to the YPD-Pi medium for 3 hr. After 3 hr, cells were grown at 37° for 1 hr prior to harvesting or cross-linking.
ChIP assay
The ChIP assay was performed as described previously (Bhaumik and Green 2003; Malik et al. 2013; Sen et al. 2014; Uprety et al. 2016). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Following sonication, cell lysate (400 µl of lysate from 50 ml of yeast culture) was precleared by centrifugation, and then 100 µl of lysate was used for each immunoprecipitation. Immunoprecipitated protein–DNA complexes were treated with proteinase K, the cross-links were reversed, and DNA was purified. Immunoprecipitated DNA was dissolved in 20 µl of TE 8.0 (10 mM Tris-HCl, pH 8.0 and 1 mM EDTA), and 1 µl of immunoprecipitated DNA was analyzed by PCR. PCR contained [α-32P] dATP (2.5 µCi for 25 µl of reaction), and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, “input” DNA was isolated from 5 µl of lysate without going through the immunoprecipitation step, and dissolved in 100 µl of TE 8.0. To compare the PCR signal arising from the immunoprecipitated DNA with the input DNA, 1 µl of input DNA was used in the PCR analysis.
The association of Esa1, Spt20, Pho2, and Pho4 with PHO84 was analyzed by modified ChIP assay, as described in our previous publication (Shukla et al. 2006a; Uprety et al. 2012, 2015). For ChIP analysis of the Rpt2 component of the proteasome, we modified the above ChIP protocol, as done previously (Uprety et al. 2012). Briefly, 1600 μl of lysate was prepared from 200 ml of yeast culture following formaldehyde-based in vivo cross-linking for 25 min. Next, 600 μl of lysate was used for each immunoprecipitation (using 10 μl of anti-Myc antibody and 100 μl of protein A/G plus agarose beads from Santa Cruz Biotechnology) and the immunoprecipitated DNA sample was dissolved in 5 μl of TE 8.0, of which 1 μl was used in PCR analysis. In parallel, PCR for input DNA was performed using 1 μl of DNA that was prepared by dissolving purified DNA from 5μl of lysate in 100 μl of TE 8.0. Primer pairs used for PCR analysis were as follows:
PHO84 (Core): 5′-GATCCACTTACTATTGTGGCTCGT-3′ and 5′-GTTTGTTGTGTGCCCTGGTGATCT-3′.
PHO84 (UAS): 5′-CCAGCACGTGGGGCGGAAATT-3′ and 5′-TTTAATCTAGCTAATAAGCAGGCAAAA-3′.
PHO12 (Core): 5′-GGAATGAGCATAAACAGCGT-3′ and 5′-GATGTTCTTGCTCTCTTTGC-3′.
PHO5 (Core): 5′-GCAAGGCATATACCCATTTGGGAT-3′ and 5′-TATTCTCATGAGAGATGAAGCCATACT-3′.
Chromosome-V (Chr.-V): 5′-GGCTGTCAGAATATGGGGCCGTAGTA-3′ and 5′-CACCCCGAAGCTGCTTTCACAATAC-3′.
Autoradiograms were scanned and quantitated by National Institutes of Health Image 1.62. Immunoprecipitated DNAs were quantitated as the ratio of immunoprecipitate to input in the autoradiogram. Three biologically independent experiments were carried out. The average ChIP signal of the biologically independent experiments is reported with SD (S.D.; Microsoft Excel). The Student’s t-test of Microsoft Excel (with tail = 2 and types = 3) was used to determine the P-values for statistical significance of the change in the ChIP signals. The changes were considered to be statistically significant at P < 0.05.
Total RNA preparation
Total RNA was prepared from yeast cell culture as described previously (Shukla et al. 2009; Durairaj et al. 2014b). Briefly, 10 ml yeast culture was harvested, and suspended in 100 µl RNA preparation buffer (500 mM NaCl, 200 mM Tris-HCl, 100 mM Na2EDTA, and 1% SDS) along with 100 µl phenol/chloroform/isoamyl alcohol and 100 µl volume equivalent of glass beads (acid washed; Sigma, St. Louis, MO). Subsequently, yeast cell suspension was vortexed with a maximum speed (10 in a VWR mini-vortexer; cat. no. 58816-121) five times (30 sec each). After vortexing, 150 µl RNA preparation buffer and 150 µl phenol/chloroform/isoamyl alcohol were added to the yeast cell suspension, followed by vortexing for 15 sec with a maximum speed on a VWR mini-vortexer. The aqueous phase was collected for isolation of total RNA by precipitation with ethanol.
Reverse transcriptase PCR analysis (RT-PCR)
RT-PCR analysis was performed as described previously (Durairaj et al. 2014b). Briefly, RNA was treated with RNase-free DNase (M610A; Promega, Madison, WI), and then reverse-transcribed into cDNA using oligo (dT) as described in the protocol supplied by Promega (A3800; Promega). PCR was performed using the synthesized first strand as template and the primer pairs targeted to the PHO84 ORF and 18S rDNA. RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. The average signal of the three biologically independent RT-PCR experiments is reported with SD (Microsoft Excel). The Student’s t-test (with tail = 2 and types = 3) was used to determine P-values for statistical significance of the change in the RT-PCR signals. The changes were considered to be statistically significant at P < 0.05. The primer pairs used in the PCR analysis of cDNAs were as follows:
PHO84: 5′-TCTGCAGACATTTTGGTCAATGGAA-3′ and 5′-AAACGTTTTTGGAACCGGCATAAC-3′.
18S rDNA: 5′-GAGTCCTTGTGGCTCTTGGC-3′ and 5′-AATACTGATGCCCCCGACC-3′.
Data availability
Yeast strains are available upon request.
Results and Discussion
SAGA is predominantly required for PIC formation at PHO84 in the presence, but not absence, of Pi in the growth medium
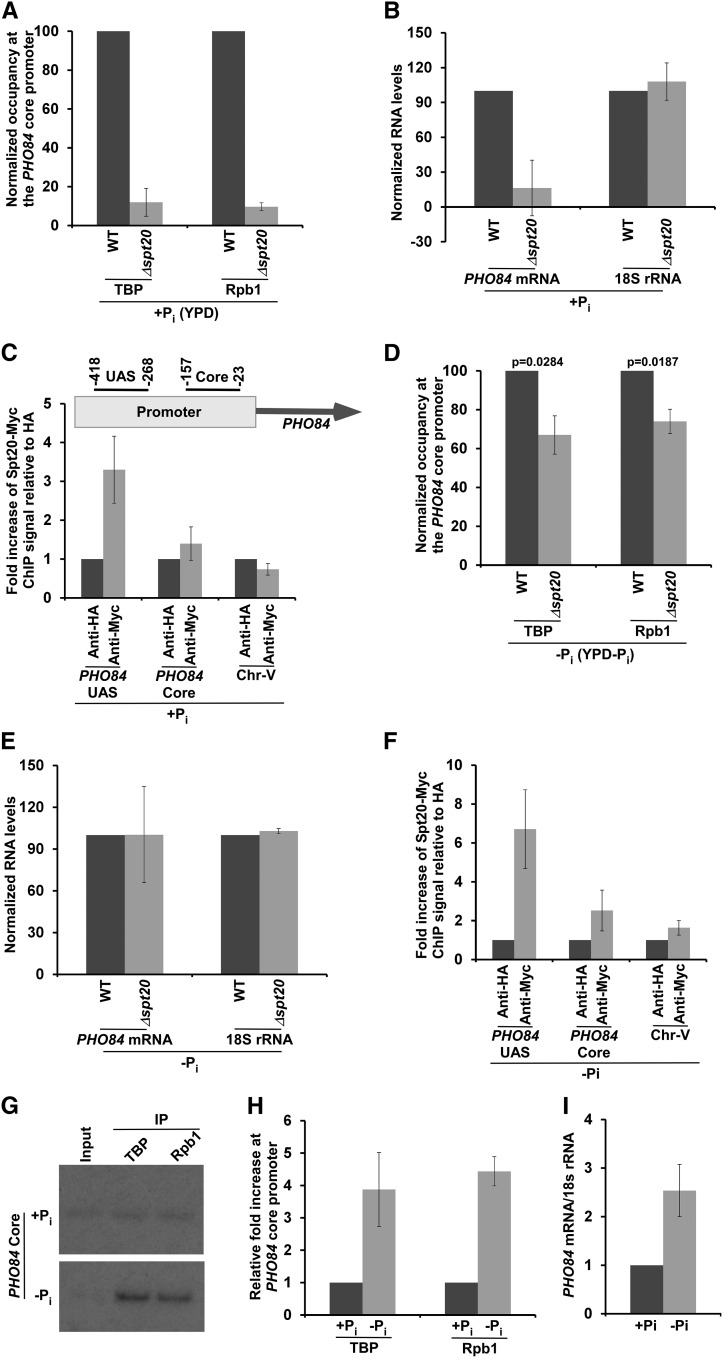
To elucidate the mechanisms of transcriptional initiation at PHO84 in response to Pi, we first analyzed the role of SAGA in the formation of the PIC at the PHO84 core promoter in the presence and absence of Pi in the growth media. Spt20 maintains the global structural and functional integrities of SAGA (Grant et al. 1997; Bhaumik and Green 2001), and thus the null mutant of Spt20 was used to determine the effect of SAGA on PIC formation at the PHO84 core promoter. PIC formation was monitored by analyzing the recruitment of TBP and RNA polymerase II to the PHO84 core promoter using the ChIP assay. TBP nucleates the assembly of general transcription factors to form the PIC, and RNA polymerase II joins toward the end of PIC formation (Bhaumik and Malik 2008; Bhaumik 2011). The largest subunit of RNA polymerase II, Rpb1, is essential to maintain the structural and functional integrities of RNA polymerase II, and thus served as a representative component for our ChIP analysis of RNA polymerase II association with the PHO84 core promoter. Our ChIP analysis revealed that the recruitment of the PIC components TBP and RNA polymerase II to the PHO84 core promoter was greatly impaired in the absence of Spt20 in YPD that contains Pi (or +Pi) (Figure 1A). In agreement with impaired PIC formation in the Δspt20 strain in YPD, transcription of PHO84 was also reduced in the absence of Spt20 (Figure 1B). As a control, the level of the 18S rRNA was monitored, since SAGA does not regulate transcription of RNA polymerase I genes or rDNA. We found that the 18S rRNA level did not change in the Δspt20 strain (Figure 1B). These results support the role of Spt20 or SAGA in the stimulation of PIC formation (and hence transcriptional initiation) at the PHO84 promoter in YPD. Therefore, Spt20 or SAGA is likely to be targeted to the UAS of PHO84 to promote PIC formation at the core promoter in initiating transcription in YPD, analogous to the SAGA-dependent transcriptional initiation mechanism at the GAL1 gene (Bhaumik and Green 2001; Larschan and Winston 2001; Bhaumik et al. 2004; Bhaumik and Malik 2008; Bhaumik 2011). To test this, we analyzed the recruitment of the Spt20 component of SAGA to the PHO84 UAS (Wippo et al. 2009) in YPD. To do this, we tagged Spt20 with Myc epitope at the C-terminus in its chromosomal locus, and then performed the ChIP assay using an anti-Myc antibody against Myc epitope-tagged Spt20. Our ChIP analysis revealed that Spt20 was recruited to the UAS of PHO84 in YPD (Figure 1C). An inactive region within Chr.-V was used as a nonspecific DNA control in the ChIP analysis, similar to our previous studies (Lee et al. 2007; Sen et al. 2016). Further, an anti-HA was used as a nonspecific antibody control in the ChIP assay. These controls did not show specific ChIP signals. However, we observed specific association of the Spt20 component of SAGA with the UAS of PHO84 (Figure 1C). Thus, our results demonstrate that SAGA is recruited to the PHO84 UAS (Figure 1C) to promote PIC formation at the core promoter (Figure 1A), and hence transcriptional initiation (Figure 1B), in the presence of Pi in the growth medium (or YPD).
Figure 1.
SAGA is required for PIC formation at the PHO84 core promoter to initiate transcription in the presence of inorganic phosphate (Pi) in the growth medium. (A) ChIP analysis of TBP and RNA polymerase II (Rpb1) at the PHO84 core promoter in the Δspt20 and WT strains in the growth medium with Pi (i.e., YPD or +Pi). The ChIP signal of the WT strain was set to 100, and the ChIP signal of the mutant strain was normalized with respect to 100. (B) RT-PCR analysis of PHO84 mRNA and 18S rRNA levels in the WT and Δspt20 strains in YPD. (C) ChIP analysis for the recruitment of the Spt20 component of SAGA to the PHO84 promoter in YPD. Upper panel: Schematic diagram showing the locations of the primer pairs at the PHO84 promoter for ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). (D) ChIP analysis of TBP and Rpb1 at the PHO84 core promoter in the Δspt20 and WT strains in the growth medium without Pi (i.e., YPD-Pi or –Pi). (E) RT-PCR analysis of PHO84 mRNA and 18S rRNA levels in the WT and Δspt20 strains in growth medium without Pi. (F) ChIP analysis for recruitment of the Spt20 component of SAGA to the PHO84 promoter in the absence of Pi in the growth medium. (G) ChIP analysis of TBP and Rpb1 at the PHO84 core promoter in the presence and absence of Pi in the growth media. (H) Results of (G) are plotted in the form of a histogram. (I) Relative PHO84 mRNA levels in the presence and absence of Pi in the growth media. ChIP, chromatin immunoprecipitation; Pi, inorganic phosphate; PIC, preinitiation complex; UAS, upstream activating sequence; WT, wild-type; Chr. -V, Chromosome V; and TBP, TATA-box binding protein.
Next, we analyzed whether SAGA plays a similar role in the stimulation of PIC formation at the PHO84 core promoter in the absence of Pi in the growth medium (i.e., YPD-Pi or -Pi). We found that the recruitment of TBP and RNA polymerase II to the PHO84 core promoter was modestly decreased in the Δspt20 strain in comparison to the wild-type equivalent in the absence of Pi in the growth medium (Figure 1D). However, such a modest defect in PIC formation at the PHO84 core promoter in the Δspt20 strain in the YPD-Pi medium did not result in an impaired steady-state level of PHO84 mRNA (Figure 1E), as a small defect in PIC formation may not be reflected in the steady-state levels of certain mRNAs with long half-life (or high stability). Taken together, our results support the idea that SAGA is predominantly required for PIC formation (and hence transcriptional initiation) at the PHO84 core promoter in the presence, but not absence, of Pi in the growth medium. However, SAGA is found to be present at the PHO84 UAS in the absence of Pi (Figure 1F), but has a modest effect on PIC formation (Figure 1D). This could be due to different chromatin structure at the PHO84 promoter in the YPD-Pi medium in comparison to that in the YPD medium; and such, chromatin structure might not predominantly require SAGA’s function, but may rather be dependent on NuA4 for PIC formation (and hence transcriptional initiation), as described below.
TFIID is predominantly required for transcriptional initiation of PHO84 in the absence, but not presence, of Pi in the growth medium
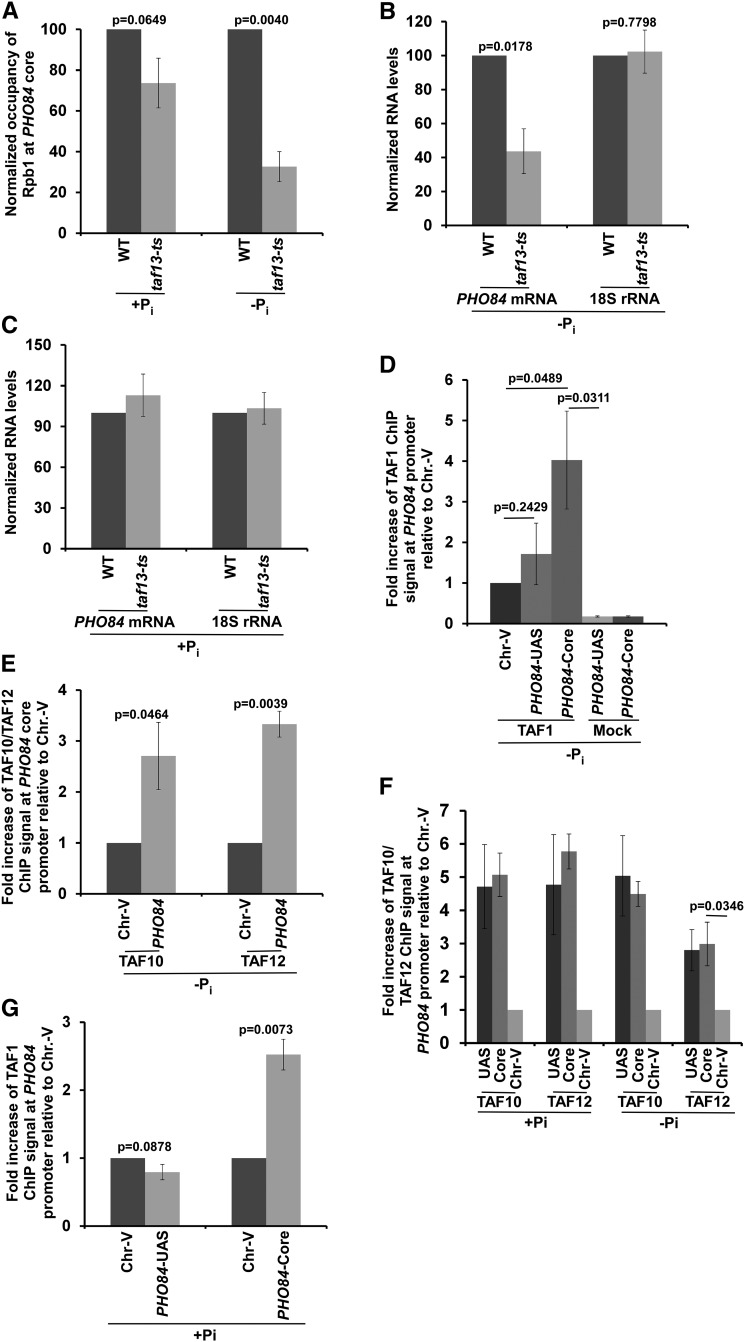
We found that SAGA is modestly required for the recruitment of TBP and RNA polymerase II to the PHO84 core promoter in the absence of Pi in the growth medium. However, recruitment of TBP and RNA polymerase II to the PHO84 core promoter (and hence transcription) is significantly higher in the absence of Pi in comparison to the presence of Pi in the growth media (Figure 1, G–I), consistent with previous studies (Springer et al. 2003). Thus, other factor(s) might promote PIC formation at the PHO84 core promoter in the absence of Pi in the growth medium. It is likely that TFIID is involved in promoting PIC formation at the PHO84 core promoter in the absence of Pi, since ∼90% RNA polymerase II genes are regulated by TFIID (Lee et al. 2000; Bhaumik 2011). To test this, we analyzed the recruitment of the RNA polymerase II component of the PIC to the PHO84 core promoter in the wild-type and ts mutant strains of the Taf13 subunit of TFIID in the absence of Pi in the growth medium. The taf13-ts mutant impairs recruitment of TAF11, and hence TFIID assembly and transcription (Shen et al. 2003). Intriguingly, we found that the recruitment of RNA polymerase II to the PHO84 core promoter was greatly decreased in the taf13-ts mutant strain in comparison to the wild-type equivalent in YPD-Pi medium (Figure 2A), thus supporting a role of TFIID in the stimulation of PIC formation at the PHO84 core promoter in the absence of Pi in the growth medium. Consistently, transcription of PHO84 was impaired in the taf13-ts mutant strain in the YPD-Pi medium (Figure 2B). The 18S rRNA level was monitored as a control, since Taf13/TFIID does not regulate transcription of the RNA polymerase I genes or rDNA. Thus, our results demonstrate that the Taf13 subunit of TFIID is required for recruitment of the RNA polymerase II component of the PIC to the PHO84 core promoter and, consequently, transcription of PHO84. These results support the role of TFIID in the stimulation of PIC formation at the PHO84 core promoter (and hence transcriptional initiation) in the absence of Pi in the growth medium.
Figure 2.
TFIID is required for PIC formation at the PHO84 core promoter to initiate transcription in the absence of Pi in the growth medium. (A) ChIP analysis of Rpb1 at the PHO84 core promoter in the taf13-ts mutant and WT strains in the growth medium with or without Pi. (B) RT-PCR analysis of PHO84 mRNA and 18S rRNA levels in the WT and taf13-ts mutant strains in the growth medium without Pi. (C) RT-PCR analysis of PHO84 mRNA and 18S rRNA levels in the WT and taf13-ts mutant strains in the growth medium with Pi. (D) ChIP analysis for the recruitment of TAF1 to the PHO84 UAS, core promoter, and Chr.-V in the absence of Pi in the growth medium. Mock ChIP control was performed without using an antibody. (E) ChIP analysis for the recruitment of TAF10 and TAF12 to the PHO84 core promoter and Chr.-V in the absence of Pi in the growth media. (F) ChIP analysis for the recruitment of TAF10 and TAF12 to the PHO84 core promoter, PHO84 UAS, and Chr.-V in the absence and presence of Pi in the growth. (G) ChIP analysis for the recruitment of TAF1 to the PHO84 core promoter, PHO84 UAS, and Chr.-V in the presence of Pi in the growth medium. ChIP, chromatin immunoprecipitation; Chr.-V, chromosome-V; Pi, inorganic phosphate; UAS, upstream activating sequence; WT, wild-type.
Since PHO84 transcriptional initiation occurs predominantly via SAGA in the presence of Pi, TFIID is not likely to participate (or is likely to have minimal role) in transcriptional initiation of PHO84 in YPD (or +Pi). To test this, we next analyzed the role of Taf13 in the regulation of PIC formation at the PHO84 core promoter in the YPD growth medium. To this end, we analyzed the recruitment of RNA polymerase II to the PHO84 core promoter in the wild-type and ts mutant strains of the Taf13 component of TFIID in the YPD medium. We found that the recruitment of RNA polymerase II to the PHO84 core promoter was not significantly decreased in the taf13-ts mutant strain in comparison to the wild-type equivalent in the YPD growth medium (Figure 2A). Further, the steady-state level of PHO84 mRNA was not found to be altered in the taf13-ts mutant strain in the YPD growth medium (Figure 2C). Taken together, our results support the idea that TFIID is predominantly required for PIC formation at the PHO84 core promoter (and hence transcriptional initiation) in the absence, but not presence, of Pi in the growth medium.
Since TFIID is predominantly required for PIC formation at the PHO84 core promoter in the absence, but not presence, of Pi in the growth medium, it is likely to be recruited to the PHO84 core promoter in the absence of Pi. To test this, we analyzed the recruitment of a TFIID-specific component, TAF1, to the PHO84 core promoter in the absence of Pi in the growth medium. We found that the TFIID-specific TAF1 was recruited to the PHO84 core promoter in the absence of Pi in the growth medium (Figure 2D). An inactive region within Chr.-V was used as a nonspecific DNA control (Figure 2D). Further, ChIP without antibody was performed as a mock control (Figure 2D). These results support the recruitment of TFIID to the PHO84 core promoter in the absence of Pi in the growth medium. Likewise, we also observed recruitment of the TAF10 and TAF12 components of TFIID to the PHO84 core promoter in the absence of Pi in the growth medium (Figure 2E). However, unlike TAF1 (Figure 2D), TAF10 and TAF12 were also recruited to the PHO84 UAS, most likely with SAGA (Figure 2F), as SAGA is recruited to the PHO84 UAS in the absence of Pi in the growth medium (Figure 1F). Likewise, TAF10 and TAF12 were predominantly recruited to the PHO84 UAS, as bona fide components of the SAGA coactivator, in the presence of Pi in the growth medium (Figure 2F), which was consistent with our results presented in Figure 1C and previous publications (Bhaumik and Green 2001, 2002; Bhaumik et al. 2004; Shukla et al. 2006a,b, 2012). However, we also observed recruitment of TAF10 and TAF12 to the PHO84 core promoter in the presence of Pi in the growth medium (Figure 2F). Such recruitment of TAF10 and TAF12 to the PHO84 core promoter in the presence of Pi in the growth medium could be mediated via TFIID, but not SAGA, as Spt20 is not recruited to the PHO84 core promoter, while TFIID-specific TAF1 is associated with the PHO84 core promoter in YPD medium (Figure 2G). Taken together, our results support the idea that TFIID is recruited to the PHO84 core promoter (Figure 2D) to stimulate PIC formation (and hence transcriptional initiation) in the absence of Pi in the growth medium (Figure 2, A and B). On the other hand, SAGA is recruited to the PHO84 UAS to promote TFIID-independent PIC formation at the core promoter in facilitating transcription in the presence of Pi in the growth medium (Figure 1, A–C and Figure 2, A and C). However, SAGA and TFIID are recruited to PHO84 under both growth conditions (i.e., +Pi and −Pi), but predominantly functional in the presence and absence of Pi, respectively, in the growth media. Such distinct functional requirements of SAGA and TFIID could be due to different chromatin structures at the PHO84 promoter in the presence and absence of Pi in the growth media, as described below.
NuA4 HAT is recruited to the PHO84 UAS to promote PIC formation at the core promoter in the absence of Pi in the growth medium
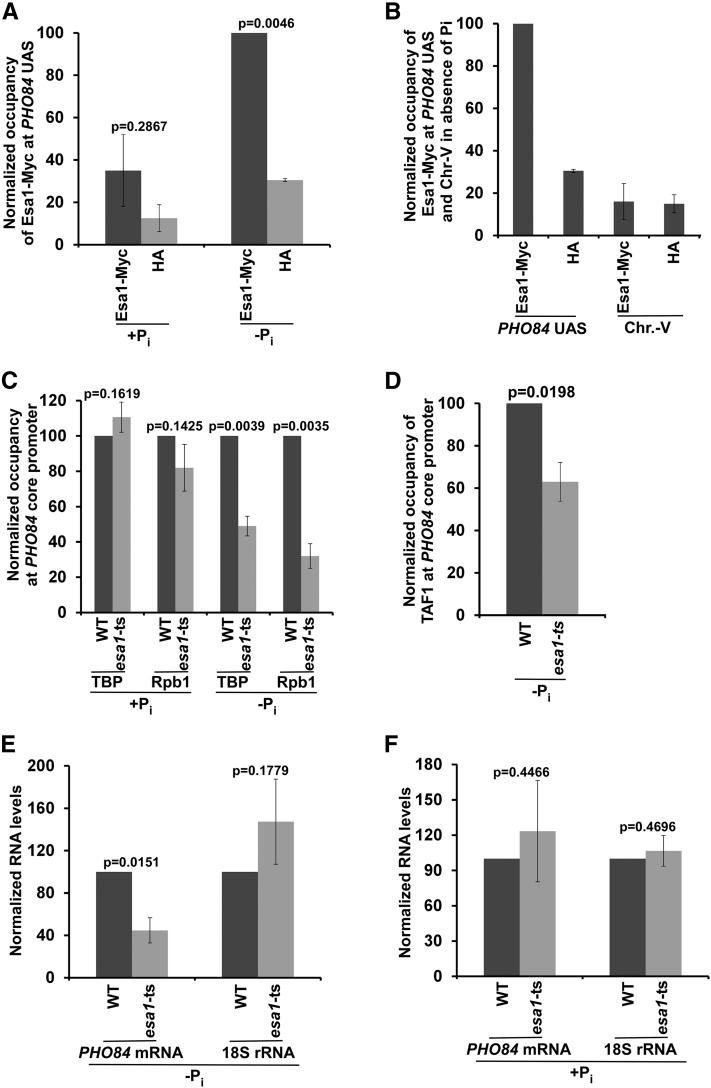
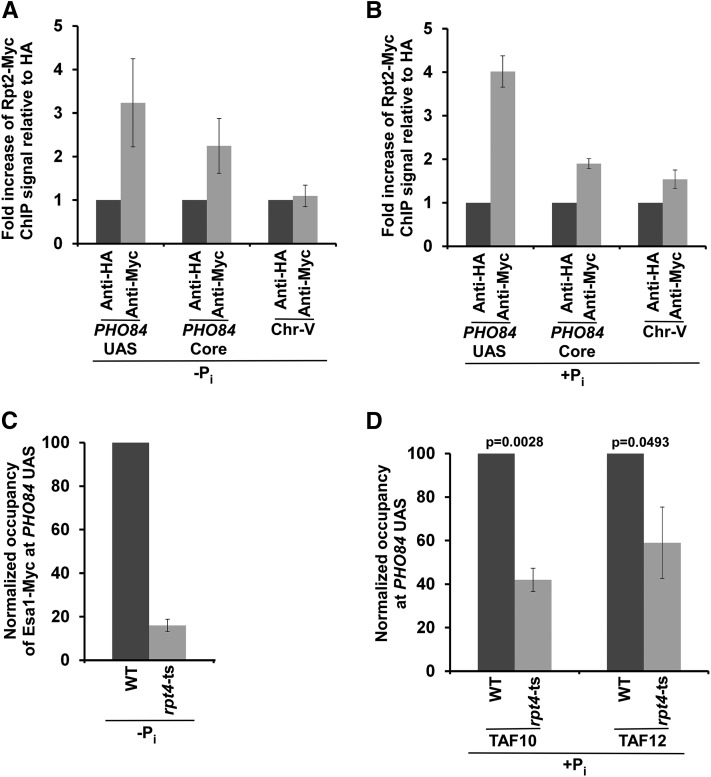
We have recently demonstrated that NuA4 HAT is targeted to the UASs of the ribosomal protein genes to facilitate the recruitment of TFIID at the core promoters to enhance transcriptional initiation (Uprety et al. 2012, 2015), thus supporting a role of NuA4 HAT in the transcriptional initiation of TFIID-regulated genes. Therefore, NuA4 HAT is likely to be recruited to the PHO84 UAS in the absence of Pi in the growth medium to promote TFIID-dependent PIC formation at the core promoter in facilitating transcription. However, it has not yet been analyzed whether NuA4 HAT is required for TFIID recruitment at nonribosomal protein genes. To address this, we analyzed the recruitment of NuA4 HAT to the PHO84 UAS and its role in PIC formation at the PHO84 core promoter (and hence transcription). To this end, we tagged the HAT-containing Esa1 subunit of NuA4 with Myc epitope, and then performed the ChIP assay at the PHO84 UAS using an anti-Myc antibody against Myc-tagged Esa1. Our ChIP analysis reveals that Esa1 is recruited to the PHO84 UAS in the absence of Pi in the growth medium (Figure 3, A and B). Consistently, NuA4 HAT enhances recruitment of the TBP and RNA polymerase II components of the PIC to the PHO84 core promoter in the absence of Pi in the growth medium (Figure 3C). Further, recruitment of the TFIID-specific TAF1 to the PHO84 core promoter is decreased in the esa1-ts mutant strain at the nonpermissive temperature in the absence of Pi in the growth medium (Figure 3D). The esa1-ts mutant impairs histone H4 acetylation, which is implicated in facilitating TFIID recruitment (Clarke et al. 1999; Reid et al. 2000; Mencía et al. 2002; Durant and Pugh 2007; Uprety et al. 2012, 2015). In agreement with these results, transcription of PHO84 is also impaired in the esa1-ts mutant strain at the nonpermissive temperature in the absence of Pi in the growth medium (Figure 3E). On the other hand, NuA4 HAT is not found to be predominantly associated with the PHO84 UAS in the presence of Pi in the growth medium, in comparison to the growth medium without Pi (Figure 3A). This is as expected, since PIC formation at the PHO84 core promoter is stimulated by SAGA in the presence of Pi (Figure 1, A–C), and the targeting of SAGA is independent of NuA4 (Reid et al. 2000; Uprety et al. 2012, 2015). Consistently, NuA4 HAT is dispensable for PIC formation at the PHO84 core promoter in the presence of Pi in the growth medium (Figure 3C). Hence, transcription of PHO84 is also not impaired in the esa1-ts mutant strain in the presence of Pi in the growth medium (Figure 3F). Thus, our results demonstrate that NuA4 HAT is recruited to the PHO84 UAS to promote TFIID-dependent PIC formation at the core promoter in the stimulation of transcriptional initiation in the absence of Pi in the growth medium (Figure 2, A, B, D, and E and Figure 3, A–E).
Figure 3.
NuA4 HAT is required for PIC formation at the PHO84 core promoter (and hence transcription) in the absence of Pi in the growth medium. (A and B) ChIP analysis of Esa1 at the PHO84 UAS in the presence and absence of Pi in the growth media. (C) ChIP analysis of TBP and Rpb1 at the PHO84 core promoter in the esa1-ts mutant and WT strains in the presence or absence of Pi in the growth medium. (D) Esa1 is required for recruitment of a TFIID-specific TAF1 to the PHO84 core promoter in the absence of Pi in the growth medium. (E) RT-PCR analysis of PHO84 mRNA and 18S rRNA levels in the WT and esa1-ts mutant strains in the absence of Pi in the growth medium. (F) RT-PCR analysis of PHO84 mRNA and 18S rRNA levels in the WT and esa1-ts mutant strains in the presence of Pi in the growth medium. ChIP, chromatin immunoprecipitation; Chr.-V, chromosome-V; Pi, inorganic phosphate; PIC, preinitiation complex; UAS, upstream activating sequence; WT, wild-type.
Esa1 HAT is present in both NuA4 and PicNuA4 (which contains Esa1, Epl1, and Yng2). While NuA4 is recruited to the active gene for targeted histone H4 acetylation, picNuA4 is involved in global histone H4 acetylation (Reid et al. 2000; Mencía et al. 2002; Uprety et al. 2012, 2015). Targeted histone H4 acetylation by NuA4 promotes transcription (Reid et al. 2000; Uprety et al. 2015). Recruitment of NuA4 for targeted histone H4 acetylation at the active gene is detected by the ChIP assay (Reid et al. 2000; Uprety et al. 2015). On the other hand, association of picNuA4 with the genome for global nontargeted histone H4 acetylation is not detected by the ChIP assay, as described previously (Reid et al. 2000; Uprety et al. 2015). Further, we find here that Esa1 is predominantly associated with PHO84 UAS in the absence of Pi in the growth medium. However, similar recruitment of Esa1 to the PHO84 UAS was not observed in the presence of Pi in the growth medium. Moreover, nonspecific DNA (Chr.-V) and nonspecific antibody (i.e., anti-HA) control signals are much lower than the Esa1 ChIP signal at the PHO84 UAS in the absence of Pi in the growth medium (Figure 3B). These results indicate that Esa1 associated with NuA4 is targeted to the PHO84 UAS in the absence of Pi in the growth medium, in agreement with previous studies (Reid et al. 2000; Uprety et al. 2015). Thus, our results are consistent with the established role of NuA4 in gene activation.
The 19S RP is required for transcriptional initiation of PHO84 in the presence, as well as absence, of Pi in the growth media
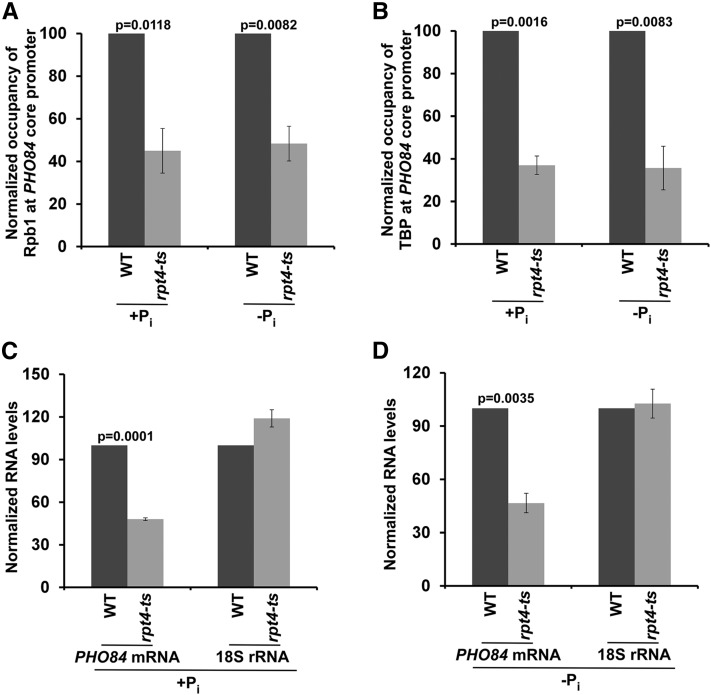
So far, our results demonstrate that NuA4-TFIID and SAGA predominantly facilitate PIC formation at the PHO84 core promoter in the absence and presence of Pi in the growth media, respectively. Our previous studies have demonstrated that both SAGA- and TFIID-dependent genes are regulated by the 19S RP (Malik et al. 2009; Bhaumik 2011; Uprety et al. 2012). Thus, TFIID and SAGA-dependent transcriptional initiations of PHO84 are likely to be regulated by the 19S RP in the presence and absence of Pi in the growth media. To test this, we analyzed the recruitment of the RNA polymerase II component of the PIC to the PHO84 core promoter in the wild-type and ts mutant strains of the Rpt4 subunit of the 19S RP, which is essential for its structural and functional integrity, and is degraded at the nonpermissive temperature (Park et al. 2009). We found that the recruitment of RNA polymerase II to the PHO84 core promoter was impaired in the rpt4-ts mutant strain at the nonpermissive temperature in the presence and absence of Pi in the growth media (Figure 4A). Likewise, recruitment of the TBP component of the PIC to the PHO84 core promoter was also impaired in the growth medium with or without Pi (Figure 4B). Thus, the 19S RP promotes the formation of the PIC at the PHO84 core promoter during both SAGA- and TFIID-dependent transcriptional initiations. Consistently, transcription of PHO84 was found to be impaired in the rpt4-ts mutant strain at the nonpermissive temperature, in the presence and absence of Pi in the growth media (Figure 4, C and D). The level of 18S rRNA was monitored as a control, since transcription of rDNA or RNA polymerase I genes is not regulated by the 19S RP. Hence, the 19S RP facilitates both SAGA- and TFIID-dependent transcriptional initiations of PHO84.
Figure 4.
The 19S RP is required for recruitment of RNA polymerase II and TBP to the PHO84 core promoter, and hence transcription, in the presence as well as absence of Pi in the growth media. (A and B) ChIP analysis for association of RNA polymerase II and TBP with the PHO84 core promoter in the WT and rpt4-ts mutant strains in the growth medium with or without Pi. (C and D) RT-PCR analysis of PHO84 mRNA and 18S rRNA levels in the WT and rpt4-ts mutant strains in the presence or absence of Pi in the growth medium. ChIP, chromatin immunoprecipitation; Chr.-V, chromosome-V; Pi, inorganic phosphate; RP, regulatory particle; UAS, upstream activating sequence; WT, wild-type.
Since the 19S RP facilitates both SAGA- and TFIID-dependent transcriptional initiations of PHO84, it is likely to be present at the PHO84 promoter in the presence as well as absence of Pi in the growth media. In support of this, our previous studies demonstrated that the 19S RP is predominantly targeted to the UASs of TFIID- and SAGA-regulated genes to enhance the recruitment of the NuA4 and SAGA coactivators, respectively, for the stimulation of PIC formation at the core promoters, and hence transcriptional initiation (Malik et al. 2009; Bhaumik 2011; Uprety et al. 2012). To test whether the 19S RP is recruited to the PHO84 UAS to enhance the targeting of NuA4 and SAGA, we first analyzed the association of the 19S RP with the PHO84 promoter in the presence and absence of Pi in the growth media, and then analyzed its role in targeting SAGA and NuA4. To this end, we tagged the Rpt2 component of the 19S RP with Myc epitope and then performed the ChIP assay. Our results revealed predominant recruitment of the 19S RP (Rpt2-Myc) to the PHO84 UAS in the presence and absence of Pi in the growth media (Figure 5, A and B). Thus, our results support the idea that the 19S RP is recruited to the PHO84 UAS (Figure 5, A and B), and promotes PIC formation (and hence transcription) at the core promoter in growth medium with or without Pi (Figure 4, A–D). Such stimulation of PIC formation at the PHO84 core promoter by the 19S RP could be mediated via facilitated recruitment of the coactivators SAGA and NuA4 to the PHO84 UAS, as the 19S RP is known to promote the targeting of SAGA and NuA4 to the UASs to enhance PIC formation at the two distinct sets of genes (Lee et al. 2005; Malik et al. 2009; Bhaumik 2011; Uprety et al. 2012). To test this, we analyzed the recruitment of NuA4 (Esa1-Myc; Uprety et al. 2012, 2015) and SAGA (TAF10 and TAF12, since they are recruited to the PHO84 UAS as SAGA components and TFIID is not recruited to the PHO84 UAS; Figure 2, D–G; Bhaumik and Green 2001) to the PHO84 UAS in the absence and presence of Pi, respectively, in the growth media in the wild-type and rpt4-ts mutant strains. Our ChIP analysis reveals that the 19S RP promotes the targeting of coactivators SAGA and NuA4 to the PHO84 UAS in the presence and absence of Pi, respectively, in the growth media (Figure 5, C and D). Taken together, our results support the idea that the 19S RP promotes the targeting of SAGA and NuA4 to the PHO84 UAS (Figure 5) to facilitate PIC formation at the core promoter (and hence transcriptional initiation) in the presence and absence of Pi in the growth media, respectively (Figure 1, Figure 2, Figure 3, and Figure 4).
Figure 5.
The 19S RP is predominantly recruited to the PHO84 UAS to promote the targeting of NuA4 HAT and SAGA to the PHO84 UAS in the absence and presence of Pi in the growth media, respectively. (A and B) ChIP analysis of Rpt2 at the PHO84 promoter and an inactive region within Chr.-V in the growth medium with or without Pi. (C) ChIP analysis of NuA4 HAT at the PHO84 UAS in the WT and rpt4-ts mutant strains in the growth medium lacking Pi. (D) ChIP analysis of TAF10 and TAF12 at the PHO84 UAS in the WT and rpt4-ts mutant strains in the growth medium containing Pi. ChIP, chromatin immunoprecipitation; Chr.-V, chromosome-V; Pi, inorganic phosphate; RP, regulatory particle; UAS, upstream activating sequence; WT, wild-type.
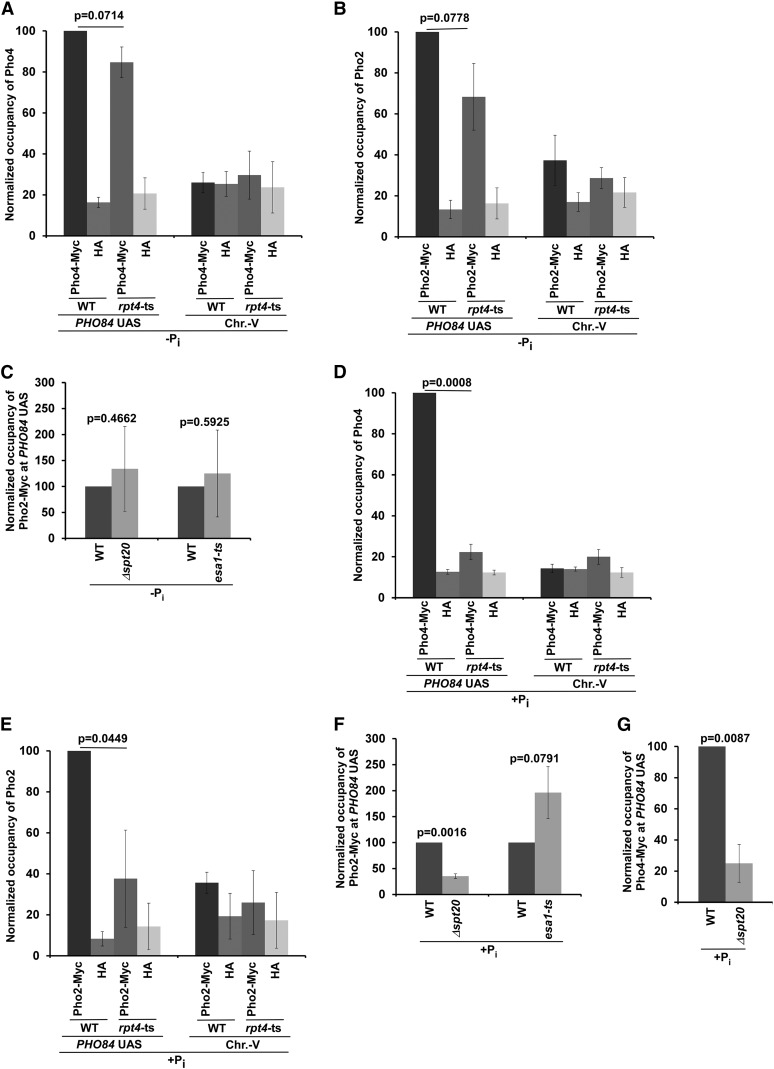
The 19S RP does not regulate the recruitment of Pho2 and Pho4 to the PHO84 UAS in the absence of Pi in the growth medium
We find that 19S RP promotes the targeting of NuA4 HAT for the recruitment of TFIID at the PHO84 core promoter in the absence of Pi in the growth medium, consistent with our recent results at the ribosomal protein genes (Uprety et al. 2012). Next, we analyzed whether the 19S RP facilitates the targeting of NuA4 HAT to the PHO84 UAS by enhancing the recruitment of the activators Pho2 and Pho4 (which are recruited together in Pho2-dependent manner; Springer et al. 2003) to the PHO84 UAS in the growth medium lacking Pi. To this end, we added Myc epitope tags to the C-termini of the Pho2 and Pho4 activators in the rpt4-ts mutant and wild-type strains, and then performed the ChIP assay. We found that the 19S RP did not promote the targeting of the activators Pho2 and Pho4 to the PHO84 UAS (Figure 6, A and B), but rather facilitated the recruitment of NuA4 HAT (Figure 5C). These results support the idea that the 19S RP facilitates the targeting of NuA4 HAT, but not activator, to the PHO84 UAS in the absence of Pi in the growth medium, similar to our results at the ribosomal protein genes (Uprety et al. 2012). Since 19S RP promotes the targeting of NuA4 HAT, but not activator, to the PHO84 UAS in the absence of Pi in the growth medium, NuA4 HAT is not likely to regulate the recruitment of activator under this growth condition, similar to the results obtained in the rpt4-ts mutant strain (Figure 6, A and B). Indeed, we find that activator recruitment to the PHO84 UAS is not altered in the esa1-ts mutant stain in the growth medium without Pi (Figure 6C). Likewise, SAGA does not regulate activator recruitment to the PHO84 UAS in the absence of Pi in the growth medium (Figure 6C). Thus, like the results at the ribosomal protein genes (Uprety et al. 2012), the 19S RP promotes the targeting of NuA4 HAT, but not activator, for PIC formation for transcriptional initiation at PHO84 in the growth medium lacking Pi. On the other hand, we find that the recruitment of Pho2 and Pho4 to the PHO84 UAS is significantly decreased in the rpt4-ts mutant in the presence of Pi in the growth medium (Figure 6, D and E). Likewise, recruitment of Pho2 and Pho4 to the PHO84 UAS is impaired in the Δspt20 strain in the growth medium containing Pi (Figure 6, F and G). However, NuA4 HAT does not regulate activator recruitment to the PHO84 UAS under this growth condition (Figure 6F), in agreement with the fact that NuA4 HAT is not predominantly associated with the PHO84 UAS for transcriptional initiation in the presence of Pi in the growth medium (Figure 3, A, C, and F). Thus, SAGA and 19S RP, but not NuA4 HAT, are required for activator recruitment to the PHO84 UAS in the growth medium containing Pi.
Figure 6.
ChIP analysis of Pho2 and Pho4 at the PHO84 UAS in the presence and absence of Pi in the growth media. (A and B) ChIP analysis of Pho2 and Pho4 at the PHO84 UAS in the rpt4-ts mutant and WT strains in the growth medium lacking Pi. Maximum ChIP signal was set to 100 and other ChIP signals were normalized with respect to 100. (C) ChIP analysis of Pho2 at the PHO84 UAS in the Δspt20 and esa1-ts mutants and their isogenic WT equivalents in the absence of Pi in the growth medium. Statistical analysis was performed using four sets of biologically independent experiments. (D and E) ChIP analysis of Pho2 and Pho4 at the PHO84 UAS in the rpt4-ts mutant and WT strains in the growth medium containing Pi. (F) ChIP analysis of Pho2 at the PHO84 UAS in the Δspt20 and esa1-ts mutants and their isogenic WT strains in the presence of Pi in the growth medium. (G) ChIP analysis of Pho4 at the PHO84 UAS in the Δspt20 mutant and its isogenic WT equivalent in the growth medium with Pi. ChIP, chromatin immunoprecipitation; Chr.-V, chromosome-V; Pi, inorganic phosphate; UAS, upstream activating sequence; WT, wild-type.
Previous studies (Magbanua et al. 1997b; Wippo et al. 2009) have demonstrated that two of the four Pho2/Pho4 activator-binding sites are within nucleosomes in the presence of Pi in the growth medium, and thus that SAGA might alter the chromatin structure to favor activator recruitment under this growth condition. Indeed, SAGA is required for activator recruitment to the PHO84 UAS (Figure 6, F and G) for transcriptional initiation in the presence of Pi in the growth medium (Figure 1, A–C). Since SAGA recruitment to the PHO84 UAS is impaired in the rpt4-ts mutant in the growth medium containing Pi (Figure 5D), activator impaired to the PHO84 UAS is thus impaired in the rpt4-ts mutant under this growth condition (Figure 6, D and E). However, SAGA and 19S RP are not required for activator recruitment to the PHO84 UAS in the absence of Pi in the growth medium (Figure 6, A–C), since all Pho2/Pho4 activator-binding sites are accessible (or not within nucleosomes) under this growth condition (Magbanua et al. 1997b; Wippo et al. 2009).
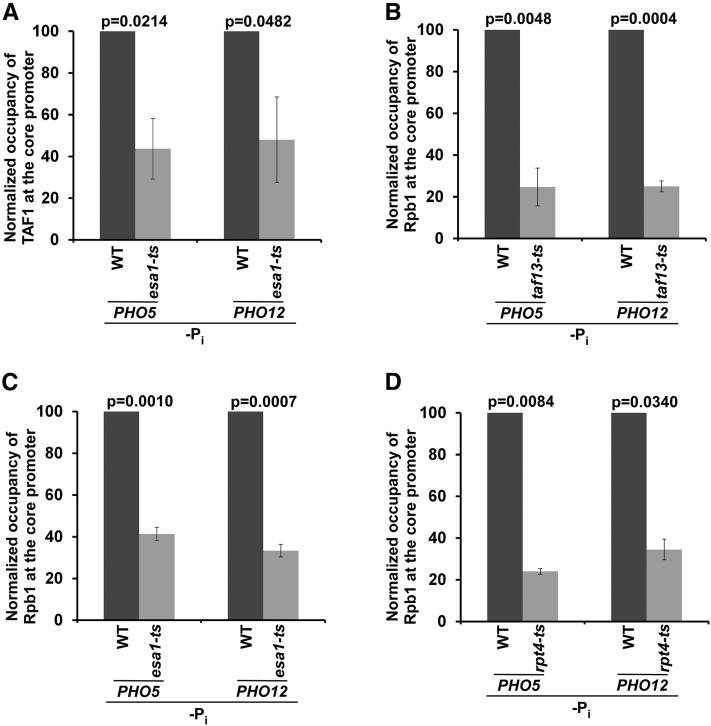
In summary, we demonstrate here that both TFIID and SAGA participate in the stimulation of PIC formation (and hence transcriptional initiation) at the PHO84 promoter under different growth conditions. We find that transcriptional initiation of PHO84 is predominantly dependent on SAGA in the presence of Pi in the growth medium (Figure 1, A–C), and that such regulation is independent of NuA4 HAT (Figure 3, A, C, and F). In the absence of Pi in the growth medium, PHO84 becomes TFIID-dependent (Figure 2, A, B, D, and E). Further, NuA4 HAT promotes the TFIID-dependent PIC formation at the PHO84 core promoter (Figure 2, A, B, D, and E and Figure 3, A–D). Therefore, PHO84 is differentially regulated by NuA4-TFIID and SAGA in response to Pi in the growth media. Such distinct transcriptional regulatory pathways are further facilitated by the 19S RP (Figure 4 and Figure 5). In the absence of Pi in the growth medium, the 19S RP promotes the targeting of NuA4 HAT (Figure 5C), but not activator (Figure 6, A and B), to facilitate TFIID-dependent transcriptional initiation of PHO84 (Figure 2, A, B, D, and E and Figure 3, A–D). In the growth medium containing Pi, the 19S RP facilitates PIC formation at the PHO84 core promoter (and hence transcriptional initiation) by enhancing activator recruitment (Figure 4, A–C, Figure 5B, and Figure 6, D and E). Thus, our results reveal here that transcriptional initiation of the Pi transporter gene PHO84 is predominantly SAGA-dependent in the presence of Pi and NuA4-TFIID-dependent in the absence of Pi in the growth medium, and that these two distinct regulatory pathways are stimulated by the 19S RP. Unlike PHO84, the PHO5 and PHO12 genes are not expressed in YPD, but rather in the absence of Pi in the growth medium (Springer et al. 2003). Similar to the results at PHO84 in the growth medium lacking Pi, transcriptional initiation at PHO5 and PHO12 occurs via the NuA4-TFIID pathway (Figure 7). We find that Esa1 is required for recruitment of the TFIID-specific TAF1 to the PHO5 and PHO12 core promoters (Figure 7A) for the recruitment of RNA polymerase II (Figure 7, B and C), and that such RNA polymerase II recruitment is further facilitated by the 19S RP (Figure 7D). Collectively, our results provide significant mechanistic insights into eukaryotic gene regulation in response to nutrient signaling. Since NuA4, SAGA, TFIID, and 19S RP are highly conserved from yeast to humans, similar regulatory mechanisms are likely to exist in higher eukaryotes.
Figure 7.
ChIP analysis of RNA polymerase II recruitment at the PHO5 and PHO12 core promoters in the esa1-ts, taf13-ts, and rpt4-ts mutants and their isogenic WT equivalents in the absence of Pi in the growth medium. (A) Esa1 is required for TAF1 recruitment to the PHO5 and PHO12 core promoters in the growth medium lacking Pi. (B–D) TAF13, Esa1, and Rpt4 are required for recruitment of Rpb1 to the PHO5 and PHO12 core promoters in the growth medium without Pi. ChIP, chromatin immunoprecipitation; Pi, inorganic phosphate; WT, wild-type.
Acknowledgments
We thank Michael R. Green, Fred Winston, Loraine Pillus, Thomas Kodadek, and Stephen Johnston for yeast strains; Michael R. Green for antibodies; and Bhawana Uprety, Arpan Roy, and Vaibhavi Gujar for technical assistance. The work in the Bhaumik laboratory was supported by grants from the National Institutes of Health (1R15GM088798-01 and 2R15GM088798-02), the American Heart Association (15GRNT25700298), and Southern Illinois University School of Medicine.
Footnotes
Communicating editor: A. Hinnebusch
Literature Cited
- Bergwitz C., Juppner H., 2009. Disorders of phosphate homeostasis and tissue mineralisation. Endocr. Dev. 16: 133–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergwitz C., Rasmussen M. D., DeRobertis C., Wee M. J., Sinha S., et al. , 2012. Roles of major facilitator superfamily transporters in phosphate response in Drosophila. PLoS One 7: e31730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington A., Kemp G. J., Graham R., Russell G., 1992. Phosphate-sensitive enzymes: possible molecular basis for cellular disorders of phosphate metabolism. Clin. Chem. Enzym. Comms. 4: 235–257. [Google Scholar]
- Bhaumik S. R., 2011. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim. Biophys. Acta 1809: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R., 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15: 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R., 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22: 7365–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R., 2003. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 370: 445–454. [DOI] [PubMed] [Google Scholar]
- Bhaumik S. R., Malik S., 2008. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit. Rev. Biochem. Mol. Biol. 43: 419–433. [DOI] [PubMed] [Google Scholar]
- Bhaumik S. R., Raha T., Aiello D. P., Green M. R., 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringhurst F. R., Leder B. Z., 2006. Regulation of calcium and phosphate homeostasis, pp. 805–843 in Endocrinology, Ed. 5, edited by DeGroot L. J., Jameson J. L. W.B. Saunders Co., Philadelphia. [Google Scholar]
- Bun-Ya M., Nishimura M., Harashima S., Oshima Y., 1991. The PH084 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11: 3229–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A. S., Lowell J. E., Jacobson S. J., Pillus L., 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19: 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durairaj G., Sen R., Uprety B., Shukla A., Bhaumik S. R., 2014a Sus1p facilitates pre-initiation complex formation at the SAGA-regulated genes independently of histone H2B de-ubiquitylation. J. Mol. Biol. 426: 2928–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durairaj G., Lahudkar S., Bhaumik S. R., 2014b A new regulatory pathway of mRNA export by an F-box protein, Mdm30. RNA 20: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant M., Pugh B. F., 2007. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 27: 5327–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Pugh B. F., 2011. Sequential recruitment of SAGA and TFIID in a genomic response to DNA damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 31: 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. A., Duggan L., Côté J., Roberts S. M., Brownell J. E., et al. , 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11: 1640–1650. [DOI] [PubMed] [Google Scholar]
- Hirst K., Fisher F., McAndrew P. C., Goding C. R., 1994. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 13: 5410–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T. J., Aken B. L., Beal K., Ballester B., Caccamo M., et al. , 2007. Ensembl 2007. Nucleic Acids Res. 35: D610–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K. L., Pugh B. F., 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 3: 573–585. [DOI] [PubMed] [Google Scholar]
- Kaffman A., Herskowitz I., Tjian R., O’Shea E. K., 1994. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science 263: 1153–1156. [DOI] [PubMed] [Google Scholar]
- Komeili A., O’Shea E. K., 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284: 977–980. [DOI] [PubMed] [Google Scholar]
- Komeili A., O’Shea E. K., 2000. Nuclear transport and transcription. Curr. Opin. Cell Biol. 12: 355–360. [DOI] [PubMed] [Google Scholar]
- Kuras L., Kosa P., Mencia M., Struhl K., 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288: 1244–1248. [DOI] [PubMed] [Google Scholar]
- Larschan E., Winston F., 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15: 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Ezhkova E., Li B., Pattenden S. G., Tansey W. P., et al. , 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123: 423–436. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Shukla A., Schneider J., Swanson S. K., Washburn M. P., et al. , 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096. [DOI] [PubMed] [Google Scholar]
- Lee T. I., Causton H. C., Holstege F. C., Shen W. C., Hannett N., et al. , 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704. [DOI] [PubMed] [Google Scholar]
- Li X. Y., Bhaumik S. R., Green M. R., 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288: 1242–1244. [DOI] [PubMed] [Google Scholar]
- Magbanua J. P., Ogawa N., Harashima S., Oshima Y., 1997a The transcriptional activators of the PHO regulon, Pho4p and Pho2p, interact directly with each other and with components of the basal transcription machinery in Saccharomyces cerevisiae. J. Biochem. 121: 1182–1189. [DOI] [PubMed] [Google Scholar]
- Magbanua J. P., Fujisawa K., Ogawa N., Oshima Y., 1997b The homeodomain protein Pho2p binds at an A/T-rich segment flanking the binding site of the basic-helix-loop-helix protein Pho4p in the yeast PHO promoters. Yeast 13: 1299–1308. [DOI] [PubMed] [Google Scholar]
- Malik S., Shukla A., Sen P., Bhaumik S. R., 2009. The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 284: 35714–35724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Durairaj G., Bhaumik S. R., 2013. Mechanisms of antisense transcription initiation from the 3′ end of the GAL10 coding sequence in vivo. Mol. Cell. Biol. 33: 3549–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencía M., Moqtaderi Z., Geisberg J. V., Kuras L., Struhl K., 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9: 823–833. [DOI] [PubMed] [Google Scholar]
- O’Neill E. M., Kaffman A., Jolly E. R., O’Shea E. K., 1996. Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin-CDK complex. Science 271: 209–212. [DOI] [PubMed] [Google Scholar]
- Oshima Y., 1997. The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 72: 323–334. [DOI] [PubMed] [Google Scholar]
- Park S., Roelofs J., Kim W., Robert J., Schmidt M., et al. , 2009. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459: 866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. L., Iyer V. R., Brown P. O., Struhl K., 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297–1307. [DOI] [PubMed] [Google Scholar]
- Roberts S. M., Winston F., 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 3206–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. J., Johnston S. A., 2001. Evidence that proteolysis of Gal4 cannot explain the transcriptional effects of proteasome ATPase mutations. J. Biol. Chem. 276: 9825–9831. [DOI] [PubMed] [Google Scholar]
- Schneider K. R., Smith R. L., O’Shea E. K., 1994. Phosphate-regulated inactivation of the kinase PHO80–PHO85 by the CDK inhibitor PHO81. Science 266: 122–126. [DOI] [PubMed] [Google Scholar]
- Sen R., Malik S., Frankland-Searby S., Uprety B., Lahudkar S., et al. , 2014. Rrd1p, an RNA polymerase II-specific prolyl isomerase and activator of phosphoprotein phosphatase, promotes transcription independently of rapamycin response. Nucleic Acids Res. 42: 9892–9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R., Ferdoush J., Kaja A., Bhaumik S. R., 2016. Fine-tuning of FACT by the ubiquitin proteasome system in regulation of transcriptional elongation. Mol. Cell. Biol. 36: 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. C., Bhaumik S. R., Causton H. C., Simon I., Zhu X., et al. , 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22: 3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S. R., 2006a Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 26: 3339–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Bajwa P., Bhaumik S. R., 2006b SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 34: 6225–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Durairaj G., Schneider J., Duan Z., Shadle T., et al. , 2009. Stimulation of mRNA export by an F-box protein, Mdm30p, in vivo. J. Mol. Biol. 389: 238–247. [DOI] [PubMed] [Google Scholar]
- Shukla A., Lahudkar S., Durairaj G., Bhaumik S. R., 2012. Sgf29p facilitates the recruitment of TATA box binding protein but does not alter SAGA’s global structural integrity in vivo. Biochemistry 51: 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Wykoff D. D., Miller N., O’Shea E. K., 2003. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A., Tanaka K., Uesono Y., Wickner R. B., 1988. PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol. Gen. Genet. 214: 162–164. [DOI] [PubMed] [Google Scholar]
- Uprety B., Lahudkar S., Malik S., Bhaumik S. R., 2012. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res. 40: 1969–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprety B., Sen R., Bhaumik S. R., 2015. Eaf1p is required for recruitment of NuA4 in targeting TFIID to the promoters of the ribosomal protein genes for transcriptional initiation in vivo. Mol. Cell. Biol. 35: 2947–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprety B., Kaja A., Ferdoush J., Sen R., Bhaumik S. R., 2016. Regulation of antisense transcription by NuA4 histone acetyltransferase and other chromatin regulatory factors. Mol. Cell. Biol. 36: 992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C. J., van Teeffelen H. A., Timmers H. T., 2005. Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol. Cell. Biol. 25: 4863–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K., Horz W., Hinnen A., 1989. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 9: 2050–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A., Kinne R. K., 2001. Evolution of the Na-P(i) cotransport systems. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280: R301–R312. [DOI] [PubMed] [Google Scholar]
- Wippo C. J., Krstulovic B. S., Ertel F., Musladin S., Blaschke D., et al. , 2009. Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol. Cell. Biol. 29: 2960–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff D. D., O’Shea E. K., 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kruk J. A., Reese J. C., 2008. Dissection of coactivator requirement at RNR3 reveals unexpected contributions from TFIID and SAGA. J. Biol. Chem. 283: 27360–27368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Yeast strains are available upon request.