The maize abnormal chromosome 10 (Ab10) meiotic drive system causes its own preferential transmission through females, yet it is found at low frequencies...
Keywords: preferential segregation, segregation distortion, meiotic drive, maize, haplotype, K10, Ab10, knobs, heterochromatin, kinesin, deleterious alleles
Abstract
Meiotic drive describes a process whereby selfish genetic elements are transmitted at levels greater than Mendelian expectations. Maize abnormal chromosome 10 (Ab10) encodes a meiotic drive system that exhibits strong preferential segregation through female gametes. We performed transmission assays on nine Ab10 chromosomes from landraces and teosinte lines and found a transmission advantage of 62–79% in heterozygotes. Despite this transmission advantage, Ab10 is present at low frequencies in natural populations, suggesting that it carries large negative fitness consequences. We measured pollen transmission, the percentage of live pollen, seed production, and seed size to estimate several of the possible fitness effects of Ab10. We found no evidence that Ab10 affects pollen transmission, i.e., Ab10 and N10 pollen are transmitted equally from heterozygous fathers. However, at the diploid (sporophyte) level, both heterozygous and homozygous Ab10-I-MMR individuals show decreased pollen viability, decreased seed set, and decreased seed weight. The observed fitness costs can nearly but not entirely account for the observed frequencies of Ab10. Sequence analysis shows a surprising amount of molecular variation among Ab10 haplotypes, suggesting that there may be other phenotypic variables that contribute to the low but stable equilibrium frequencies.
IN the vast majority of cases, meiosis provides each allele with an equal opportunity of being passed on to the next generation. However, a number of systems are known that cause specific alleles or chromosomes to be transmitted at rates > 50%, a process known as meiotic drive (Sandler and Novitski 1957). Meiotic drive systems have independently arisen many times and involve widely diverse mechanisms. One of best understood is the Segregation Distorter (SD) system in Drosophila [reviewed in Larracuente and Presgraves (2012)]. The Sd locus encodes two elements in linkage: a truncated form of RanGAP, which is a protein involved in nuclear transport, and a region of satellite DNA called Responder (Rsp) (Wu et al. 1988; Merrill et al. 1999). In the absence of Rsp, the truncated Sd-RanGAP is mislocalized and spermatocytes fail to mature (Kusano et al. 2001), whereas sperm develop normally in the presence of Rsp. The t-haplotype in mice functions similarly; several inversions and multiple distorter loci that reduce sperm motility are linked to a responder locus that provides immunity to these effects (Herrmann et al. 1999). Other meiotic drive systems directly affect the mechanics of meiotic chromosome segregation to favor their own transmission. These include the D locus in Mimulus (Fishman and Willis 2005), the biased segregation of Robertsonian translocations in mice (Chmatal et al. 2014), and the preferential segregation of abnormal chromosome 10 (Ab10) in Zea mays (maize), the subject of this study.
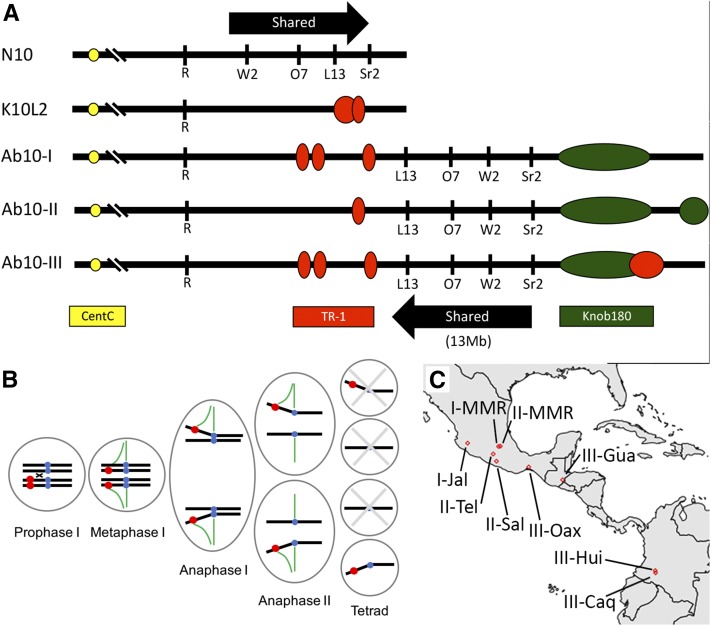
Maize has two forms of chromosome 10: a common form without any knobs referred to as normal chromosome 10 (N10) and a rarer Ab10 with large heterochromatic knobs at the end of the long arm (Longley 1938). Similar knobs are found on the arms of other chromosomes and are made up of two different repeats: a major 180-bp repeat and a 360-bp TR-1 repeat (Peacock et al. 1981; Ananiev et al. 1998). In addition to knobs, the long arm of Ab10 contains a set of genes that are shared with N10 (Figure 1A). The shared region contains several inversions relative to N10 that prevent recombination in this region (Rhoades and Dempsey 1985b; Mroczek et al. 2006). Rhoades showed that the Ab10 haplotype displays strong preferential segregation over N10 chromosomes when crossed as a female, such that Ab10 is reliably transmitted to 70–75% of progeny (Rhoades 1942). Further studies demonstrated that Ab10 not only causes the preferential segregation of itself, but all loci linked to knobs on other chromosomes as well, as long as the knob is heterozygous and Ab10 is present (Longley 1945). Ab10 chromosomes from different accessions can be visibly different (Figure 1A) and have been classified based on their knob patterning: Ab10-I, Ab10-II, and Ab10-III. In addition, there is another chromosome 10 variant called K10L2 that contains a large TR-1 knob but does not show strong meiotic drive (Kanizay et al. 2013a).
Figure 1.
The abnormal chromosome 10 (Ab10) haplotype in Z. mays. (A) Chromosome models of different haplotypes of chromosome 10 in Z. mays. The locations of centromeres (yellow), TR-1 (red), and Knob-180 (green) repeats are colored as shown. The locations of marker loci Colored1 (R1), White seedling2 (W2), Opaque endosperm7 (O7), Luteus13 (L13), and Striate leaves2 (Sr2) are listed below their relative positions. Note that in the Ab10-I, Ab10-II, and Ab10-III haplotypes, the relative ordering of W2, O7, and L13 is inverted in what is known as the shared region. K10L2 contains these same genes but their order and position relative to its knob is unknown. N10 indicates normal chromosome 10. (B) The Rhoades model of neocentromere-mediated meiotic drive. Knobs are shown in red (representing both the 180-bp knob repeats and TR-1 repeats), centromeres are in blue, and spindle fibers are in green. Meiotic drive requires that recombination occurs between the centromere and the Ab10 haplotype (and between centromeres and knobs on other chromosomes). Neocentromere activity, encoded by genes on Ab10, causes Ab10 and other knobs to move along microtubules, creating a polar arrangement that persists through metaphase I, anaphase I, metaphase II (data not shown), and anaphase II. The result is that Ab10 and other knobs end up in the upper and lower cells of the linear tetrad. As only the bottom-most cell will become a viable gamete, Ab10 and other knobs are preferentially transmitted. (C) Map of where the Ab10 haplotypes used in this study were originally collected.
Knobs are typically inert during meiosis, but when Ab10 is present they become “neocentromeres” that actively move along meiotic spindles ahead of centromeres (Rhoades and Vilkomerson 1942). This dramatic behavior led Marcus Rhoades to propose that neocentromere activity provides the mechanism for driving knobs to the fertile egg cell in female meiosis (Rhoades 1952) (Figure 1B). As knobs tend to be positioned in midarm positions, free recombination is expected to create a meiosis II (MII) segregation pattern—where one chromatid has a knob and the other does not–in two-thirds of the meioses. The MII arrangement allows neocentromere activity to consistently drive knobs to the fertile egg cell. In the remaining one-third of meioses, a meiosis I segregation pattern occurs and knobs are randomly transmitted. Thus, a driving knob can be preferentially transmitted via this mechanism to, at most, 2/3 × 1 + 1/3 × 1/2 = 5/6 (83%) of female gametes. Genetic and cell biological data show that several loci are required for meiotic drive (Hiatt and Dawe 2003), that neocentromeres do not recruit typical centromere/kinetochore proteins (Dawe and Hiatt 2004), and that knobs containing 180-bp repeats and those containing TR-1 repeats are activated to form neocentromeres by separate factors on Ab10 (Hiatt et al. 2002). Most recently, it has been shown that a novel kinesin motor protein on the distal tip of Ab10 called Kinesin driver (Kindr) is necessary for 180-bp neocentromere activity and meiotic drive (Lowry 2015; R. K. Dawe, E. G. Lowry, J. I. Gent, M. C. Stitzer, and D. M. Higgins, unpublished data).
While meiotic drive systems often have large transmission advantages in heterozygotes, those that are polymorphic, and thus observable, within populations have failed to reach fixation, suggesting that they have fitness costs that prevent them from fixing. In general, for such stable meiotic drive systems to occur, they must be advantageous when rare, at least when they first arise, but have fitness costs that prevent their fixation when they become common (Burt and Trivers 2006). Due to their associated fitness costs, segregating meiotic drivers cause genomic conflict because they distort their own segregation at the expense of the fitness of the rest of the genome. This genomic conflict sets the stage for both the evolution of unlinked suppressors of drive and linked modifiers that increase drive. Fixation of the SD system in flies is prevented by its effects on male fertility and by multiple unlinked suppressors that reduce the effectiveness of meiotic drive (Hartl 1970b; Lyttle 1979). The t-haplotype is homozygous male sterile and sometimes homozygous lethal (Ardlie 1998). As a result, the t-haplotype can never go to fixation, and there is a direct relationship between its frequency and the survival of offspring in populations that carry it. Population surveys have shown that Ab10 is present in 18–22% of landraces of maize, while within those populations the average frequency of individuals carrying Ab10 is ∼30% (Kanizay et al. 2013b). The low frequency of Ab10, despite its high segregation advantage in heterozygotes, suggests that there are deleterious fitness consequences associated with Ab10, although the nature of these defects has not been previously determined.
To better understand the evolution of the Ab10-driving chromosome in maize, we measured the level of drive in nine Ab10 haplotypes. The results reveal variable, but large, segregation advantages through ovules in the heterozygous condition for all Ab10 haplotypes. For five Ab10 haplotypes, we measured transmission through pollen in heterozygotes, and for one Ab10 haplotype we measured its effects on pollen viability, seed viability, and seed size in both heterozygotes and homozygotes. We found that Ab10 does not exhibit reduced transmission through pollen but does reduce pollen viability, seed viability, and seed weight, especially when homozygous. Mathematical modeling of the system indicates that the observed fitness costs can nearly but not entirely explain the frequency of Ab10 observed in natural populations. Sequence analysis of expressed genes in the shared regions of the Ab10 haplotypes failed to identify conserved deleterious SNPs, suggesting that the fitness defects may be caused by structural polymorphisms or properties of the drive system itself.
Materials and Methods
Seed stocks
The reference forms of Ab10-I and Ab10-II (Ab10-I-MMR and Ab10-II-MMR) were originally obtained from Marcus M. Rhoades (the “MMR” notation reflects his initials). The K10L2 chromosome was originally isolated from the inbred CI66 (Kanizay et al. 2013a). Seven additional Ab10 isolates were extracted from landraces and teosintes obtained from the North Central Plant Introduction Research Station (Ames, IA) based on prior results (Kanizay et al. 2013b). We identified Ab10 by PCR and crossed and backcrossed the Ab10 chromosome at least four times into standard maize tester stocks. The chromosomes were then named based on their morphology (Figure 1A) and place of origin: Ab10-I-Jal (PI 628445, Jalisco, Mexico), Ab10-II-Sal (Ames 21826, El Salado Guerrero, Mexico), Ab10-II-Tel (Ames 21826, Teloloapan-Arcelia Road, Guerrero, Mexico), Ab10-III–Hui (PI 444834, Huila, Colombia), Ab10-III-Caq (PI 444296, Caqueta, Colombia), Ab10-III-Oax (Ames 19980, Oaxaca, Mexico), and Ab10-III-Gua (PI490825, Guatemala).
Transmission assays
The transmission rates of the two original Ab10 haplotypes and seven newly extracted Ab10 haplotypes was inferred using the marker gene colored1 (R1). R1 is located near the end of the long arm of maize chromosome 10 and is in tight linkage with the Ab10 haplotype (Rhoades 1942). Kernels homozygous for the recessive allele (r1) are yellow while those with the dominant allele (R1) are purple. The Ab10-I-MMR and Ab10-II-MMR haplotypes are linked to R1, and all other Ab10 haplotypes are linked to r1. The Ab10 chromosomes were crossed to appropriately marked N10 chromosomes [R1-N10, r1-N10, or R1-nj-N10 (R1-nj causes only the tops of kernels to be purple)], and the resulting F1 plants crossed to r1-N10/r1-N10 testers to measure transmission. The transmission of each Ab10 chromosome was estimated by scoring kernels on 2–7 and 7–15 plants for ovule and pollen transmission, respectively (Supplemental Material, Table S1 and Table S2 in File S1). The experiment quantifying the transmission of Ab10 as a male parent was conducted during the summer of 2014 in Athens, GA. The experiment quantifying the transmission of Ab10 as a female parent was conducted during the summer of 2016 in Athens, GA.
Pollen viability assays
Pollen viability of Ab10-I-MMR was calculated using a modified version (Peterson et al. 2010) of the Alexander stain protocol (Alexander 1969) as described previously (Higgins et al. 2016). Five-hundred pollen grains from each plant were scored for viability based on staining color. Between 4 and 12 plants per genotype were scored (Table S3 in File S1).
Measuring seed set and weight
The effect of Ab10 on seed set and weight was measured using an Ab10-I-MMR line that was backcrossed into the B73 background for six generations and self-crossed. Sibling plants either homozygous (R-Ab10/R-Ab10) or heterozygous for Ab10 (R-Ab10/r-N10), or homozygous for r-N10 (the B73 inbred itself was used), were pollinated by B73 pollen. These plants were grown in Ames, IA, during the summer of 2016. The number of seeds per ear was counted manually and the mass of seeds from each ear was measured using a balance. Between 46 and 83 ears per genotype were measured (each plant had one ear; Table S4 in File S1).
Whole-transcriptome sequencing
Whole-transcriptome sequencing (RNA-seq) from young anthers was employed to identify SNPs and measure Kindr expression. Meiotically-staged anthers were dissected from tassels and RNA extracted as described previously (Lowry 2015; R. K. Dawe, E. G. Lowry, J. I. Gent, M. C. Stitzer, and D. M. Higgins, unpublished data). Sequencing libraries were generated using the KAPA Stranded RNA-Seq Kit (Kapa Biosystems, Wilmington, MA). Paired-end 75 bp sequencing was completed on an Illumina NextSeq at the Georgia Genomics Facility (Athens, GA). We sequenced mRNA from anthers of 2–5 plants as biological replicates for each Ab10 chromosome. All plants were heterozygous for Ab10 and therefore also expressed genes from an N10 tester chromosome (the tester chromosomes in this case are from nonspecific backgrounds and contain different R1 alleles). For Ab10-I-MMR and Ab10-II-Tel, we sequenced RNA from three plants containing the Ab10 chromosome, as well as three sibling plants that lacked the Ab10 chromosome but contained the same N10 chromosome (allowing us to remove SNPs from N10, see below).
SNP mapping and analysis
Reads were cleaned using Trimmomatic to remove adapters and 5′ and 3′ ends with low-quality scores < Phred 5 (Bolger et al. 2014). Cleaned reads were aligned to the maize reference genome (B73 AGPv4; Jiao et al. 2017) using the program STAR under the default settings (Dobin et al. 2012). Variants were called using the software package Genome Analysis Toolkit (GATK) (McKenna et al. 2010) following the Best Practices Workflow using HaplotypeCaller (Van der Auwera et al. 2013). SNPs that were not present in each of the replicates (n = 2–5) were removed using vcftools (Danecek et al. 2011). To identify SNPs unique to the shared region (coordinates 10:139,790,276–10:150,982,314 on B73 RefGen_v4) in Ab10-I-MMR and Ab10-II-Tel, we removed all SNPs identified in wild-type siblings from the same cross (which contained the same N10 tester chromosome as the Ab10 plants). Similarly, all the other Ab10 haplotypes (Ab10-I-Jal, Ab10-II-MMR, Ab10-II-Sal, Ab10-II-Oax, Ab10-III–Hui, Ab10-III-Caq, and Ab10-III-Gua) were heterozygous with one of the known N10 tester chromosomes (linked to R1-nj), and the SNPs from this N10 chromosome were removed prior to analysis.
To estimate the effects of Ab10-specific SNPs, we used the Ensembl Variant Effect Predictor (McLaren et al. 2016) with default settings. Those variants that resulted in a stop codon, a frameshift, or a missense substitution within the shared region distal to R1 were analyzed using the PROVEAN Protein webtool to estimate the biological impact of the substitutions (Choi and Chan 2015).
Expression of Kindr
To quantify the expression of Kindr in the 10 different accessions (all nine Ab10 haplotypes and K10L2), cDNA generation, high-throughput sequencing, and read processing were carried out as described above on heterozygous plants. A modified version of the maize B73 genome (AGPv3.27, Schnable et al. 2009) was used as the genomic reference for read alignment: The sequence of the 10 maize chromosomes was appended with an additional 100 kb of sequence from the Ab10 distal tip centered on the E9 copy of Kindr (Lowry 2015). Reads were aligned using TopHat version 2.0.14 (Kim et al. 2013), and differential expression of transcripts including Kindr was calculated using Cufflinks and Cuffdiff version 2.2.1 (Trapnell et al. 2010) following the previously published protocol (Trapnell et al. 2012).
Data availability
Transcriptome reads are available for download at the National center for Biotechnology Information Sequence Read Archive (ncbi.nlm.nih.gov/sra) under project accession numbers SRP082328, SRP058857, and SRP082329. The supplemental material provides primary data on the meiotic drive and male transmission of different Ab10 haplotypes; primary data on the effects of Ab10 on pollen viability, seed count, and seed weight; and a partial listing of missense mutations observed the shared region of Ab10.
Results
Isolation of seven new Ab10 haplotypes and measurements of Kindr expression
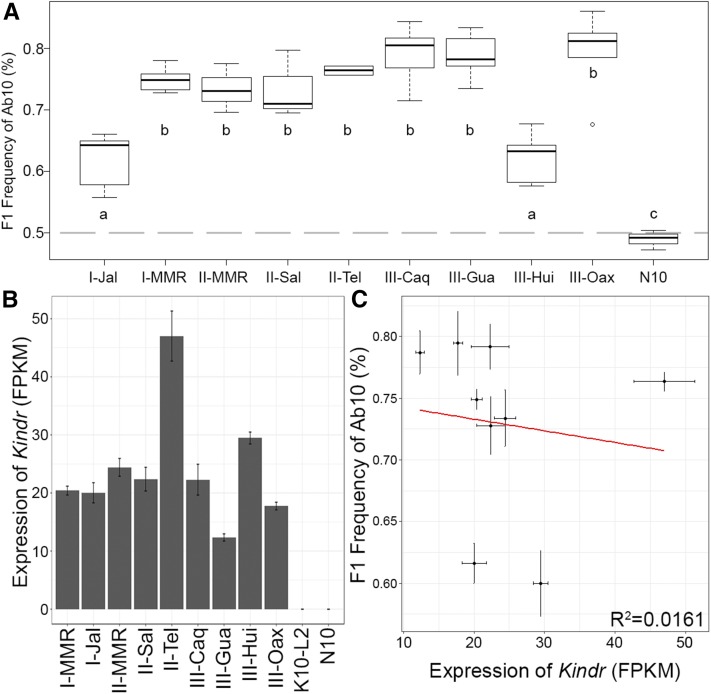
We extracted seven new Ab10 chromosomes from maize landraces and teosintes from across Central and South America (Figure 1C). These were initially identified in tropical maize or teosinte backgrounds (Kanizay et al. 2013b) and crossed into standard corn-belt maize lines. Previously published measurements of meiotic drive by the Ab10-I-MMR and Ab10-II-MMR haplotypes are 70–79% (Rhoades 1942; Kanizay et al. 2013a). Measurements of meiotic drive from the seven new haplotypes revealed a broader range (61.6–79.5% transmission; Figure 2A and Table S1 in File S1). While many were not significantly different from the transmission rate of Ab10-I-MMR ( = 74.9%) by ANOVA and Tukey’s honest significant difference test (α= 0.05), two haplotypes, Ab10-I-Jal ( = 61.6%) and Ab10-III-Hui ( = 60.0%), displayed significantly lower levels of meiotic drive (but were significantly higher than the N10 control, = 48.9%).
Figure 2.
Rates of meiotic drive and Kindr expression in different haplotypes. (A) The female transmission of abnormal chromosome 10 (Ab10) was calculated using a standard testcrossing scheme (see Figure 3A). Groups of statistical significance are listed with letters (a, b, and c) as determined by ANOVA and Tukey’s honest significant difference test. N10 indicates normal chromosome 10. (B) Expression of Kindr in each Ab10 haplotype measured in fragments per kilobase of transcript per million mapped reads (FPKM). Error bars represent SE as calculated by Cuffdiff. (C) When plotted against one another, the two variables of Kindr expression and meiotic drive show no correlation (R2 = 0.0161). Error bars represent SE for both variables.
Kindr is a kinesin motor protein, discovered on the distal tip of the Ab10-I-MMR haplotype, which has been implicated in neocentromere activity and meiotic drive (Lowry 2015; R. K. Dawe, E. G. Lowry, J. I. Gent, M. C. Stitzer, and D. M. Higgins, unpublished data). Analysis of Kindr expression in the nine haplotypes revealed quantitative variation in expression (Figure 2B), where with the Ab10-II-Tel haplotype showed approximately twice the Kindr expression of Ab10-I-MMR, and Ab10-III-Gua having approximately half the Kindr expression of Ab10-I-MMR. K10L2, the Ab10 variant that lacks the large 180-bp knob (Figure 1A) and fails to show strong meiotic drive (Kanizay et al. 2013a), showed no Kindr expression. There was no significant correlation between Kindr expression and the degree of preferential segregation across Ab10 haplotypes (R2 = 0.0161, Figure 2C).
Ab10 has no transmission disadvantage through the male
As meiotic drive only occurs when Ab10 is crossed as a female, it is possible that the fitness advantage through females is counterbalanced by fitness disadvantages in the male. The original study by Rhoades (1942) provides support for this view, showing that the transmission of Ab10-I-MMR through pollen was 42%, significantly below the expected 1:1 ratio. However, an alternative finding in a later study indicated that male transmission of Ab10-II-MMR was roughly Mendelian (Rhoades and Dempsey 1988), leaving open the question as to whether there are consistent effects of Ab10 on pollen transmission.
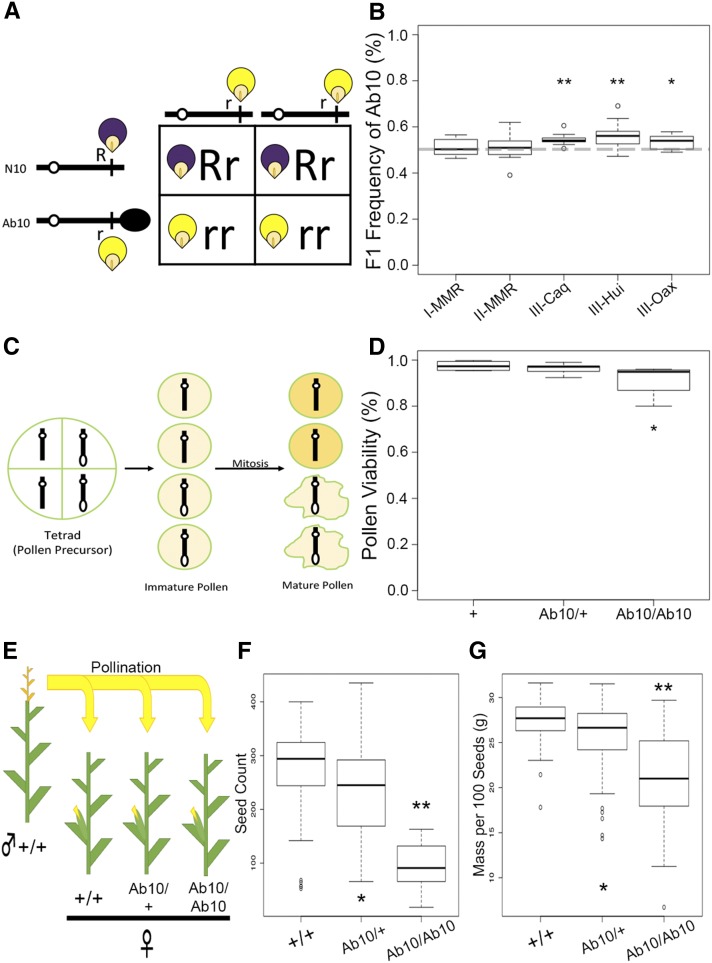
The male transmission of Ab10-I-MMR, Ab10-II-MMR, and three new isolates of Ab10-III (-Hui, -Caq, and -Oax) was measured by crossing each as heterozygotes to a recessive tester line. The presence of Ab10 was scored using alleles of the linked marker gene colored1 (R1) (Figure 2A). The average of all five haplotypes combined was > 50% (50.4–57.1%) (Figure 2B and Table S2 in File S1). Three of the five showed significantly higher transmission than expected for a 1:1 ratio by Fisher’s Exact Test (Ab10-III-Caq and Ab10-III-Hui, P < 0.0001 and Ab10-III-Oax, P = 0.01). Although male transmission is presumably influenced by genetic background and environment, our data suggest that Ab10 does not have a transmission disadvantage in heterozygotes, and might actually have a slight advantage. However, we are hesitant to draw too much from this apparent overrepresentation of Ab10 in male progeny. Two different N10 tester chromosomes were used in this analysis; Ab10-I-MMR and Ab10-II-MMR were linked to R and were heterozygous for a r1-N10 tester chromosome, while Ab10-III-Caq, Ab10-III-Hui, and Ab10-III-Oax were linked to r1 and the tester chromosome was R1-nj. The apparent differences between these two groups may be an effect of deleterious alleles on the R1-nj chromosome or poor penetrance of the R1-nj allele, which only pigments the cap of the kernel.
Ab10 reduces pollen viability
Plant gametes are known as gametophytes since they undergo mitotic divisions in the haploid state before they mature and are ready for fertilization. In the male gametophyte, each pollen mother cell undergoes a complex differentiation process involving two mitotic cell divisions. Cells carrying Ab10 may be less likely to survive this process (Figure 3C). To test if this is true, we estimated the percentage of live pollen in plants heterozygous and homozygous for Ab10 and homozygous for N10 using a modified version of Alexander’s stain (an indirect measure of pollen viability that identifies intact protoplasm). We scored ∼500 pollen grains per plant and found no differences in Alexander staining between N10/N10 homozygotes (viability = 0.97, n = 4) and Ab10/N10 heterozygotes (viability = 0.96, n = 12) as determined by a Student’s t-test (P = 0.649; Figure 3D and Table S3 in File S1). However, Alexander staining in Ab10/Ab10 homozygotes (viability = 0.91, n = 8) was significantly lower than both N10/N10 homozygotes (P = 0.022) and Ab10/N10 heterozygotes (P = 0.013). These data support the transmission results, which indicated that Ab10 pollen survival is not reduced, since pollen showed no significant difference in viability in N10/N10 and Ab10/N10 plants. We note that we had the statistical power to detect pollen viability differences as small as 2.2% in the Ab10/N10 heterozygote relative to the N10/N10 homozygote, given our sample sizes. The observed 1% reduction in Ab10/N10 pollen viability was below this difference and thus could not have reached statistical significance. The reduction in healthy pollen observed in Ab10/Ab10 homozygotes suggests a defect at the level of anther development and/or pollen maturation.
Figure 3.
Phenotypic consequences of Ab10-I-MMR. (A) Punnett square diagramming the testcrossing scheme used to measure inheritance of Ab10. In this case, the Ab10 haplotype is linked to the recessive allele of r1, resulting in yellow kernels. The dominant allele R1 is linked to N10 and conditions purple kernels. (B) Quantification of the transmission rates of five Ab10 haplotypes as a male parent. None of the medians were lower than 50%. Transmission of three of the haplotypes was significantly higher than expected as determined by Fisher’s Exact Test (* P < 0.01 and ** P < 0.0001). (C) Pollen development involves two rounds of mitosis in the haploid phase, where Ab10 could have negative phenotypic consequences (here shown as crinkled, dead pollen). (D) Pollen viability in different Ab10-I-MMR genotypes. Pollen viability in N10/N10 homozygotes ( = 0.97) and Ab10/N10 heterozygotes ( = 0.96) were not significantly different, whereas Ab10/Ab10 homozygotes had significantly lower pollen viability ( = 0.91) than both N10/N10 and Ab10/N10 (P = 0.049 and 0.009, respectively). (E) Crossing scheme for measuring the effects of Ab10 on seed count and mass. All genotypes were in the B73 background. (F) Seed count for Ab10/Ab10 homozygotes ( = 95) and Ab10/N10 heterozygotes ( = 233) was significantly reduced compared to N10/N10 wild-types ( = 271). Statistical significance is shown (* P < 0.05 and ** P < 0.001) as measured by Student’s t-test. (G) Mass per 100 seeds in Ab10 homozygotes ( = 20.8 g) and Ab10 heterozygotes ( = 25.7 g) is significantly reduced compared to N10 wild-types ( = 27.3 g). Ab10, abnormal chromosome 10; N10, normal chromosome 10.
Ab10 reduces seed set and seed size
To reduce the yield variability that typically occurs when creating a segregating population, we first backcrossed Ab10-I-MMR to the standard B73 inbred for six generations and self-crossed to generate plants that were heterozygous and homozygous for Ab10. For N10 controls, we used the standard B73 inbred. These plants were pollinated by B73 pollen (Figure 3E) and the seeds produced were counted and weighed.
The data derived from 176 ears and over 36,000 seeds show that homozygosity for Ab10 caused a dramatic 64.9% reduction in seed count (Figure 3F and Table S4 in File S1, P < 0.001). In addition, homozygosity for Ab10 caused a 23.7% reduction in seed weight (Figure 3G and Table S4 in File S1, P < 0.001). Ab10 heterozygotes showed an intermediate phenotype that was more similar to wild-type, with a 13.8% reduction in seed count (P = 0.023) and 5.9% reduction in seed weight (P = 0.013).
Extensive molecular variation among Ab10 haplotypes
The shared region is conserved between N10 and Ab10, and contains essential genes necessary for plant growth and development. However, several inversions prevent the two haplotypes from recombining at a high frequency (Figure 1A) (Rhoades and Dempsey 1985a; Mroczek et al. 2006). This region provides a likely environment for the accumulation of deleterious alleles within the Ab10 haplotype. To assess sequence diversity in the shared region, we first identified sequence variants (differing from the B73 reference genome) in the Ab10-I-MMR and Ab10-II-Tel haplotypes using an RNA-seq approach. The plants used were heterozygous for Ab10/N10 and SNPs derived from the (non-B73) N10 chromosome were filtered out using RNA-seq data from wild-type siblings. We found 1120 sequence variants in the shared region of Ab10-I-MMR, and 803 in the shared region of Ab10-II-Tel. Of the 228 sequence variants found in both haplotypes, 72 resulted in missense mutations (Table S5 in File S1), eight of which were predicted to be deleterious by PROVEAN software (Choi et al. 2012). These numbers, revealing that around one-third of the SNPs result in missense mutations, is consistent with the overall genome average (Mezmouk and Ross-Ibarra 2014). We then asked whether the 228 SNPs common to both Ab10-I-MMR and Ab10-II-Tel were present in plants containing the remaining seven Ab10 haplotypes (Ab10-I-Jal, Ab10-II-MMR, Ab10-II-Sal, Ab10-II-Oax, Ab10-III–Hui, Ab10-III-Caq, and Ab10-III-Gua). Surprisingly, only six of the 228 SNPs were found in all nine Ab10 haplotypes, and none of these were predicted to be deleterious by PROVEAN (Table S5 in File S1).
Discussion
In this study, we assessed the diversity and phenotypic consequences of abnormal chromosome 10 with the aim of shedding light on the question of why it is observed at low frequencies in natural populations. Data from other systems suggest that deleterious alleles linked to meiotic drive elements negatively impact their frequency within populations. For instance, the D locus in Mimulus, another female-specific meiotic drive system in plants, displays a 19% reduction in pollen viability (Fishman and Saunders 2008) and a 21% reduction in seed production (Fishman and Kelly 2015) in homozygotes. Here, we describe seven new Ab10 haplotypes and measure meiotic drive, male transmission, and molecular polymorphism. In addition, we performed the first detailed analysis of pollen viability, seed viability, and seed size in heterozygotes and homozygotes for the original Ab10-I-MMR accession.
Analysis of Ab10 chromosomes from different geographical locations revealed two haplotypes (Ab10-I-Jal and Ab10-III-Hui) with distinctly lower levels of meiotic drive (∼61%) than had been previously described. Lower transmission rates could be caused by features that reduce recombination between the centromere and the Ab10 haplotype [Ab10 is known to encode a factor(s) that affects recombination (Hiatt and Dawe 2003)], inefficiencies in neocentromere activity or other processes required for meiotic drive, or could be caused by selective abortion of female gametophytes carrying Ab10 (we did not measure seed set for these haplotypes). In contrast to prior results (Rhoades 1942), we found no evidence indicating that the transmission of Ab10 is reduced in male crosses; instead, we found some evidence that Ab10 can be moderately favored through the male (Figure 3B). We observed substantial deleterious effects in Ab10 homozygotes, including pollen viability, seed set, and seed mass (Figure 3, D, F, and G). The observed reduction in pollen viability could be caused by defects in the development and function of anthers, which provide not only the structural support for pollen formation but many proteins and carbohydrates necessary for the formation of the outer wall (exine) of mature pollen grains (Bedinger 1992). Reductions in seed set could be caused by failures at any level of pollination and fertilization, including silk development and the capacity of silks to support pollen tube development. The observed reductions in seed mass suggest that there may also be defects in the conducting tissue that supplies seeds with carbohydrates. Low seed weight can be associated with low seedling emergence under poor soil conditions (Graven and Carter 1990) or reduced plant size (Hunter and Kannenberg 1972), although these effects tend to be minor and situation-specific, at least in cultivated maize.
An important question is whether the reductions in live pollen, seed viability, and seed size can explain the observed frequency of Ab10 in natural populations. We developed a population genetic model to address this question in a companion study (Hall and Dawe 2017). Using estimates of the parameter values obtained above, and assuming seed size is directly proportional to seed viability, we found that the frequency of Ab10 observed in nature (0–33%) is only predicted when the level of meiotic drive is toward the low end of what we observed (58.6–69.1%). This result holds even if we use the lowest estimates of all measured fitness components within our C.I.s (meiotic drive range only increases to 60–72%). Seven of the nine haplotypes studied here showed substantially higher levels of meiotic drive than this predicted range (Figure 2A). These results suggest that the fitness consequences elucidated in this study are not sufficient on their own to maintain Ab10 at current frequencies. Two obvious factors that have not been measured are genotype-specific germination rate and survival to adulthood. We also have not accounted for phenotypes that may be revealed in less ideal environments, such as biotic and abiotic stresses, and competition from other plants. Finally, Ab10 causes the meiotic drive of knobs throughout the genome (Buckler et al. 1999), placing it in linkage disequilibrium with many unlinked loci that may be deleterious alone or in combination with alleles on Ab10.
The shared region of Ab10 is a likely place for deleterious mutations to accumulate because it contains hundreds of essential genes and recombination with N10 is reduced due to multiple inversions (Mroczek et al. 2006). Generally, small effect deleterious mutations are purged from a population. However, in the absence of recombination, Muller’s ratchet, genetic hitchhiking, and background selection can all lead to the accumulation of deleterious alleles on nonrecombining chromosomes (Charlesworth and Charlesworth 1998). Our analysis identified deleterious sequence variants in the shared region, but at frequencies similar to what has been observed in other maize lines (Mezmouk and Ross-Ibarra 2014). Further, very few of the SNPs in the Ab10 haplotype are conserved; only six sequence variants in the shared region are common to all Ab10 haplotypes. The high level of polymorphism on Ab10 is consistent with an ancient origin and a high level of gene flow among Ab10 and N10 chromosomes, similar to the sex-ratio system in Drosophila neotestacea (Pieper and Dyer 2016).
The lack of conserved SNPs in the shared region raises the question of what genetic lesions underlie the observed fitness defects in homozygotes. Our inability to identify conserved deleterious SNPs may be a result of the fact that the RNA-seq strategy only samples genes expressed in the tissue used to prepare the RNA (young anthers). Deeper sampling of multiple tissues may have revealed additional, perhaps conserved and deleterious SNPs. Another explanation is that Ab10 is simply missing some of the genes normally contributed by the shared region on N10. Presence–absence variation is common in maize, to the extent that any two inbreds may differ in gene content by as much as 3.4% (Hirsch et al. 2016). It is likely that the inversions that isolated the Ab10 haplotype from N10 included gene absence polymorphisms that either themselves confer negative fitness consequences or do so when combined with other polymorphisms in the genome. Unlike SNPs, which can flow among chromosomes by gene conversion, repairing a deletion using N10 as a template would require double recombination, which is extremely rare in nested inversions such as found on Ab10. Our RNA-seq strategy would not have detected missing genes and, likewise, would have missed genes unique to Ab10 that are not present in the B73 reference. It is also formally possible that the drive system itself, which involves specialized kinesin proteins, may interfere with cell division to cause some of the observed fitness defects. Full sequencing and assembly of the Ab10 chromosome will help fill the gaps in our understanding of Ab10.
Taken together, our data show that Ab10 has negative fitness consequences as a homozygote and that these phenotypes limit the spread of Ab10 in nature, consistent with observations from other meiotic drive systems that encompass large haplotypes (Ardlie 1998; Burt and Trivers 2006; Fishman and Saunders 2008). While our modeling suggests that Ab10 can be stabilized at its observed frequency, these data only apply to Ab10-I-MMR in the B73 genetic background when grown and hand pollinated in the fields of Iowa. Other Ab10 types with different natural levels of meiotic drive (Figure 2A) and different deleterious alleles grown in other natural environments may be expected to stabilize at different frequencies, fix, or be purged from populations. For instance, prior analyses suggest that Ab10-III is most frequent in cultivated maize and Ab10-II is most commonly observed in teosinte (Kanizay et al. 2013b). These differences may represent historical contingencies or differential adaptation of Ab10 to the environmental and genetic backgrounds of their respective populations, allowing Ab10 to maintain a low but consistent frequency across the entire geographical range of Z. mays.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300060/-/DC1.
Acknowledgments
We thank Amy Hodges for technical help and Jeffrey Ross-Ibarra for providing useful comments on an early version of the manuscript. This work was supported by a grant from the National Science Foundation (1412063) to R.K.D.
Note added in proof: See Hall and Dawe (pp. 123–130) in G3: Genes|Genomes|Genetics for a related work.
Footnotes
Communicating editor: S. Wright
Literature Cited
- Alexander M., 1969. Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Ananiev E., Phillips R., Rines H., 1998. A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: are chromosome knobs megatransposons? Proc. Natl. Acad. Sci. USA 95: 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie K. G., 1998. Putting the brake on drive: meiotic drive of t haplotypes in natural populations of mice. Trends Genet. 14: 189–193. [DOI] [PubMed] [Google Scholar]
- Bedinger P., 1992. The remarkable biology of pollen. Plant Cell 4: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E. S. I., Phelps-Durr T. L., Buckler C. S. K., Dawe R. K., Doebley J. F., et al. , 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., Trivers R. L., 2006. Genes in Conflict: The Biology of Selfish Genetic Elements. Harvard University Press, Cambridge, MA. [Google Scholar]
- Charlesworth B., Charlesworth D., 1998. Some evolutionary consequences of deleterious mutations. Genetica 102–103: 3–19. [PubMed] [Google Scholar]
- Chmatal L., Gabriel S. I., Mitsainas G. P., Martinez-Vargas J., Ventura J., et al. , 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 24: 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Chan A. P., 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31: 2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Sims G. E., Murphy S., Miller J. R., Chan A. P., 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe R. K., Hiatt E. N., 2004. Plant neocentromeres: fast, focused, and driven. Chromosome Res. 12: 655–669. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., et al. , 2012. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Kelly J. K., 2015. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution 69: 1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Saunders A., 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322: 1559–1562. [DOI] [PubMed] [Google Scholar]
- Fishman L., Willis J. H., 2005. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics 169: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graven L. M., Carter P. R., 1990. Seed size/shape and tillage system effect on corn growth and grain-yield. J. Prod. Agric. 3: 445–452. [Google Scholar]
- Hall, D. W., and R. K. Dawe, 2017 Modeling the evolution of female meiotic drive in maize. G3 (Bethesda). 8: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., 1970b Meiotic drive in natural populations of Drosophila melanogaster. IX. Suppressors of segregation distorter in wild populations. Can. J. Genet. Cytol. 12: 594–600. [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Koschorz B., Wertz K., McLaughlin K. J., Kispert A., 1999. A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature 402: 141–146. [DOI] [PubMed] [Google Scholar]
- Hiatt E. N., Dawe R. K., 2003. Four loci on abnormal chromosome 10 contribute to meiotic drive in maize. Genetics 164: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt E. N., Kentner E. K., Dawe R. K., 2002. Independently-regulated neocentromere activity of two classes of satellite sequences in maize. Plant Cell 14: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. M., Nannas N. J., Dawe R. K., 2016. The maize divergent spindle-1 (dv1) gene encodes a kinesin-14A motor protein required for meiotic spindle pole organization. Front. Plant Sci. 7: 1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C. N., Hirsch C. D., Brohammer A. B., Bowman M. J., Soifer I., et al. , 2016. Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28: 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter R. B., Kannenberg L., 1972. Effects of seed size on emergence, grain yield, and plant height in corn. Can. J. Plant Sci. 52: 252–256. [Google Scholar]
- Jiao Y., Peluso P., Shi J., Liang T., Stitzer M. C., et al. , 2017. Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanizay L. B., Albert P. S., Birchler J. A., Dawe R. K., 2013a Intragenomic conflict between the two major knob repeats of maize. Genetics 194: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanizay L. B., Pyhajarvi T., Lowry E. G., Hufford M. B., Peterson D. G., et al. , 2013b Diversity and abundance of the abnormal chromosome 10 meiotic drive complex in Zea mays. Heredity (Edinb) 110: 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano A., Staber C., Ganetzky B., 2001. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev. Cell 1: 351–361. [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Presgraves D. C., 2012. The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics 192: 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley A. E., 1938. Chromosomes of maize from Native American Indians. J. Agric. Res. 56: 177–195. [Google Scholar]
- Longley A. E., 1945. Abnormal segregation during megasporogenesis in maize. Genetics 30: 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, E. G., 2015 The meiotic drive mechanism of a selfish chromosome in Zea mays. Ph.D. Thesis, University of Georgia, Athens. [Google Scholar]
- Lyttle T. W., 1979. Experimental population genetics of meiotic drive systems II. Accumulation of genetic modifiers of segregation distorter (SD) in laboratory populations. Genetics 91: 339–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S. E., Riat H. S., Ritchie G. R. S., et al. , 2016. The Ensembl variant effect predictor. Genome Biol. 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill C., Bayraktaroglu L., Kusano A., Ganetzky B., 1999. Truncated RanGAP encoded by the segregation distorter locus of Drosophila. Science 283: 1742–1745. [DOI] [PubMed] [Google Scholar]
- Mezmouk S., Ross-Ibarra J., 2014. The pattern and distribution of deleterious mutations in maize. G3 (Bethesda) 4: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek R. J., Melo J. R., Luce A. C., Hiatt E. N., Dawe R. K., 2006. The maize Ab10 meiotic drive system maps to supernumerary sequences in a large complex haplotype. Genetics 174: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock W., Dennis E., Rhoades M., Pryor A., 1981. Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78: 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R., Slovin J. P., Chen C., 2010. A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 1: 66–69. [Google Scholar]
- Pieper K. E., Dyer K. A., 2016. Occasional recombination of a selfish X-chromosome may permit its persistence at high frequencies in the wild. J. Evol. Biol. 29: 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. M., 1942. Preferential segregation in maize. Genetics 27: 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. M., 1952. Preferential segregation in maize, pp. 66–80 in Heterosis, edited by Gowen J. W. Iowa State College Press, Ames, IA. [Google Scholar]
- Rhoades M. M., Dempsey E., 1985a Structural heterogeneity of chromosome 10 in races of maize and teosinte, pp. 1–18 in Plant Genetics, edited by Freeling M. Alan R. Liss, New York. [Google Scholar]
- Rhoades M. M., Dempsey E., 1985b Structural heterogeneity of chromosome 10 in races of maize and teosinte, pp. 1–18 in Plant Genetics: Proceedings of the Third Annual ARCO Plant Cell Research Institute-UCLA Symposium on Plant Biology, edited by Freeling M. A.R. Liss, New York. [Google Scholar]
- Rhoades M. M., Dempsey E., 1988. Structure of K10-II chromosome and comparison with K10-I. Maize Genet. Coop. News Lett. 62: 33. [Google Scholar]
- Rhoades M. M., Vilkomerson H., 1942. On the anaphase movement of chromosomes. Proc. Natl. Acad. Sci. USA 28: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Novitski E., 1957. Meiotic drive as an evolutionary force. Am. Nat. 91: 105–110. [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera, G. A., M. O. Carneiro, C. Hartl, R. Poplin, G. del Angel et al., 2013 From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current Protocols in Bioinformatics. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-I., Lyttle T. W., Wu M.-L., Lin G.-F., 1988. Association between a satellite DNA sequence and the responder of segregation distorter in D. melanogaster. Cell 54: 179–189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome reads are available for download at the National center for Biotechnology Information Sequence Read Archive (ncbi.nlm.nih.gov/sra) under project accession numbers SRP082328, SRP058857, and SRP082329. The supplemental material provides primary data on the meiotic drive and male transmission of different Ab10 haplotypes; primary data on the effects of Ab10 on pollen viability, seed count, and seed weight; and a partial listing of missense mutations observed the shared region of Ab10.