Abstract
Histone post-translational modifications (PTMs) and differential incorporation of variant and canonical histones into chromatin are central modes of epigenetic regulation. Despite similar protein sequences, histone variants are enriched for different suites of PTMs compared to their canonical counterparts. For example, variant histone H3.3 occurs primarily in transcribed regions and is enriched for “active” histone PTMs like Lys9 acetylation (H3.3K9ac), whereas the canonical histone H3 is enriched for Lys9 methylation (H3K9me), which is found in transcriptionally silent heterochromatin. To determine the functions of K9 modification on variant vs. canonical H3, we compared the phenotypes caused by engineering H3.3K9R and H3K9R mutant genotypes in Drosophila melanogaster. Whereas most H3.3K9R, and a small number of H3K9R, mutant animals are capable of completing development and do not have substantially altered protein-coding transcriptomes, all H3.3K9R H3K9R combined mutants die soon after embryogenesis and display decreased expression of genes enriched for K9ac. These data suggest that the role of K9ac in gene activation during development can be provided by either H3 or H3.3. Conversely, we found that H3.3K9 is methylated at telomeric transposons and that this mark contributes to repressive chromatin architecture, supporting a role for H3.3 in heterochromatin that is distinct from that of H3. Thus, our genetic and molecular analyses demonstrate that K9 modification of variant and canonical H3 have overlapping roles in development and transcriptional regulation, though to differing extents in euchromatin and heterochromatin.
Keywords: Drosophila, histone variant, H3, transcription, heterochromatin
DNA interacts with histones and other proteins to establish chromatin environments that affect all DNA-dependent processes. The establishment of chromatin environments is accomplished through multiple mechanisms that collectively comprise the bulk of epigenetic regulation found in eukaryotes. In particular, post-translational modification (PTM) of histones influences DNA/histone interactions and also provides binding sites for recruitment of chromatin modulators that influence gene expression, DNA replication and repair, and chromosome segregation during cell division (Wallrath et al. 2014). In addition to histone PTMs, epigenetic regulation is modulated by the type of histone protein deposited onto DNA. There are two major categories of histone proteins: the canonical histones and the closely related histone variants (Talbert and Henikoff 2010, 2017). These two histone categories are distinguished by the timing of their expression during the cell cycle and their mechanism of deposition onto DNA. Canonical histones are encoded by multiple genes (e.g., ∼55 in humans and ∼500 in flies), organized into clusters that are highly expressed during S-phase of the cell cycle, and are deposited onto DNA by the histone chaperone CAF-1 in a replication-coupled manner (Verreault et al. 1996; Marzluff et al. 2002; Tagami et al. 2004). In contrast, variant histones are typically encoded by one or two genes, are expressed throughout the cell cycle, and can be deposited onto DNA independently of replication by histone chaperones other than CAF-1 (Tagami et al. 2004; Henikoff and Ahmad 2005; Szenker et al. 2011). Variant histones are often deposited at specific genomic locations and have functions that can differ from canonical histones. For example, two histone H2A variants, H2AX and H2A.Z, play critical roles in DNA repair (Scully and Xie 2013; Price and Andrea 2014), and the histone H3 variant CENP-A localizes to centromeres and is essential for kinetochore formation (Blower and Karpen 2001; Mellone and Allshire 2003; Henikoff and Ahmad 2005).
The major histone H3 variant in animal genomes is H3.3, which in both mice and Drosophila is encoded by two different genes (H3.3A and H3.3B) that produce identical proteins. Variant histone H3.3 differs from canonical H3.2 and H3.1 by only four or five amino acids, respectively (Szenker et al. 2011). In each case, three of these different amino acids are located in the globular domain of H3.3 and are necessary and sufficient for interaction with the replication-independent chaperones HIRA and ATRX-DAXX (Ahmad and Henikoff 2002; Tagami et al. 2004; Goldberg et al. 2010; Lewis et al. 2010). In H3.2, the only replication-dependent histone in Drosophila, the fourth amino acid difference occurs at position 31 in the unstructured N-terminal tail (Szenker et al. 2011). Histones H3.2 and H3.1 (collectively hereafter referred to as H3), along with H3.3, are some of the most conserved proteins in all eukaryotes (Malik and Henikoff 2003). The conservation of amino acid differences between H3 and H3.3 during evolution strongly suggests that these proteins perform distinct functions. Indeed, H3.3 and H3 are deposited in different genomic regions in a variety of species (Allis and Wiggins 1984; Mito et al. 2005; Schwartz and Ahmad 2005; Tamura et al. 2009; Jin et al. 2011; Kraushaar et al. 2013). Also, the level of enrichment for particular PTMs differs between H3.3 and H3 (McKittrick et al. 2004; Hake et al. 2006), and H3.3-containing nucleosomes can be less stable than those with H3 (Jin and Felsenfeld 2007; Xu et al. 2010). Although the epigenetic PTM signature on variant and canonical H3 histones is distinct, the degree to which particular histone PTMs found on both H3 and H3.3 can compensate for one another is not fully understood. Here, we explore the common and distinct functions of variant and canonical H3K9 function during Drosophila development.
H3.3 is associated with transcriptionally active regions of the genome with high nucleosome turnover, consistent with H3.3 being enriched in “activating” histone PTMs and depleted in “repressing” histone PTMs (McKittrick et al. 2004; Hake et al. 2006). One of the histone PTMs enriched on H3.3 relative to H3 is acetylation of lysine nine (K9ac), a mark associated with accessible chromatin (McKittrick et al. 2004; Hake et al. 2006). Previous studies have identified K9ac at promoters of genes and in regions of high transcriptional activity (Liang et al. 2004; Bernstein et al. 2005; Roh et al. 2005; Kharchenko et al. 2011). Additionally, mutation of H3K9 acetyltransferases results in compromised transcriptional output, suggesting that K9ac contributes to or is a consequence of gene expression activation (Georgakopoulos and Thireos 1992; Kuo et al. 1998; Wang et al. 1998). Importantly, H3K9 acetyltransferases target other histone residues and have nonhistone substrates as well (Glozak et al. 2005; Spange et al. 2009), indicating that one cannot deduce the function of K9ac solely by mutation of H3K9 acetyltransferases. For example, whereas mutation of the H3K9 acetyltransferase Rtt109 in budding yeast results in sensitivity to DNA-damaging agents, H3K9R mutants, which cannot be acetylated by Rtt109, are insensitive to DNA-damaging agents (Fillingham et al. 2008). Direct investigation of K9ac function in vivo therefore requires mutation of H3K9 itself. Previously, we used a Drosophila histone gene replacement platform (McKay et al. 2015) to generate a canonical H3K9R mutant, and found no significant changes in gene expression at regions of the genome enriched in K9ac (Penke et al. 2016). This observation raises the possibility that H3.3K9ac functions in gene regulation and can compensate for the absence of H3K9ac.
H3.3 is also found at transcriptionally inactive, heterochromatic regions of the genome (Goldberg et al. 2010; Lewis et al. 2010; Wong et al. 2010). Heterochromatin is enriched in H3K9 di- and trimethylation (me2/me3), modifications that recruit Heterochromatin Protein 1 (HP1) and are essential for heterochromatin function (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001; Penke et al. 2016). DNA within heterochromatin is composed of repeated sequence elements, many of which are transcriptionally silent and consist of immobile transposons or transposon remnants. Using H3.3 mutants, it was recently demonstrated that H3.3 is essential for the repression of endogenous retroviral elements and that H3.3 can be methylated at lysine nine (Elsässer et al. 2015). H3.3K9me3 is also important for heterochromatin formation at mouse telomeres (Udugama et al. 2015). These studies did not assess the contribution of canonical H3K9 because strategies for mutating all replication-dependent H3 genes in mammalian cells have not been developed. We recently showed in Drosophila that mutation of canonical H3K9 causes defects in heterochromatin formation and transposon repression (Penke et al. 2016), similar to phenotypes observed in Caenorhabditis elegans in the absence of H3K9 methyltransferases (Zeller et al. 2016). In addition, we detected low levels of K9me2/me3 in H3K9R mutants. Combined, these data suggest that methylated H3.3K9 plays a role in heterochromatin formation and can compensate for the absence of canonical H3K9. However, the extent of functional overlap between variant and canonical H3K9 and the intriguing possibility that identical modifications on variant or canonical histones have distinct functions has yet to be fully investigated.
To better understand the functions of H3 and H3.3 and to compare the functions of the variant and canonical H3K9 residues, we used clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 to generate a variant K9R substitution mutation (H3.3K9R) in Drosophila and combined this with our previously described canonical H3K9R mutant (Penke et al. 2016). By comparing the individual mutant phenotypes of H3.3K9R and H3K9R to the combined H3.3K9R H3K9R mutants using a variety of genomic and cell biological assays, we demonstrate that variant and canonical versions of H3K9 can compensate for each other, although to substantially different extents in euchromatin vs. heterochromatin. H3K9 plays a more substantial role than H3.3K9 in heterochromatin formation and in the repression of transposons, whereas they compensate for each other in controlling euchromatic gene expression, particularly in regions enriched in the activating modification, K9ac.
Materials and Methods
Generation of K9R mutant genotypes
Variant H3.3K9R mutants generated by the cross scheme illustrated in Supplemental Material, Figure S1A in File S1 were selected by the absence of GFP fluorescence and/or the presence of straight wings. First instar larvae from the variant and canonical H3.3K9R H3.2K9R cross described in Figure S1B in File S1 were selected based on the presence of GFP fluorescence. Only larvae that receive the H3HWT or H3K9R transgene will survive embryogenesis, as this transgene provides the only source of canonical histone genes. In Table 2, rows one and two indicate progeny from the cross yw; H3.32x1/CyO, twiGFP × yw; Df(2L)BSC110/CyO, twiGFP. Rows three and four indicate progeny from the cross H3.3BK9R; H3.32x1/CyO, twiGFP × H3.3BK9R; Df(2L)BSC110/CyO, twiGFP. In these crosses, the expected ratio of heterozygous to homozygous H3.3A2x1 animals is 2:1, as CyO, twiGFP/CyO, twiGFP animals do not eclose as adults.
Table 2. H3.3K9R mutants are viable but sterile.
| H3.3B | H3.3Aa | Observed | Expected | Pb | Fertile |
|---|---|---|---|---|---|
| WT | ∆/+ | 535 | 535.3 | n.s. | Yes |
| WT | ∆ | 268 | 267.7 | n.s. | Yes |
| K9R | ∆/+ | 400 | 438 | < 0.005 | Yes |
| K9R | ∆ | 257c | 219 | < 0.005 | Nod |
WT, wild-type; n.s., not significant.
H3.3A2x1 deletion allele.
P-value calculated with a χ2 test.
The higher than expected number of observed H3.3BK9R H3.3ANull animals is presumably due to nonspecific detrimental effects caused by the presence of a balancer chromosome in siblings with the H3.3BK9R mutation and balancer-derived wild-type H3.3A.
Both males and females.
CRISPR/Cas9 mutagenesis and transgene integration
A single guide RNA targeting H3.3B near the K9 residue was inserted into pCFD3 and co-injected with a 2-kb homologous repair template containing the H3.3BK9R substitution (File S2). Constructs were injected into embryos expressing Cas9 from the nanos promoter (nanos-cas9; Kondo and Ueda 2013). Recovered H3.3BK9R alleles were subsequently crossed into H3.3A null backgrounds [H3.3A2x1 over deficiency Df(2L)BSC110]. Independent H3.3BK9R CRISPR alleles were used to generate trans-heterozygous animals for all experiments. To generate H3.3B rescue constructs, a 5-kb genomic sequence containing the entire wild-type H3.3B transcription unit was PCR amplified from genomic DNA of nanos-cas9 flies and cloned into pATTB (File S2). Gibson assembly (Gibson et al. 2009) using primers containing K9R or K9Q substitutions was used to generate mutated versions of H3.3B, and all three constructs were integrated into the 86FB attP landing site by ΦC31-mediated recombination.
Immunofluorescence
Salivary gland preparations stained using anti-H3K9me2, anti-H3K9me3, anti-H3K9ac, or anti-HP1a were performed as previously described (Cai et al. 2010). Antibody sources and concentrations are included in File S2. First instar larval brains were prepared similarly to imaginal wing disc preparations described in Estella et al. (2008).
Western blots
ImageJ densitometry analysis was used to determine K9me2, K9ac, or H3 band intensity (See File S2). Histone modification signal was normalized to corresponding H3 loading control signal. Normalized signals from different titrations of the same genotype were averaged and consequent values were set relative to the wild-type value. This process was completed for two biological replicates for both K9me2 and K9ac.
Sample preparation and sequence data analysis
Formaldehyde Assisted Isolation of Regulatory Elements followed by whole-genome sequencing (FAIRE-seq) and RNA sequencing (RNA-seq) samples were prepared from wandering third instar imaginal wing discs as previously described (McKay and Lieb 2013). Sequencing reads were aligned to the dm6 (6.04) reference genome using Bowtie2 (FAIRE) and TopHat (RNA) default parameters (Langmead and Salzberg 2012; Trapnell et al. 2014). FAIRE peaks were called with MACS2 using a shift size of 110 bp and a stringency cutoff of 0.01 (Zhang et al. 2008). Transcripts were assembled with Cufflinks (Trapnell et al. 2014). Bedtools was used to determine read coverage at peaks and transcripts (Quinlan and Hall 2010) and DESeq2 was used to determine statistical significance (P < 0.05) (Love et al. 2014). The following modENCODE third instar larval chromatin immunoprecipitation sequencing (ChIP-seq) data sets were used: K9me2 = GSE47260 and K9me3 = GSE47258. K9ac ChIP-seq data from imaginal wings discs was generated by Pérez-Lluch et al. (2015) (GSM1363590).
Chromatin state analysis was performed using data from Kharchenko et al. (2011), which assigns small regions of the genome into one of nine different chromatin states. FAIRE peaks were classified as one or more chromatin states based on overlap with regions defined by Kharchenko et al. (2011). Of all the peaks in a particular chromatin state, we determined the percentage of peaks that had significantly different FAIRE signals in mutant compared to wild-type samples. RNA chromatin state analysis was performed in a similar fashion.
Data availability
See Supplemental Experimental Procedures in File S2 for a detailed description of the methods. Strains are available upon request. Sequencing data are available at the Gene Expression Omnibus under accession number GSE106192.
Results
H3.3K9R mutant animals are viable but sterile
To investigate the role of H3.3K9 in Drosophila development and compare it to the role of H3K9, we first generated an H3.3K9R animal by introducing a K9R substitution at the endogenous H3.3B locus using CRISPR/Cas9 and then combining recovered H3.3BK9R mutant alleles with a previously generated H3.3A null allele (H3.3A/B combined genotype denoted hereafter as H3.3K9R; see Figure S1A in File S1, Table 1, and Table S1 in File S1 for histone genotype nomenclature) (Sakai et al. 2009). These H3.3K9R mutants, which contain the full complement of endogenous canonical H3 genes, eclose as adults at the expected Mendelian ratios (Table 2) and appear morphologically normal. Therefore, canonical H3 can provide all of the H3K9 function during Drosophila development. This result is consistent with a previous study that found that flies without any H3.3 protein could be propagated as a stock if canonical H3.2 was expressed from a transgene using the H3.3B promoter (Hödl and Basler 2012). Our results are also in-line with a previous report in which H3.3A and H3.3B null animals containing an H3.3AK9R transgene were viable (Sakai et al. 2009). However, whereas these H3.3AK9R transgenic animals were fertile (Sakai et al. 2009), we found that animals with an endogenous H3.3BK9R mutation and the same H3.3A null allele used by Sakai et al. (2009) were sterile. The sterility of our H3.3K9R animals was rescued in both males and females by a transgene containing the wild-type H3.3B gene ectopically integrated into the genome, suggesting that the relative abundance of H3.3K9R causes sterility (Figure S2 in File S1). We conclude that H3.3K9 plays an essential role during gametogenesis and speculate that different amounts of H3.3K9R histones from H3.3A or H3.3B promoters may account for the differences between our observations and those of Sakai et al. (2009).
Table 1. Genotype description of H3.3 and H3 K9R mutants.
| Canonical histone genotypes | Variant histone genotypes | |||
|---|---|---|---|---|
| Nomenclature | Endogenous | Transgenic | H3.3B | H3.3A |
| WT | WT | — | WT | WT |
| H3.3BK9R | WT | — | K9R | WT |
| H3.3ANull | WT | — | WT | Δ |
| H3.3K9R | WT | — | K9R | Δ |
| H3HWT | Δ | WT | WT | WT |
| H3K9R | Δ | K9R | WT | WT |
| H3.3K9R H3HWT | Δ | WT | K9R | Δ |
| H3.3K9R H3K9R | Δ | K9R | K9R | Δ |
See Table S1 in File S1 for full genotypes. WT, wild-type (WT); —, no transgenic histone array; Δ, gene deletion.
H3.3K9 and H3K9 have overlapping functions during development
We previously observed that canonical H3K9R mutants could complete development, although 98% of these mutant animals died during larval or pupal stages (Penke et al. 2016). We considered the possibility that H3K9R mutant animals progressed to late larval or pupal stages of development because of compensation by H3.3K9. We therefore tested whether the H3.3K9R genotype would advance the H3K9R mutant stage of lethality by observing the development of animals in which the H3.3K9R and H3K9R mutant genotypes were combined (Figure S1B in File S1, Table 1, and Table S1 in File S1). The H3K9R genotype was generated using our previously described histone replacement platform (McKay et al. 2015; Penke et al. 2016). Briefly, the endogenous array of ∼100 canonical histone gene clusters was deleted and replaced with an ectopically located transgene encoding a BAC-based, tandem array of 12 canonical histone gene clusters in which the H3 genes contain a K9R mutation (Figure S1B in File S1). A 12× tandem array of wild-type, canonical histone genes (denoted histone wild-type or H3HWT, Figure S1B in File S1), which fully rescues deletion of the endogenous histone gene array, was used as a control (McKay et al. 2015). Similar to the H3.3K9R mutants, H3HWT animals with the H3.3K9R mutant genotype (denoted hereafter as H3.3K9R H3HWT; see Figure S1B in File S1 and Table 1) were viable (Table 3). However, only 34.6% of H3.3K9R H3HWT progeny eclosed as adults (Table 3), compared to essentially 100% of the H3.3K9R genotype that contained the full complement of endogenous, wild-type H3 genes (Table 2). This result suggests that in the presence of fewer total canonical H3 gene copies, the H3.3K9R mutation is more detrimental. Importantly, animals with the H3.3K9R H3K9R combined mutant genotype containing both the variant and canonical K9R mutation were 100% inviable, dying with high penetrance at the first instar larval stage, much earlier than the majority of H3K9R mutants. These results demonstrate that H3.3K9 can partially compensate for the absence of H3K9, indicating that H3.3K9 and H3K9 have some redundant functions.
Table 3. H3.3K9R and H3K9R mutations are synthetically lethal.
| Embryo hatching | Pupatea | Eclose | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Obs | No. | % | Pb | Obs | No. | % | P | Obs | No. | % | P |
| H3HWT | 389 | 450 | 86.4 | — | 98 | 140 | 70.0 | — | 88 | 140 | 62.9 | — |
| H3K9R | 370 | 480 | 77.1 | < 0.0005 | 183 | 285 | 64.2 | < 0.05 | 3 | 285 | 1.1 | < 0.0005 |
| H3.3K9R H3HWT | 350 | 465 | 75.3 | < 0.0005 | 279 | 462 | 60.4 | < 0.0005 | 160 | 462 | 34.6 | < 0.0005 |
| H3.3K9R H3K9R | 214 | 325 | 65.8 | < 0.0005 | 0 | 130 | 0.0 | < 0.0005 | 0 | 130 | 0.0 | < 0.0005 |
Obs, observed; No., number; —, P value not determined.
The pupation and eclosion values have an identical number of animals analyzed for each genotype because they were obtained from the same brood of animals, while the embryo hatching values were obtained from independent experiments.
P-value calculated with a χ2 test using H3HWT observed values as expected values.
H3K9 PTMs are lost in animals lacking H3.3K9 and H3K9
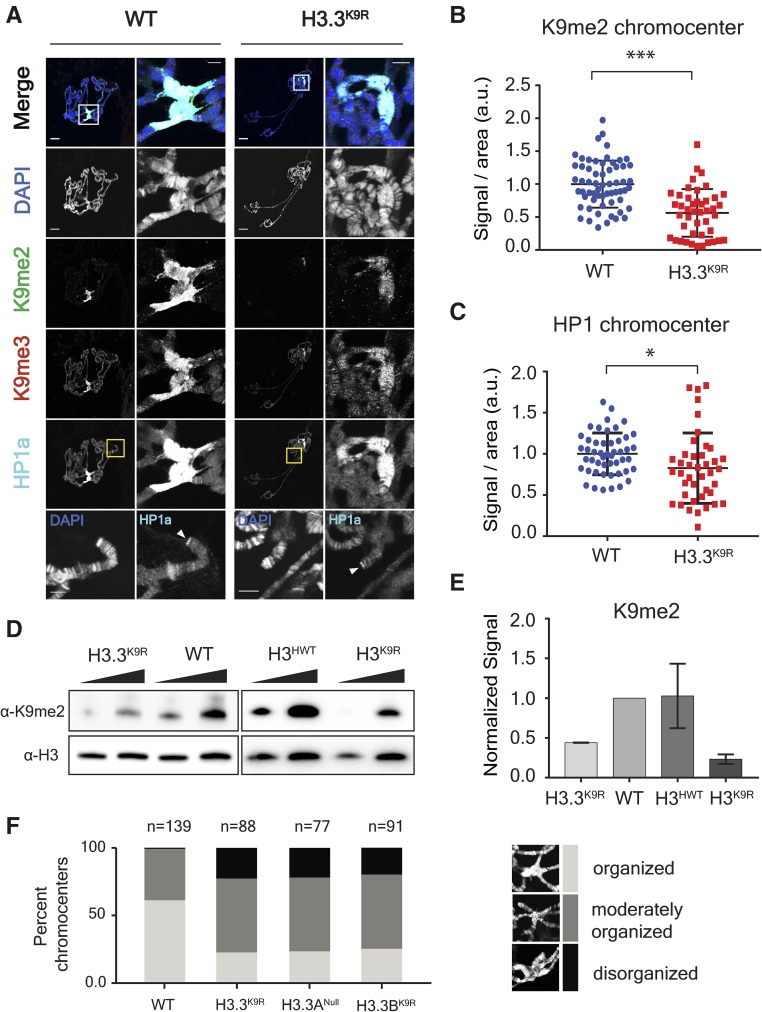
We previously found that the K9me2/me3 signal in H3K9R mutant animals is substantially reduced but not absent. Thus, a possible reason why H3.3K9R H3K9R mutants have a more severe developmental defect than H3K9R mutants is complete loss of K9me throughout the genome. We therefore assessed K9me2/me3 levels in H3.3K9R and H3.3K9R H3K9R mutants by immunofluorescence. We first assessed K9me2/me3 levels in salivary gland polytene chromosomes of H3.3K9R mutants, with the expectation that if H3.3K9 is methylated the signal will be reduced relative to controls. The salivary gland is a highly polyploid tissue (> 1000C), and the alignment of chromatids in the polytene chromosomes results in easily visible structures that provide information about levels and genomic locations of histone PTMs using immunofluorescence. H3.3K9R mutants had lower levels of both K9me2 and K9me3 compared to wild-type controls at the largely heterochromatic chromocenter, demonstrating that H3.3K9 is normally methylated in the pericentric heterochromatin of otherwise wild-type animals (Figure 1, A and B). In support of this result, western blot analysis of salivary glands demonstrated that K9me2 levels were decreased in H3.3K9R mutants compared to wild-type controls (Figure 1, D and E).
Figure 1.
K9me2/me3 and HP1a signal is decreased in H3.3K9R mutants. (A) Third instar larval salivary gland polytene chromosome spreads from wild-type (left) and H3.3K9R mutants (right) stained with anti-K9me2, anti-K9me3, anti-HP1a, and DAPI to mark DNA. Right panel for each genotype shows enlarged chromocenter indicated by white boxes. Bottom panel shows magnified view of telomere indicated by yellow boxes. Bars, 20 μm (whole polytene) and 5 microns (chromocenter/telomere). (B and C) Immunofluorescent signal of K9me2 (B) or HP1a (C) at chromocenters in wild-type (WT) and H3.3K9R mutants (a.u., arbitrary units). Values were normalized to area of the chromocenter and set relative to the average WT value from matched slides (see File S2). Significance was determined using the Student’s t-test (* P < 0.05, ** P < 0.005, and *** P < 0.0005). (D) Western blot of K9me2 from salivary glands with H3 used as loading control. (E) K9me2 signal was quantified by densitometry and normalized to corresponding H3 loading control band. Normalized values were set relative to WT normalized signal. Error bars represent SEM from two independent biological replicates (see Materials and Methods). (F) Quantification of chromocenter organization from WT, H3.3K9R, H3.3ANull, and H3.3BK9R mutants.
Because H3.3K9R mutants exhibited reduced K9me2/me3 signals at the chromocenter, we next used immunofluorescence to examine localization of HP1a, which binds K9me2/me3. In-line with reduced K9me2/me3 signal, HP1a signal at the chromocenter of H3.3K9R mutants was reduced compared to wild-type controls (Figure 1, A and C). HP1a and H3.3 also both localize to telomeres (Goldberg et al. 2010; Lewis et al. 2010). We found that HP1a localizes to telomeres in H3.3K9R mutants (Figure 1A), as it does in H3K9R mutants (Penke et al. 2016). These results are consistent with previous observations that HP1 recruitment to telomeres requires telomere-binding proteins (Badugu et al. 2003; Raffa et al. 2011; Vedelek et al. 2015) and not the H3K9 methyltransferase Su(var)3-9 (Perrini et al. 2004), suggesting that H3K9me is not required for HP1 recruitment to telomeres.
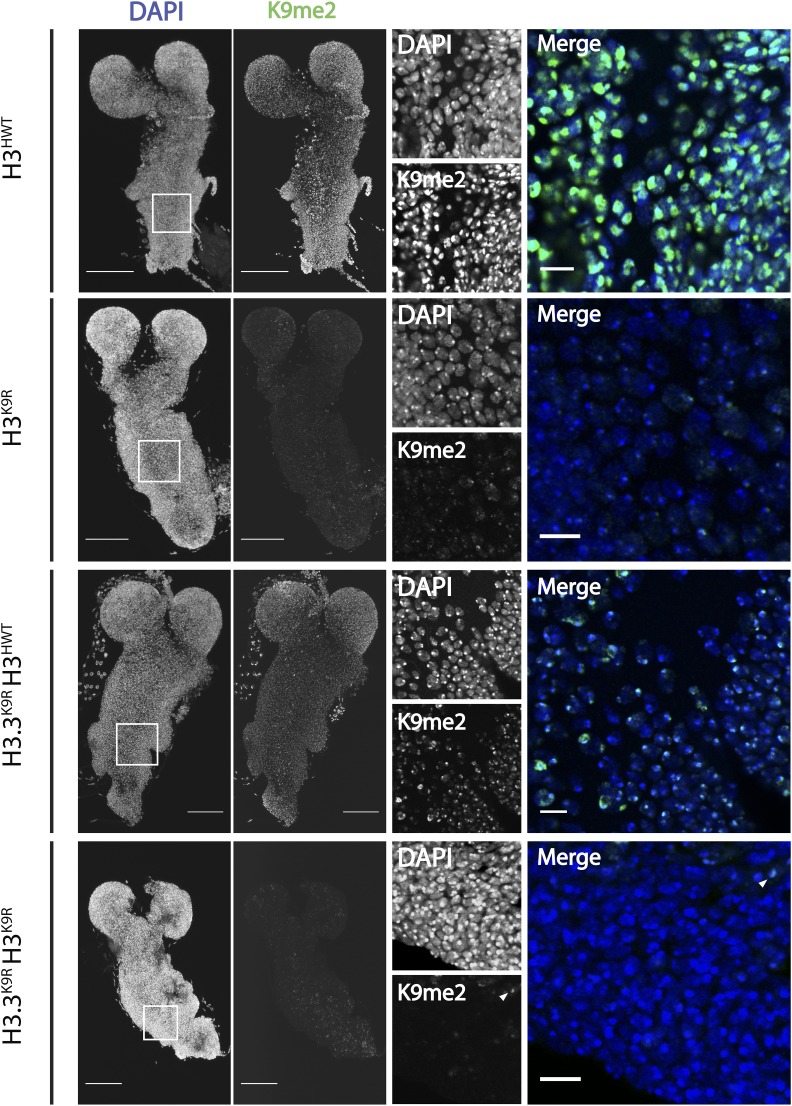
Because H3.3K9R H3K9R combined mutants do not develop to the third instar larval stage, we examined K9me2 levels in first instar larval brains. H3K9R mutants (with wild-type variant histones) and H3.3K9R H3HWT mutants (with a 12× transgenic complement of wild-type canonical histone genes) each exhibited reduced K9me2 levels by immunofluorescence compared to H3HWT controls, consistent with the polytene chromosome data (Figure 2A). In contrast, the H3.3K9R H3K9R variant and canonical combined mutant brains had undetectable levels of K9me2 in the vast majority of cells (Figure 2A). These results provide further evidence that H3.3K9 is methylated and that the total amount of K9me is derived from both H3.3 and H3.
Figure 2.
K9me2/me3 signal is diminished in K9R mutants. (A) First instar larval brains stained with anti-K9me2 and DAPI to mark DNA from H3HWT, H3K9R, H3.3K9R H3HWT, and H3.3K9R H3K9R animals. Left panel shows maximum projection of 2-μm confocal sections through the entire brain. Right panel shows a magnified, single confocal section from the area indicated by the white boxes. Arrowheads indicate cells with residual K9me2 signal in H3.3K9R H3K9R animals. Bars, 50 μm (whole brain) and 10 μm (enlarged image).
Interestingly, a small number of cells in the H3.3K9R H3K9R first instar mutant brains retained low levels of K9me2 signal at the chromocenter (arrowheads, Figure 2). Cells with residual K9me2 express ELAV, a pan-neuronal marker, and lack expression of Deadpan and Prospero, markers of proliferating neuroblasts and ganglion mother cells, respectively (circles, Figure S3 in File S1). These data indicate that cells with K9me2-positive chromocenters in H3.3K9R H3K9R mutant first instar larval brains are differentiated neurons. We suspect that the K9me2 signal in these cells reflects maternally provided wild-type H3 protein remaining in the genomes of quiescent neurons that differentiated prior to having their maternal H3 fully replaced by zygotically expressed H3K9R mutant histones. A corollary to this conclusion is that the proliferating neuroblasts and their GMC daughters have likely progressed through a sufficient number of S phases, such that replacement of maternal H3 with zygotic H3K9R eliminates detectable K9me2 signal.
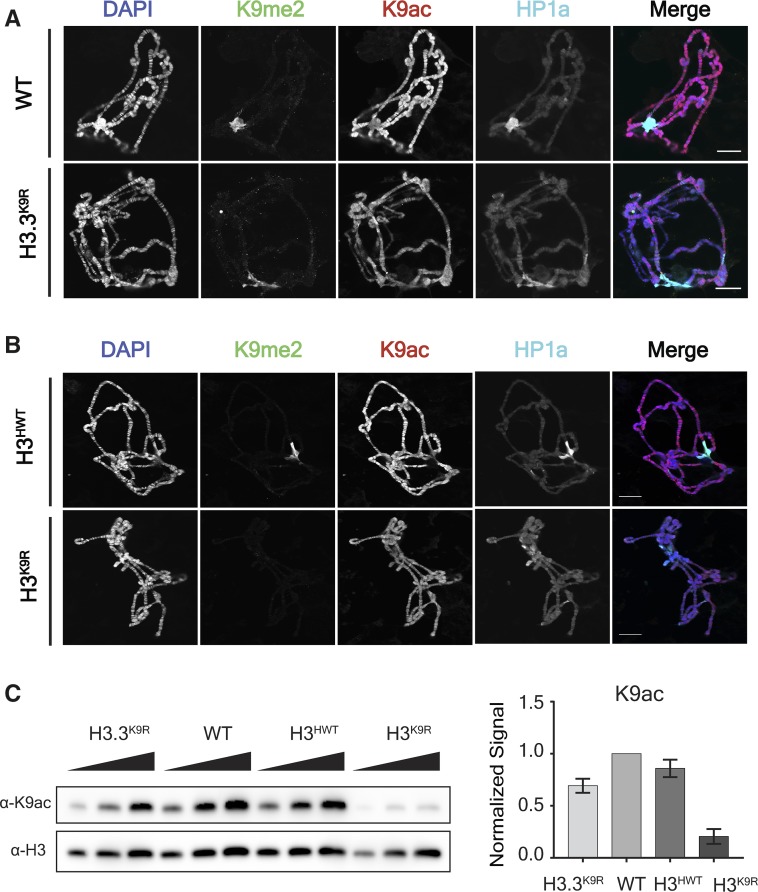
We also found that levels of H3K9 acetylation were reduced in both the H3.3K9R mutant and the H3K9R mutant relative to controls, as determined both by immunofluorescence of salivary gland polytene chromosomes (Figure 3, A and B) and by western blots of salivary gland extracts (Figure 3C). Because a substantial amount of K9ac is placed on H3.3, we considered the possibility that a lack of K9ac was responsible for the fertility defects of H3.3K9R mutants and the early lethality of H3.3K9R H3K9R mutants. To address this question, we integrated either an H3.3BK9, an H3.3BK9R, or an H3.3BK9Q transgene into the same genomic position to determine if a K9Q acetyl mimic could restore fertility to H3.3K9R mutants. Animals with only an H3.3BK9R mutation at the endogenous locus (i.e., containing a wild-type H3.3A gene) and carrying either an H3.3BK9R or H3.3BK9Q transgene were sterile, precluding us from constructing the genotype to test if these transgenes could rescue the sterility of H3.3K9R mutant adults (Figure S2 in File S1). This result suggests that both the H3.3BK9R and H3.3BK9Q transgenes acted dominantly to compromise fertility. Furthermore, these data imply that H3.3BK9R and H3.3BK9Q histones are incorporated into chromatin.
Figure 3.
K9ac signal is decreased in H3.3K9R mutants. (A and B) Polytene chromosome spreads from wild-type (WT) and H3.3K9R mutants (A) or H3HWT and H3K9R mutants (B) stained with anti-K9me2, anti-K9ac, anti-HP1a, and DAPI to mark DNA. Bar, 20 μm. (C) Western blot of K9ac from salivary glands with H3 used as loading control. K9ac signal was quantified by densitometry and normalized to corresponding H3 loading control band. Normalized values were set relative to WT normalized signal. Error bars represent SEM from two independent biological replicates (see Materials and Methods).
H3.3K9 regulates chromatin organization at the chromocenter, telomeres, and transposons
We next asked if the reduction of K9me2/me3 in H3.3K9R mutants affected chromatin organization by cytological examination of salivary gland polytene chromosomes using DAPI staining of DNA. As we found previously in H3K9R mutants (Penke et al. 2016), in some H3.3K9R mutants polytene chromosome spreads, the chromocenter appeared abnormal and not fully condensed (Figure 1F). The cause of this phenotype is unclear but may reflect altered chromatin organization or defects in the underreplication of salivary gland pericentric heterochromatin (Belyaeva et al. 1998; Zhimulev et al. 2003). Based on their cytology, we binned chromocenters into three categories: organized, moderately organized, and disorganized (Figure 1F). We categorized chromocenters from four genotypes: wild-type (i.e., with the endogenous canonical histone genes), an H3.3A null mutant (H3.3ANull), an H3.3B K9R substitution mutant (H3.3BK9R), and the H3.3BK9R; H3.3ANull double mutant in which all H3.3 contains the K9R substitution (H3.3K9R) (Figure S1A in File S1, Table 1, and Table S1 in File S1). Whereas the majority of wild-type chromocenters were organized (60% organized vs. 40% moderately organized), both the H3.3BK9R and the H3.3ANull single mutants had increased percentages of moderately organized and disorganized chromocenters (Figure 1F). For example, ∼22% of chromocenters in the various H3.3 mutants were disorganized compared to < 1% of wild-type chromocenters. These results indicate that H3.3 contributes to chromocenter structure. Interestingly, the H3.3BK9R; H3.3ANull double mutant had the same proportion of moderately organized and disorganized chromocenters as either single mutant. This result suggests that either reducing H3.3 gene dose (i.e., the H3.3ANull allele) or expressing K9R mutant H3.3 histones (i.e., the H3.3BK9R mutation) can prevent normal H3.3 function at pericentric heterochromatin.
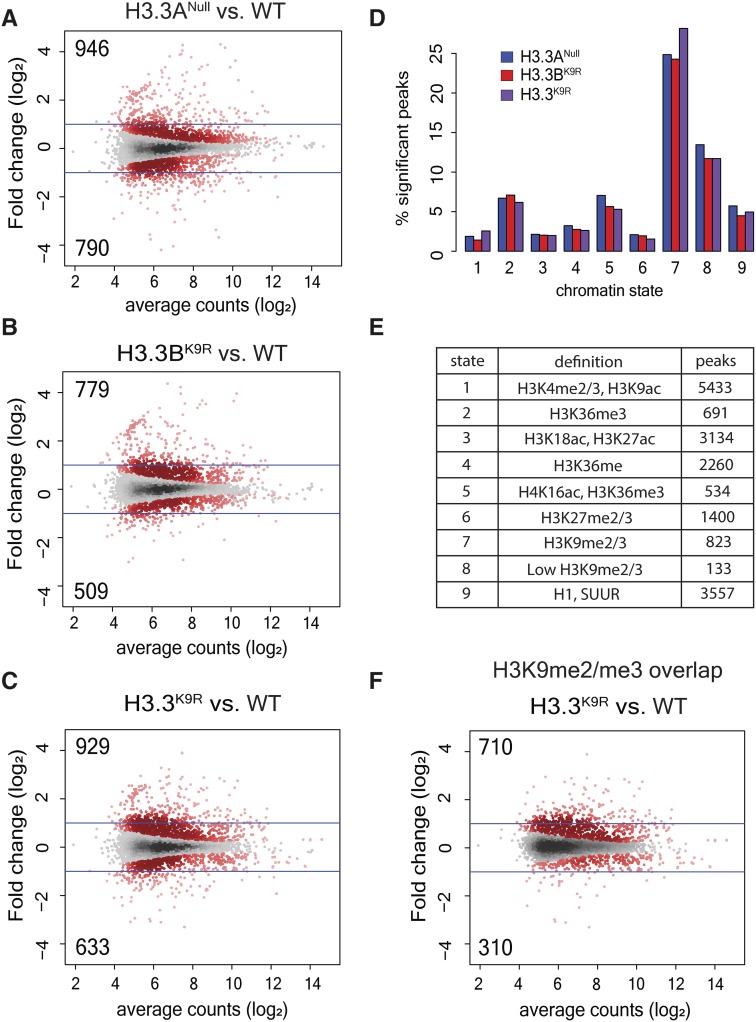
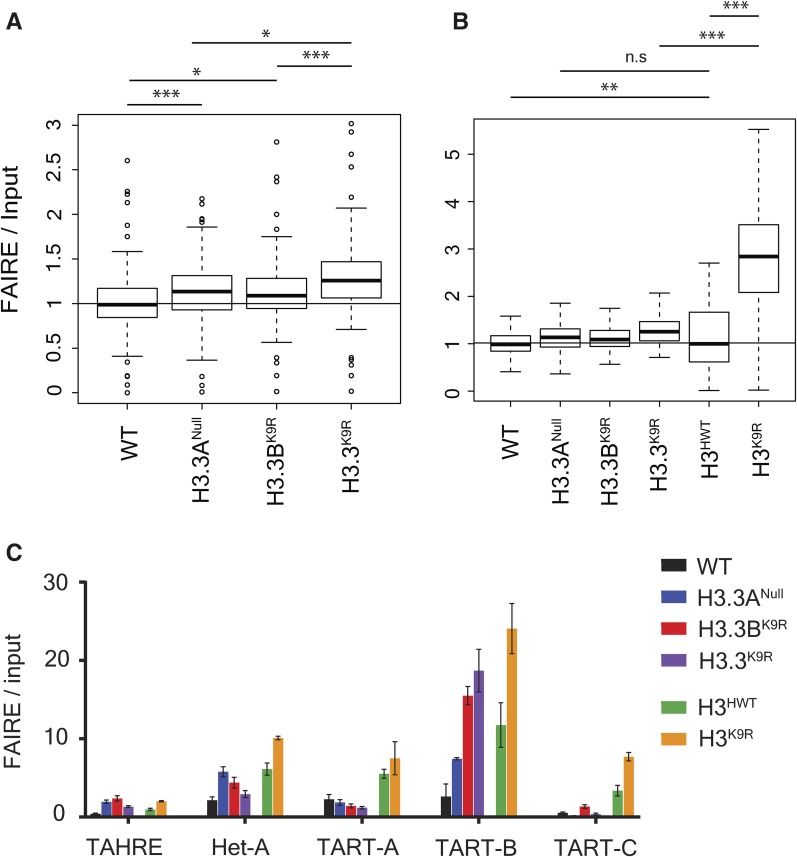
Given the disrupted chromocenter structure in H3.3K9R mutants, we next examined chromatin structure genome-wide by performing FAIRE-seq. FAIRE-seq provides a measure of local nucleosome occupancy across the genome, revealing regions of “open” chromatin that are relatively depleted of nucleosomes (Simon et al. 2013). Using this technique, we previously found that regions of heterochromatin enriched in K9me, particularly pericentromeric heterochromatin, were more open in canonical H3K9R mutants relative to H3HWT controls (Penke et al. 2016). To determine if variant H3.3K9R mutants had a similar phenotype, we performed FAIRE-seq in triplicate on imaginal wing discs from wandering third instar larvae in wild-type, H3.3ANull, H3.3BK9R, and H3.3BK9R; H3.3ANull (H3.3K9R) double mutant genotypes. Sequencing reads were aligned to the genome, and peaks were called on each of the three replicates and combined into a merged peak set. Called peaks were consistent across replicates and read coverage across peaks was highly correlated (R ≥ 0.96) (Figure S4, A and B in File S1). Additionally, wild-type FAIRE data were consistent with previously generated data from wing discs (McKay and Lieb 2013) (Figure S4D in File S1). H3.3ANull, H3.3BK9R, and H3.3K9R mutants each had a similar percentage of peaks with a significantly altered FAIRE signal when compared to wild-type: 8.8, 6.5, and 7.9%, respectively (Figure 4, A–C). Moreover, significantly changed peaks across the three mutants exhibited a high degree of overlap. Of the 2660 significantly changed peaks across all mutants, 21% were shared among all three and 52% by at least two mutants (Figure S5A in File S1). FAIRE signals at significantly changed peaks also displayed similar fold changes in mutants compared to wild-type and were not exacerbated in the double mutant compared to either single mutant (Figure S5B in File S1). These data suggest that H3.3A and H3.3BK9 both function to regulate chromatin architecture.
Figure 4.
H3.3K9 regulates chromatin architecture in regions of K9me. (A–C) Mutant: wild-type (WT) ratio of H3.3Anull (A), H3.3BK9R (B), or H3.3K9R (C). Formaldehyde Assisted Isolation of Regulatory Elements (FAIRE) signal from third instar imaginal wing discs at 19,738 FAIRE peaks called by MACS2. Red dots indicate significantly different peaks (P < 0.05), and insets indicate the number of significantly increased (top) or decreased (bottom) peaks. Average counts signify average normalized reads that overlap a peak in mutant and WT samples. (D) Percentage of peaks in a particular chromatin state that have significantly different FAIRE signal in mutants vs. WT (top). (E) Summary of histone modifications or proteins that define a chromatin state and the number of FAIRE peaks assigned to a given chromatin state. (F) Plot from (C) showing only those peaks that overlap a K9me2 or K9me3 peak from modENCODE chromatin immunoprecipitation sequencing data.
We next asked if the changes in FAIRE signals that we observed in H3.3 mutants were characterized by a particular chromatin signature. We assigned each called FAIRE peak to one of nine different chromatin states characterized by different combinations of histone PTMs, as defined by Kharchenko et al. (2011). We then calculated the percentage of FAIRE peaks that changed between an H3.3 mutant and wild-type within each chromatin state. Regions of K9me2/me3 showed the highest percentage of changes in FAIRE signals in H3.3ANull, H3.3BK9R, and the H3.3K9R mutant compared to wild-type, supporting the idea that H3.3K9 is methylated and plays a necessary role in regulating chromatin architecture (Figure 4D). Changes in FAIRE signal was also more likely to occur in regions of H3K36me3, a mark that is enriched along gene bodies that are themselves enriched for H3.3 (Bannister et al. 2005; Szenker et al. 2011). Finally, we used modENCODE K9me2 and K9me3 ChIP-seq data to complement the chromatin state analysis. Of the FAIRE peaks significantly increased or decreased in H3.3K9R mutants compared to wild-type, 76.4% and 49.0% overlapped a K9me2 or K9me3 peak, respectively (Figure 4F). These results demonstrate that altered FAIRE signals in H3.3K9R mutants occurred in regions normally occupied by K9me.
We also observed increased FAIRE signal at telomeres in all three H3.3 mutant genotypes, particularly on chromosomes 2R and 3L (Figure S5C in File S1), suggesting that H3.3 regulates telomeric chromatin architecture. In Drosophila, telomeres are composed of retrotransposons enriched in K9me2/me3 (Levis et al. 1993; Cenci et al. 2005). H3.3 plays a similar role in the mouse, in which H3.3 null mutant embryonic stem cells exhibit an increase in transcripts from transposons (Elsässer et al. 2015) and telomeres (Udugama et al. 2015). Additionally, we previously observed transposon activation and mobilization in canonical H3K9R mutants (Penke et al. 2016). For these reasons, we examined FAIRE signal at transposons in our H3.3 mutants using the piPipes pipeline, which avoids ambiguity in aligning reads to repetitive transposons by mapping to transposon families (Han et al. 2015). Both H3.3ANull and H3.3BK9R mutants resulted in significantly increased FAIRE signal at transposons, and H3.3K9R mutants had on average even higher increased FAIRE signal at transposons (Figure 5, A and B). Moreover, FAIRE signal at some telomeric transposons, particularly TART-B, was increased in H3.3 mutants (Figure 5C). However, the extent of increase in H3.3K9R mutants was not as severe as previously observed for H3K9R mutants (Penke et al. 2016) (Figure 5B). These results support a role for H3.3K9 in chromatin-mediated transposon repression, though to a lesser extent than H3K9.
Figure 5.
Imaginal wing disc FAIRE signal of H3.3 mutants is increased at telomeres and transposons. (A) Boxplot of average FAIRE enrichment determined by piPipes pipeline across 126 transposon families (Han et al. 2015). Genomic DNA from Drosophila embryos used as input control. (B) Boxplots in (A) shown alongside FAIRE enrichment for H3HWT and H3K9R mutants from a separate experiment (Penke et al. 2016). (C) FAIRE enrichment of H3.3 and H3K9R mutants at telomeric transposons. Error bars indicate SD from three replicates for each genotype. Statistical significance determined by paired t-test (* P < 0.05, ** P < 0.005, and *** P < 0.0005,). FAIRE, Formaldehyde Assisted Isolation of Regulatory Elements, n.s., not significant; WT, wild-type.
H3.3K9 and H3K9 functions overlap in regions of K9ac and partially in regions of K9me
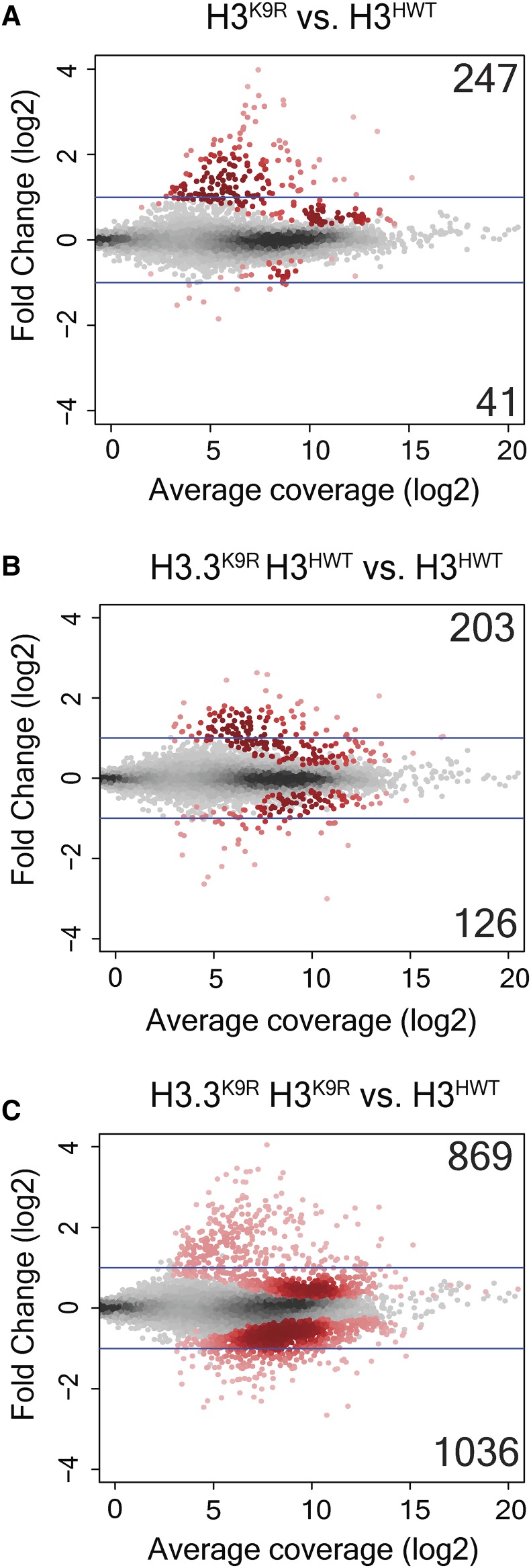
To investigate the cause of lethality when both variant and canonical H3 histones contain the K9R mutation, we performed RNA-seq of first instar larvae from four genotypes: H3HWT, H3K9R, H3.3K9R H3HWT, and H3.3K9R H3K9R (Table 1 and Table S1 in File S1). Larvae of the correct genotype were identified by GFP fluorescence (see Materials and Methods). RNA sequencing reads were aligned to the genome using TopHat, transcript assembly was performed by Cufflinks, and DESeq2 was used for statistical analysis (Trapnell et al. 2014; Love et al. 2014). Each genotype was verified by examination of RNA-seq reads mapping to the K9 codon of variant and canonical histones. Correlation analysis demonstrated that transcript abundance across all assembled transcripts was highly similar among replicates, and was also similar to previously generated data from wild-type first instar larvae (Figure S6 in File S1) (Graveley et al. 2011). Additionally, histone expression was similar across all genotypes, suggesting that variation in histone levels does not underlie observed phenotypes (Figure S7A in File S1). In-line with our previous analysis of H3K9R RNA-seq data from imaginal wing discs (Penke et al. 2016), the majority of significantly changed transcripts in H3K9R first instar samples was increased compared to H3HWT (247 increased vs. 41 decreased), supporting a role for H3K9me in gene silencing (Figure 6A). H3.3K9R H3HWT samples had a similar number of significantly changed transcripts, and again most transcripts showed increased signals compared to H3HWT (203 vs. 126), though fold changes were smaller than those of H3K9R mutants (Figure 6B). By contrast, the H3.3K9R H3K9R combined mutant genotype caused a much more pronounced effect on gene expression compared to either the H3.3K9R H3HWT or the H3K9R mutant genotypes (Figure 6C); 869 transcripts exhibited increased RNA signals and 1036 transcripts were decreased compared to H3HWT samples. The number of decreased transcripts in H3.3K9R H3K9R animals compared to H3HWT was therefore about 10-fold higher than either the variant or canonical K9R mutant alone. Thus, similar to our viability analysis (Table 3), these RNA-seq results demonstrated that variant and canonical versions of H3K9 compensate for each other in the regulation of gene expression.
Figure 6.
H3.3K9 and H3K9 redundantly regulate gene expression. (A–C) Mutant: wild-type ratio of H3K9R (A), H3.3K9R (B), or H3.3K9R H3K9R (C) RNA signal from first instar larvae at 10,253 transcripts assembled by Cufflinks. The y-axis indicates the log2 transformation of mutant/control signal between the genotypes being compared (indicated at the top of each plot). Red dots indicate significantly different transcripts (P < 0.05) and insets signify the number of significantly increased (top) or decreased (bottom) transcripts. Average coverage signifies the average number of normalized reads that overlap a transcript in mutant and H3HWT samples.
Because we observed increases in FAIRE signal at transposons in H3.3K9R mutants from wing disc samples, we examined RNA levels of transposon families in first instar larvae. Similar to our previous RNA-seq observations from H3K9R mutant wing discs (Penke et al. 2016), RNA signal at transposons in H3K9R first instar larvae were increased relative to the H3HWT control (Figure S7, B and C in File S1). Although, on average, transposon levels were only slightly higher in H3.3K9R H3HWT mutants compared to H3HWT, transposon levels in H3.3K9R H3K9R combined mutants were significantly higher than either H3.3K9R H3HWT or H3K9R mutants alone (Figure S7, B and C in File S1). Moreover, telomeric transposons are generally increased in all K9R mutants compared to H3HWT controls (Figure S7D in File S1). Together, these results support an overlapping role for H3.3K9 and H3K9 in regulating gene expression and transposon repression.
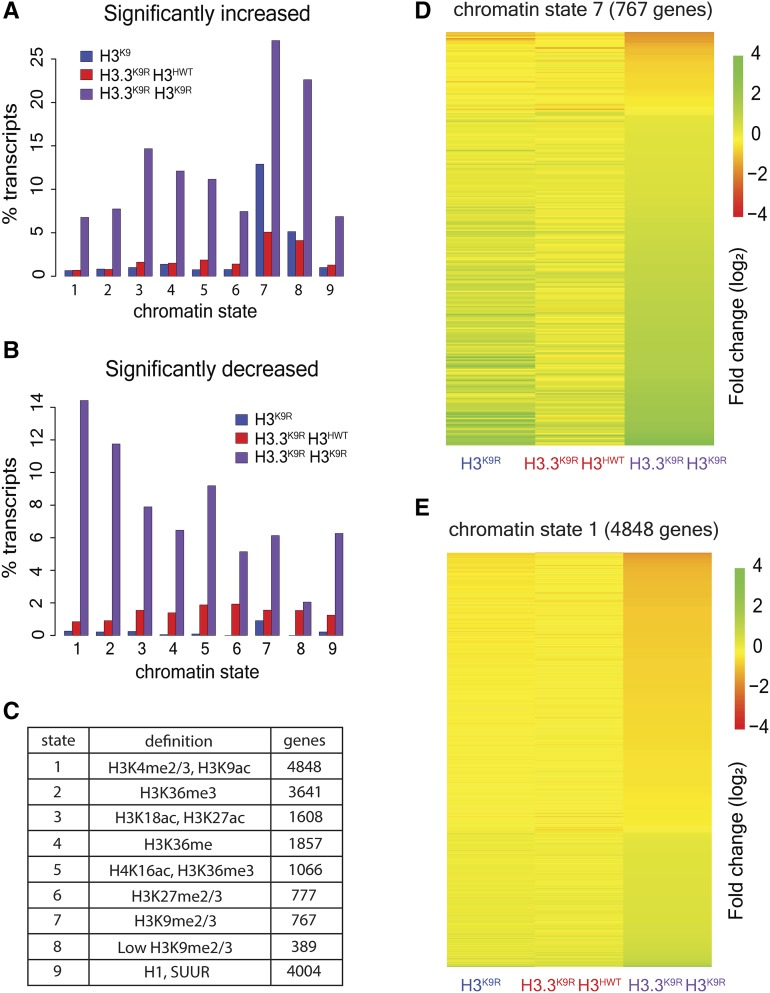
We next examined chromatin signatures of significantly altered transcripts to explore the mechanism of the observed gene expression changes. All transcripts were assigned to one or more chromatin states based on their overlap with genomic regions defined by Kharchenko et al. (2011). We then determined the percentage of transcripts within a given chromatin state that were either increased or decreased in K9R mutants relative to H3HWT controls (Figure 7, A–C). Transcripts in regions of K9me2/me3 (chromatin state 7 and 8) were the most likely to have significantly increased RNA levels in mutants compared to H3HWT. Although H3.3K9R H3K9R combined mutants had the highest percentage of chromatin state 7 transcripts that were significantly increased (∼26%), H3K9R mutants also displayed a high percentage (∼13%) of change within chromatin state 7 (Figure 7, A and D and Figure S8A in File S1). These results suggest that H3.3K9 contributes to gene repression in regions of K9me2/me3 but cannot completely compensate for the absence of H3K9.
Figure 7.
H3.3K9 and H3K9 redundancy differs in heterochromatin and euchromatin. (A and B) Percentage of transcripts in a chromatin state that have significantly increased (A) or decreased (B) RNA signal in mutants vs. H3HWT. (C) Table indicates the number of transcripts that overlap a particular chromatin state. (D and E) Heatmaps showing fold change of K9R mutants over H3HWT at chromatin state 7 regions (D) and chromatin state 1 regions (E). Each row indicates a transcript that overlaps the indicated chromatin state.
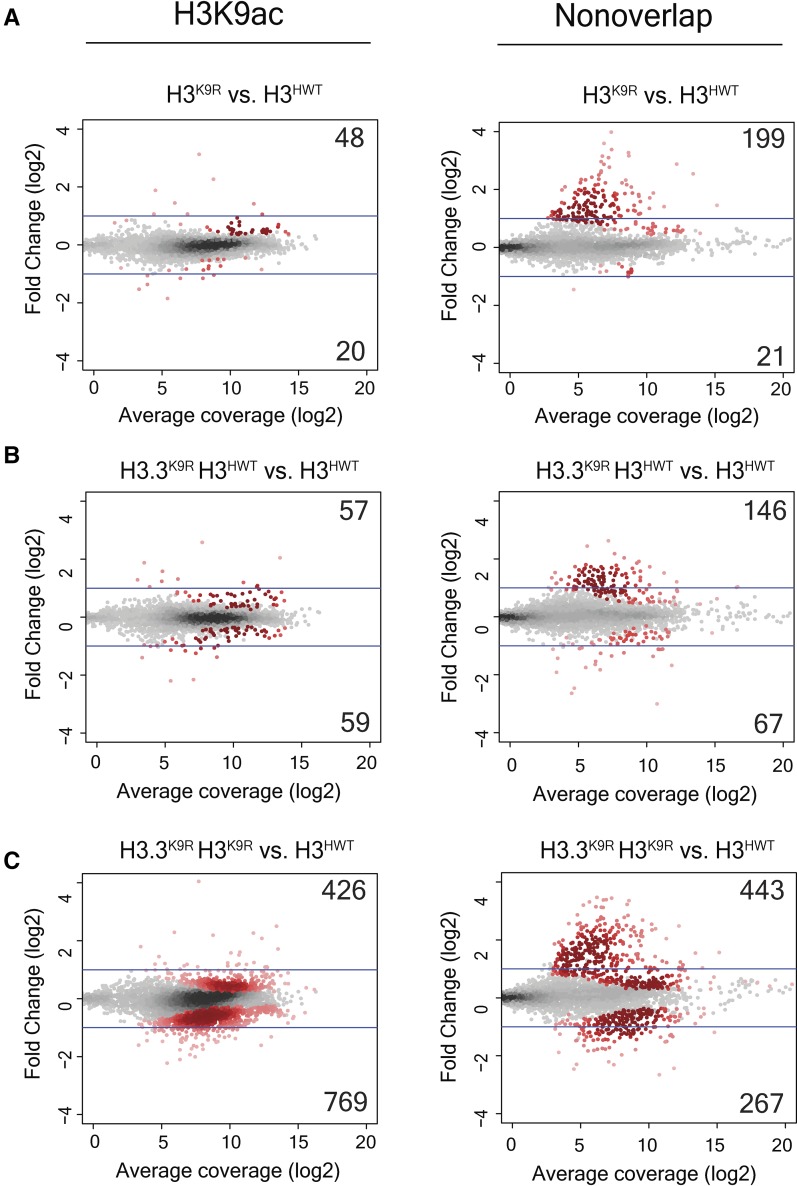
In contrast to upregulated transcripts, very few transcripts were significantly decreased in H3.3K9R H3HWT or H3K9R mutants. However, the H3.3K9R H3K9R combined mutant displayed numerous significant decreases in transcript abundance. Interestingly, transcripts in chromatin state 1, characterized by K9ac and a lack of K9me, were most likely to be decreased (Figure 7, B and E). Several other chromatin states showed elevated transcript changes, particularly in the H3.3K9R H3K9R combined mutant; however, in this analysis, transcripts can be assigned to more than one chromatin state. Indeed, many transcripts in chromatin state one also overlap other chromatin states. We therefore performed a supplementary analysis that examined only transcripts that overlap a single chromatin state. This analysis demonstrated that transcripts solely in chromatin state 1 were much more likely to change in K9R mutants than those in other chromatin states (Figure S8A in File S1). Similar results were obtained using imaginal wing disc K9ac ChIP data from Pérez-Lluch et al. (2015). Whereas few transcripts that overlapped K9ac were significantly altered in either single mutant (68 in H3K9R and 116 in H3.3K9R H3HWT), 1195 K9ac-associated transcripts exhibited changed expression levels in H3.3K9R H3K9R combined mutants (Figure 8). These data suggest that in regions of K9ac, H3.3 and H3 can completely compensate for each other. Additionally, these data provide evidence that K9ac facilitates gene expression.
Figure 8.
K9ac-associated transcripts are altered in H3.3K9R H3K9R double mutants. MA (M (log ratio) A (mean average)) plot showing fold change of normalized RNA signal in H3K9R (A), H3.3K9R H3HWT (B), and H3.3K9R H3K9R (C) mutants vs. H3HWT at all transcripts from merged transcriptome. Average coverage on x-axis represents the mean expression level of a transcript. Transcripts that overlap a K9ac peak called from chromatin immunoprecipitation sequencing data (GSM1363590; Pérez-Lluch et al. 2015) are shown in the left panel, while those that do not are shown in the right panel. Significance (shown in red) was determined using DESeq2 (Love et al. 2014) and an adjusted P-value cutoff of 0.05.
Discussion
Overlapping and distinct developmental functions of H3 and H3.3
In this study, we determined the distinct and overlapping roles that lysine nine of variant and canonical histone H3 play in gene expression and heterochromatin function during Drosophila development. Our developmental genetic analyses demonstrate that H3.3K9 is necessary for fertility but not viability in Drosophila. In addition, we find that some euchromatic functions of H3K9 can be provided by either variant H3.3 or canonical H3, whereas H3.3K9 cannot completely compensate for H3K9 in some regions of heterochromatin, as discussed below.
Several studies from multiple species have investigated the developmental functions of H3.3 and H3. In mice, single mutation of either H3.3A or H3.3B results in reduced viability and compromised fertility (Couldrey et al. 1999; Bush et al. 2013). Similarly, Drosophila H3.3A and H3.3B double mutants appear at lower than expected Mendelian ratios and are sterile (Sakai et al. 2009). H3.3 in Tetrahymena thermophila is also important for sexual reproduction, although it is not required for viability or maintenance of nucleosome density (Cui et al. 2006). Both Tetrahymena and Drosophila H3.3 and H3 can compensate for one another. In Tetrahymena, canonical H3 is dispensable if H3.3 is overexpressed (Cui et al. 2006). Similarly, in Drosophila, transgenic expression of H3 can rescue both the semilethality (Sakai et al. 2009) and infertility (Hödl and Basler 2012) of H3.3 mutants, indicating some functional redundancy between the two histones. Indeed, when expressed equivalently, Drosophila H3.3 can provide all of the developmental functions of H3 (Hödl and Basler 2012). Moreover, H3.3 is the sole H3 protein in Schizosaccharomyces pombe and Saccharomyces cerevisiae yeast (Malik and Henikoff 2003).
H3.3K9 functions in heterochromatin
We find that under endogenous expression conditions, H3.3K9 functions in heterochromatin, including pericentromeric and telomeric regions of the genome. We detected H3.3K9 methylation in pericentromeric heterochromatin, congruous with previous data demonstrating that H3.3 in Drosophila is deposited at the chromocenter of polytene chromosomes in a replication-dependent manner (Schwartz and Ahmad 2005). We also observed that H3.3K9R mutants exhibited an abnormal chromocenter structure in polytene chromosomes. Moreover, we provide evidence that H3.3K9 is required for the maintenance of telomeric chromatin architecture and repression of certain telomeric transcripts, indicating that replication-coupled expression of H3 cannot provide these particular H3K9 functions. These findings in Drosophila are consistent with studies in mouse embryonic stem cells showing that H3.3 is localized to telomeres, is methylated at K9, and functions in the repression of telomeric repeat-containing RNAs (Goldberg et al. 2010; Udugama et al. 2015). Conversely, the genetic data that we presented here and previously (Penke et al. 2016) indicate that H3K9 is essential for the repression of transposon-derived transcripts in pericentric heterochromatin, and that H3.3K9 cannot compensate for the lack of H3K9 at these regions of the genome. The role of H3.3K9 in telomere structure and function may be independent of HP1, as HP1 is recruited to telomeres via the terminin complex independently of H3K9me (Badugu et al. 2003; Raffa et al. 2011; Vedelek et al. 2015).
K9ac regulates euchromatic gene expression
Previous studies that mapped histone modifications across the genome identified K9ac as a characteristic of transcriptionally active regions (Liang et al. 2004; Bernstein et al. 2005; Roh et al. 2005; Kharchenko et al. 2011). Moreover, mutation of H3K9 acetyltransferases results in compromised transcriptional activity (Georgakopoulos and Thireos 1992; Kuo et al. 1998; Wang et al. 1998). However, H3K9 acetyltransferases have nonhistone substrates in addition to H3K9, and decreased transcriptional output may be the result of pleiotropic effects (Glozak et al. 2005; Fillingham et al. 2008; Spange et al. 2009). Our study provides evidence that K9ac, rather than nonhistone targets of H3K9 acetyltransferases, contributes to activating transcription, as H3.3K9R and H3K9R mutants exhibit reduced gene expression in regions normally enriched for K9ac. Importantly, these K9ac-rich regions with reduced gene expression are not normally enriched in K9me2 or me3, indicating that the observed phenotype is not due to changes in K9me2 or me3 and likely results from loss of K9ac. This change in gene expression was accompanied by a fully penetrant lethality early in larval development of H3.3K9R H3K9R combined mutant animals, raising the possibility that gene expression control via acetylation of H3K9 is critical for the completion of animal development. These data are also consistent with previous studies in C. elegans demonstrating that H3K9 methylation is not essential for viability (Towbin et al. 2012; Zeller et al. 2016).
Overlapping and distinct genomic functions of H3K9 and H3.3K9
Functional overlap of H3K9 and H3.3K9 appears to vary at different regions of the genome. Whereas H3.3K9 and H3K9 can perform similar functions in euchromatic regions of the genome and can fully compensate for one another, our RNA-seq data demonstrate that H3.3K9 can only partially compensate for H3K9 in regions of heterochromatin. Partial compensation by H3.3K9 in regions of K9me2/me3 is in-line with previous studies showing that H3.3 is found at heterochromatin (Goldberg et al. 2010; Lewis et al. 2010; Wong et al. 2010) and plays a role in transposon repression (Elsässer et al. 2015). In the genotypes that we analyzed, mRNAs encoding variant and canonical H3 are expressed from their native promoters. Thus, disparity in functional overlap might be due to differences in modes of expression and deposition, and thus total amounts of variant and canonical H3 histones in particular regions of the genome. For instance, H3 is normally enriched in heterochromatin compared to H3.3 (Ahmad and Henikoff 2002), which may cause H3K9R mutations to be more detrimental in these regions. However, H3.3 may be able to provide all H3 function when highly expressed in a replication-dependent manner, as a transgenic histone gene array in which the H3.2 coding region was replaced by H3.3 is nearly fully functional in larval imaginal discs (Hödl and Basler 2012). Thus, differences in expression and/or deposition into chromatin may be the only basis for functional differences between H3.3 and H3.2 that we observed.
Heterochromatin may be particularly sensitive to the incorporation of nonmodifiable K9 residues. H3K9 methylation serves as a binding site for the protein HP1, which can in turn recruit H3K9 methyltransferases (Grewal and Jia 2007; Elgin and Reuter 2013). Methylation of a neighboring nucleosome can restart the cycle and initiate the propagation of a heterochromatic configuration along the chromosome. The introduction of even a small number of H3K9R-containing nucleosomes may therefore disrupt this cycle, and prevent proper heterochromatin formation and gene repression. Incorporation of H3.3BK9R histones into regions of heterochromatin may dominantly affect chromatin structure, resulting in the observed phenotypes at pericentromeres and telomeres in H3.3K9R mutants. In contrast, incorporation of low amounts of H3K9R histones in euchromatin may not reduce K9ac levels sufficiently to disrupt gene expression. Finally, amino acid differences in variant and canonical H3 may direct distinct histone modification states on different histone types by influencing the binding of chromatin-modifying enzymes (Jacob et al. 2014). Different histone modification states on H3.3 and H3 may underlie variation in compensation at different genomic regions.
In summary, our data investigating H3.3K9 and H3K9 function provide evidence that K9ac activates gene expression, and advance our understanding of the overlapping and distinct functional roles of variant and canonical histones.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300480/-/DC1.
Acknowledgments
We thank Bhawana Bariar for assistance generating the guide RNA construct, Jeff Sekelsky for the pBlueSurf construct, and Kami Ahmad for providing the H3.3A2x1 flies. This work was supported by National Institutes of Health grants 5T32GM007092-39 and F31GM115194 to T.J.R.P. and R01DA036897 to D.J.M., B.D.S., A.G.M., and R.J.D.
Footnotes
Communicating editor: P. Geyer
Literature Cited
- Ahmad K., Henikoff S., 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Wiggins J. C., 1984. Histone rearrangements accompany nuclear differentiation and dedifferentiation in Tetrahymena. Dev. Biol. 101: 282–294. [DOI] [PubMed] [Google Scholar]
- Badugu R. K., Shareef M. M., Kellum R., 2003. Novel Drosophila heterochromatin protein I (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J. Biol. Chem. 278: 34491–34498. [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., et al. , 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Schneider R., Myers F. A., Thorne A. W., Crane-Robinson C., et al. , 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280: 17732–17736. [DOI] [PubMed] [Google Scholar]
- Belyaeva E. S., Zhimulev I. F., Volkova E. I., Alekseyenko A. A., Moshkin Y. M., et al. , 1998. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl. Acad. Sci. USA 95: 7532–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D. K., et al. , 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181. [DOI] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H., 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. M., Yuen B. T., Barrilleaux B. L., Riggs J. W., O’Geen H., et al. , 2013. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics Chromatin 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Jin Y., Girton J., Johansen J., Johansen K. M, 2010. Preparation of Drosophila polytene chromosome squashes for antibody labeling. J. Vis. Exp. 6: 1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Ciapponi L., Gatti M., 2005. The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma 114: 135–145. [DOI] [PubMed] [Google Scholar]
- Couldrey C., Carlton M. B., Nolan P. M., Colledge W. H., Evans M. J., 1999. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum. Mol. Genet. 8: 2489–2495. [DOI] [PubMed] [Google Scholar]
- Cui B., Liu Y., Gorovsky M. A., 2006. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol. Cell. Biol. 26: 7719–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. R., Reuter G., 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer S. J., Noh K. M., Diaz N., Allis C. D., Banaszynski L. A., 2015. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C., McKay D. J., Mann R. S., 2008. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into distalless during Drosophila leg development. Dev. Cell 14: 86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J., Recht J., Silva A. C., Suter B., Emili A., et al. , 2008. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol. Cell. Biol. 28: 4342–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos T., Thireos G., 1992. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11: 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Glozak M. A., Sengupta N., Zhang X., Seto E., 2005. Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23. [DOI] [PubMed] [Google Scholar]
- Goldberg A. D., Banaszynski L. A., Noh K., Lewis P. W., Elsaesser S. J., et al. , 2010. Distinct factors control histone at specific genomic regions. Cell 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S. I. S., Jia S., 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Hake S. B., Garcia B. A., Duncan E. M., Kauer M., Dellaire G., et al. , 2006. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281: 559–568. [DOI] [PubMed] [Google Scholar]
- Han B. W., Wang W., Zamore P. D., Weng Z., 2015. PiPipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome-and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics 31: 593–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., 2005. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21: 133–153. [DOI] [PubMed] [Google Scholar]
- Hödl M., Basler K., 2012. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr. Biol. 22: 2253–2257. [DOI] [PubMed] [Google Scholar]
- Jacob Y., Bergamin E., Donoghue M. T., Mongeon V., LeBlanc C., et al. , 2014. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343: 1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G., 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Zang C., Wei G., Cui K., Peng W., et al. , 2011. H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions in the human genome. Nat. Genet. 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P., Alekseyenko A., Schwartz Y. B., Minoda A., Riddle N. C., et al. , 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar D. C., Jin W., Maunakea A., Abraham B., Ha M., et al. , 2013. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3. 3. Genome Biol. 14: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.-H., Zhou J., Jambeck P., Churchill M. E. A., Allis C. D., 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12: 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W., Ganesan R., Houtchens K., Tolar L. A., Sheen F. M., 1993. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Lewis P. W., Elsaesser S. J., Noh K., Stadler S. C., Allis C. D., 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 107: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Lin J. C. Y., Wei V., Yoo C., Cheng J. C., et al. , 2004. Distinct localization of histone H3 acetylation and H3–K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA 101: 7357–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H. S., Henikoff S., 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10: 882–891. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F., Gongidi P., Woods K. R., Jin J., Maltais L. J., 2002. The human and mouse replication-dependent histone genes. Genomics 80: 487–498. [PubMed] [Google Scholar]
- McKay D. J., Lieb J. D., 2013. A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27: 306–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. J., Klusza S., Penke T. J. R., Meers M. P., Curry K. P., et al. , 2015. Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E., Gafken P. R., Ahmad K., Henikoff S., 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101: 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone B. G., Allshire R. C., 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13: 191–198. [DOI] [PubMed] [Google Scholar]
- Mito Y., Henikoff J. G., Henikoff S., 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37: 1090–1097. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I., 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Penke T. J. R., McKay D. J., Strahl B. D., Gregory Matera A., Duronio R. J., 2016. Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev. 30: 1866–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Lluch S., Blanco E., Tilgner H., Curado J., Ruiz-Romero M., et al. , 2015. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat. Genet. 47: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrini B., Piacentini L., Fanti L., Altieri F., Chichiarelli S., et al. , 2004. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell 15: 467–476. [DOI] [PubMed] [Google Scholar]
- Price B. D., Andrea A. D. D., 2014. Chromatin remodeling at DNA double strand breaks. Cell 152: 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools : a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G. D., Ciapponi L., Cenci G., Gatti M., 2011. Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus 2: 383–391. [DOI] [PubMed] [Google Scholar]
- Roh T., Cuddapah S., Zhao K., 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A., Schwartz B. E., Goldstein S., Ahmad K., 2009. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 19: 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B. E., Ahmad K., 2005. Transcriptional activation triggers deposition and removal of the histone variant H3. 3. Genes Dev. 19: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Xie A., 2013. Double strand break repair functions of histone H2AX. Mutat. Res. 750: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J. M., P. G. Giresi, I. J. Davis, and J. D. Lieb, 2013 A detailed protocol for formaldehyde-assisted isolation of regulatory elements (FAIRE). Curr Protoc Mol Biol. 102: 21.26.1–21.26.15. [DOI] [PubMed] [Google Scholar]
- Spange S., Wagner T., Heinzel T., Krämer O. H., 2009. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41: 185–198. [DOI] [PubMed] [Google Scholar]
- Szenker E., Ray-Gallet D., Almouzni G., 2011. The double face of the histone variant H3.3. Cell Res. 21: 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y., 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61. [DOI] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S., 2010. Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11: 264–275. [DOI] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S., 2017. Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18: 115–126. [DOI] [PubMed] [Google Scholar]
- Tamura T., Smith M., Kanno T., Dasenbrock H., Nishiyama A., et al. , 2009. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J. Biol. Chem. 284: 12217–12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin B. D., González-Aguilera C., Sack R., Gaidatzis D., Kalck V., et al. , 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2014. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama M., Chang F. T. M, Chan F. L., Tang M. C., Pickett H. A., et al. , 2015. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 43: 10227–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedelek B., Blastyák A., Boros I. M., 2015. Cross-species interaction between rapidly evolving telomere-specific drosophila proteins. PLoS One 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A., Kaufman P. D., Kobayashi R., Stillman B., 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87: 95–104. [DOI] [PubMed] [Google Scholar]
- Wallrath, L. L., M. W. Vitalini, and S. C. R Elgin, 2014 pp. 529–552 in Fundamentals of Chromatin, edited by J. L. Workman and S. M. Abmayr. Springer-Verlag, New York.
- Wang L., Liu L., Berger S. L., 1998. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12: 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. H., McGhie J. D., Sim M., Anderson M. A., Ahn S., et al. , 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20: 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Long C., Chen X., Huang C., Chen S., et al. , 2010. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98. [DOI] [PubMed] [Google Scholar]
- Zeller P., Padeken J., Van Schendel R., Kalck V., Tijsterman M., et al. , 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev I. F., Belyaeva E. S., Semeshin V. F., Shloma V. V., Makunin I. V., et al. , 2003. Overexpression of the SuUR gene induces reversible modifications at pericentric, telomeric and intercalary heterochromatin of Drosophila melanogaster polytene chromosomes. J. Cell Sci. 116: 169–176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
See Supplemental Experimental Procedures in File S2 for a detailed description of the methods. Strains are available upon request. Sequencing data are available at the Gene Expression Omnibus under accession number GSE106192.