Abstract
We re-examine the model of Kirkpatrick and Barton for the spread of an inversion into a local population. This model assumes that local selection maintains alleles at two or more loci, despite immigration of alternative alleles at these loci from another population. We show that an inversion is favored because it prevents the breakdown of linkage disequilibrium generated by migration; the selective advantage of an inversion is dependent on the amount of recombination between the loci involved, as in other cases where inversions are selected for as a result of their effects on recombination. We derive expressions for the rate of spread of an inversion; when the loci covered by the inversion are tightly linked, these conditions deviate substantially from those proposed previously, and imply that an inversion can then have only a small advantage.
Keywords: inversion, linkage disequilibrium, migration, recombination, selection
KIRKPATRICK and Barton (2006) proposed an influential model for the spread of an inversion that suppresses recombination between two or more loci that are under selection in a local population, in the face of the introduction of disfavored alleles by gene flow. Up to now, direct evidence for this model has been lacking, but a recent paper on the highly self-fertilizing plant Boechnera stricta claims to provide an example (Lee et al. 2017). In this case, an inversion has become common in a hybrid zone between two ecologically distinct subspecies, over an estimated time of between 1000 to 4000 generations. But it is hard to see how recombination suppression could confer a significant selective advantage when there is a high level of inbreeding within a population, where the effective recombination rate between a pair of loci is greatly reduced (Nordborg 1997), because the advantage of recombination suppression must depend on the rate of recombination in the initial population.
Equation 3 of Kirkpatrick and Barton (2006) was intended to describe the selective advantage to a rare inversion, in terms of the rate of recombination r between adjacent loci, the selective disadvantage s suffered by an immigrant allele, and the rate of migration m. For the case of two loci, this equation states that the selective advantage of an inversion, sl, is approximately equal to 2rm/(2r + ms). If ms << r, sl ≈ m, and is only weakly dependent on r. At first sight, this suggests that the B. stricta example could indeed be explained by their model, although Kirkpatrick and Barton did point out that the selective advantage of an inversion should tend to zero as the recombination rate becomes small.
This note re-examines the Kirkpatrick-Barton model. Two main conclusions are reached. First, the condition for the spread of an inversion is similar to that derived by Charlesworth and Charlesworth (1973) for the case of a randomly mating population at equilibrium under epistatic selection—linkage disequilibrium (LD) must be present among the loci subject to selection. Second, Equation 3 of Kirkpatrick and Barton is wrong, presumably because of an error in its derivation (the existence of an error in this equation was previously pointed out by Bürger and Akerman (2011)). The denominator is (2r + ms), which is dimensionally inconsistent in the continuous time version of the model, in which the coefficients r, m and s all have dimension t−1. Using the correct expression, the selective advantage of an inversion becomes very small when the effective recombination rate is small. This casts some doubt on the interpretation of their data by Lee et al. (2017).
Following Kirkpatrick and Barton (2006), we use a model of a haploid species with a focal deme subject to migration from a source deme, with migration rate m into the focal deme. Alleles at two or more loci are assumed to be at fixation in the source deme, and are disfavored by selection in the focal deme. While this model does not fully describe what happens with diploidy, it should provide a good approximation to the dynamics of a highly inbred population, where selection is predominantly among homozygous genotypes. We assume that migration is sufficiently weak in relation to selection that the disfavored alleles in the focal population are kept rare, which greatly simplifies the calculations. With this low migration assumption, the model is equivalent to the case of two demes with selection in opposite directions (Moran 1962, pp. 173–175; Charlesworth and Charlesworth 2010, p. 146). We first consider the case of a pair of selected loci, allowing for the possibility of epistasis in fitness, and then analyze the multi-locus case with additive fitness effects. A treatment that relaxes the assumption of low migration is given in the Appendix, which is equivalent to the analysis of the diploid model with no dominance or epistasis by Bürger and Akerman (2011). Our main findings for the two-locus case when epistasis is absent are equivalent to those of Bürger and Akerman with weak migration.
Results for Two Selected Loci with Low Migration
The state of the initial population
Here, we assume two loci, 1 and 2, with alleles Ai and ai at locus i. The relative fitnesses of the four haplotypes A1A2, A1a2, a1A2 and a1a2 in the focal deme are 1, 1 − s2, 1 − s1, and 1 − s1 − s2 + e, respectively, where si is the selection coefficient against the disfavored allele at locus i, and e is a measure of epistasis. Let the frequencies of the four haplotypes in the focal deme i be x1, x2, x3, and x4; the frequencies of alleles Ai and ai at locus i are pi and qi, respectively. The coefficient of LD is D = x1x4 − x2x3. Selection is assumed to be sufficiently strong compared with migration that second-order terms in the frequencies of the disfavored haplotypes can be neglected, but sufficiently weak that second-order terms in the selection parameters are negligible. We write li = m/si as a measure of the relative strength of migration and selection at locus i; given our assumptions, li << 1.
Even with an additive fitness model, the changes in allele frequencies at one locus are not independent of the frequencies of the alleles at the other locus when there is LD generated by migration. The magnitude of this LD at equilibrium is given in Equation 1 below (a simple derivation is provided in the Appendix):
| (1) |
Note that asterisks are used to denote the equilibrium values of variables.
Substituting from Equation 1 into Equations A2, we get:
| (2a) |
| (2b) |
With close linkage, such that r << s1 + s2 − e, the equilibrium frequencies of the disfavored alleles are substantially lower than the values with D* = 0. When s1 = s2 = s and e = 0, the symmetrical additive fitness case considered by Kirkpatrick and Barton (2006), these frequencies approach m/(2s) as r tends to zero. The equilibrium mean fitness of the population is greater than the value with no LD, 1 − 2m; it approaches 1 − m as r tends to zero. In general, the equilibrium mean fitness is given by:
| (3) |
Conditions for the spread of an inversion
We now consider the introduction into haplotype 1 of an inversion that completely suppresses crossing over between the two loci. Let the frequency of the inverted haplotype in the focal deme be xI. Then, assuming that the system is initially at equilibrium, and, using Equations 3 and A3a to determine its change in frequency, we obtain:
| (4) |
This is equivalent to Equation 5.2 of Bürger and Akerman (2011) for the corresponding continuous time model without epistasis. The multiplicand of xI provides a measure of the selective advantage of the inversion, sI, which corresponds to the quantity λ − 1 in Kirkpatrick and Barton (2006).
This expression shows that sI is equal to the difference between the migration load at equilibrium, and the migration load experienced by the inversion, m. This difference depends on the existence of LD in the initial population, as can be seen from the last term in Equation 4. However, with loose linkage, LD is inversely proportional to the recombination rate, and the rate of increase given by Equation 4 is nearly independent of r, and equal to m, the value for loose linkage given by Equation 1 of Kirkpatrick and Barton (2006). Positive epistasis (e > 0) increases the selective advantage of an inversion, whereas negative epistasis (e < 0) has the opposite effect. As in other situations where reduced recombination is favored by selection, a rare modifier of recombination cannot have a selective advantage in the absence of LD in the initial population, since recombination has no effect on changes in haplotype frequencies in the absence of LD (Feldman 1972; Charlesworth and Charlesworth 1973).
The selective advantage to the inversion given by the right-hand side of Equation 4 differs substantially from that given by the expression for λ in Equation 3 of Kirkpatrick and Barton (2006) for the case when s1 = s2 = s and e = 0. That equation is obviously incorrect, for the following reason. When all processes are slow (m, r, and s << 1), so that the population can be treated as evolving continuously with respect to time, m, r, and s all have dimensions of the reciprocal of time. The denominator in the equation of Kirkpatrick and Barton (2006), 2mr/(2r + ms), thus has inconsistent dimensions, and the effect of recombination relative to selection is greatly overestimated, as noted in the Introduction. For example, with r = 0.005, m = 0.01, s = 0.1, Equation 4 gives sl = 0.00024 compared with 0.0091 from the formula of Kirkpatrick and Barton. The Appendix gives the exact two-locus solution in the continuous-time limit, for the case of additive selection; essentially the same results were derived previously by Bürger and Akerman (2011) for the diploid model without dominance.
The multi-locus case with additive fitnesses:
For simplicity, only the case when each locus has the same selection coefficient, s, will be considered, as in Kirkpatrick and Barton (2006). The same notation as above is used, except that there are now n loci under selection; the recombination rate between loci i and j is rij, and the coefficient of LD between these loci in the focal deme is Dij. Equations 3 of Barton (1983) can be used to be obtain an exact solution, assuming weak selection. Here, we provide a heuristic treatment of the problem, assuming additive fitness effects.
With tight linkage, and assuming that migration is much weaker than selection, the focal population will be composed predominantly of haplotype 1, carrying the favored allele Ai at each of the n loci in the chromosomal region under consideration; the immigrant haplotypes all have allele ai at these loci. With these assumptions, haplotype 1 will be broken down by recombination at rate Rx1x0, where R is the probability of at least one recombination event in the region in question, x1 is the frequency of haplotype 1, and x0 is the frequency of the complementary, immigrant haplotype. For the initial population, we thus have:
| (5) |
At equilibrium, the selective advantage of an inversion in haplotype 1 is thus given by:
| (6a) |
But, with tight linkage, the system behaves very like a single locus with net selection coefficient S = ns, so that:
Substituting this into Equation 6a, we obtain:
| (6b) |
If the loci are equally spaced along the chromosome, with distance r between each pair, R = (n − 1)r (assuming that terms in r2 can be neglected) and:
| (6c) |
With n >> 2, sI is largely determined by the frequency of recombination between adjacent loci, and the strength of selection at each locus.
For the opposite extreme of free recombination, the procedure of neglecting LD used by Kirkpatrick and Barton (2006) can be followed. Because each locus then causes an equilibrium reduction in fitness of m, the equilibrium mean fitness of the focal population is ∼1 − nm; substituting this into Equation 6a gives their multi-locus expression for sI with loose linkage,
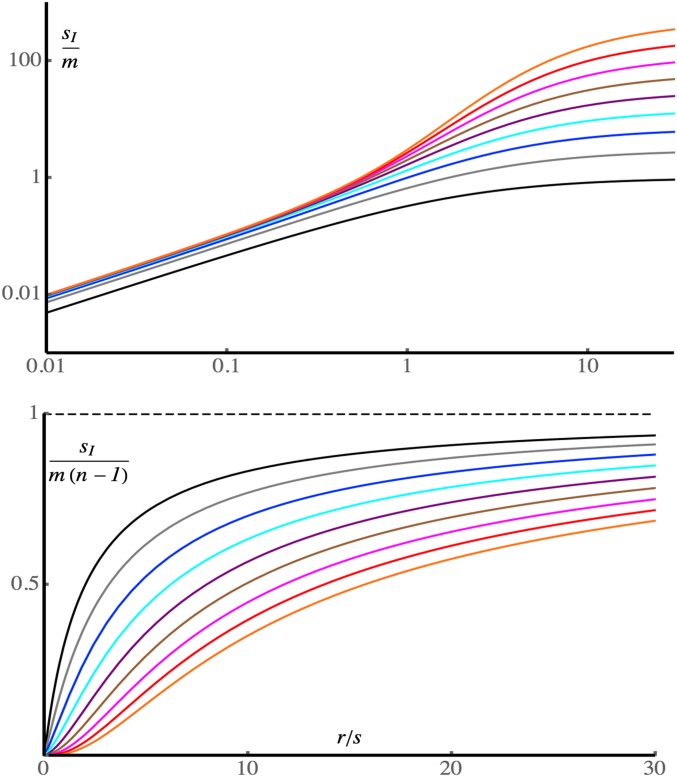
Figure 1 shows how the advantage of the inversion depends on the rate of recombination relative to selection, ρ = r/s. An exact treatment using Equations 3 of Barton (1983) was applied in order to generate the curves. When ρ < 0.5, we have sI < m and sI approaches at most mr/(s − r) as the number of loci increases. In this regime, all the loci are in strong LD, the migration load is close to m, and the inversion does not have much of an advantage. When ρ > 1, the migration load, and, hence, the advantage to the inversion, can be much larger (right-hand side of the upper panel in Figure 1), especially when many loci are involved. The advantage is now a substantial fraction of sI = m(n−1), the limiting value with loose linkage (lower panel of Figure 1).
Figure 1.
The selective advantage of an inversion, sI, in the limit of low migration, for n = 2, 4, 8, …, 256, 512 loci, plotted against r/s. The upper plot shows sI/m on a log-log scale, with the number of loci increasing from bottom to top. The lower plot shows sI /m(n−1), with the number of loci increasing from top to bottom; the dashed line indicates the selective advantage in the limit of loose linkage, m(n−1). The curves were calculated using the recursion in Equation 3 of Barton (1983).
Discussion
Here, we will briefly consider the implications of these results for the interpretation of the results of Lee et al. (2017) on the inversion polymorphism of B. stricta. Equations 4, 6c, and A8 imply that, when there is a high rate of self-fertilization in the population, this mechanism can provide only a weak selective advantage for an inversion. This follows from the fact that, with an inbreeding coefficient of f resulting from a departure from random mating within a population, the effective recombination rate that replaces r is (1 − f)r (Nordborg 1997; Charlesworth and Charlesworth 2010, p. 382). Song et al. (2006) obtained an estimate of f = 0.90 for B. stricta, using data on microsatellite genotypes. The effective recombination rate for this example is thus about one-tenth of the rate of recombination per meiosis, and the advantage of the inversion would be ∼0.1 mr/s if a moderate number of loci were covered by it.
Because extensive differentiation between the two subspecies at loci under selection requires m < < s, the selective advantage to an inversion is likely to be substantially <0.001 if the selected loci are <10 cM apart. This implies a chance of only ∼0.002 that a single new inversion would become established in the population as a result of selection (Haldane 1927), so that many independent mutational events generating an inversion in the chromosomal region in question would be needed before one succeeded in spreading to a high frequency. While Lee et al. (2017) provide convincing evidence that the inversion is associated with locally adaptive alleles, their interpretation in terms of the Kirkpatrick and Barton (2006) model thus has little or no advantage over a scenario in which an inversion that spread to an intermediate frequency by drift happened to pick up a selectively favorable mutation, and was then driven to a high frequency by hitchhiking.
Acknowledgments
We thank the Edinburgh Evolutionary Genetics Laboratory Group, especially Ben Jackson, for useful comments, and an anonymous reviewer, Reinhard Bürger, Mark Kirkpatrick, and Thomas Mitchell-Olds for their comments on the draft manuscript.
Appendix
Two-Locus Results: Approximation for Weak Migration Relative to Selection
The difference in the frequency of allele A2 at locus 2 between carriers of alleles A1 and a1 is equal to D/p1q1 (Charlesworth and Charlesworth 2010, p. 410), so that the effect on fitness of substituting A1 for a1 at locus 1 through its associated effect on locus 2 is equal to (s2 − e)D/p1q1; this term can be added to the direct fitness effect of the substitution s1. If q1 << 1, the net selective advantage of A1 over a1 is given by:
| (A1a) |
Similarly, the selective advantage of A2 over a2 is:
| (A1b) |
The change in qi due to selection is approximately equal to qi δwi; the change due to migration is approximately equal to m when m << si, as is assumed here. At equilibrium (denoted by asterisks), the equilibrium frequencies of the disfavored alleles are given by:
| (A2a) |
| (A2b) |
The recursion relations for each haplotype can be used to determine D*. If the source deme is fixed for haplotype 4, the haplotype recursion relations with weak selection are as follows:
| (A3a) |
| (A3b) |
| (A3c) |
| (A3d) |
where is the population mean fitness.
We have:
| (A4) |
Writing the haplotype frequencies as x1 = p1p2 + D, x2 = p1q2 − D, x3 = q1p2 − D, x4 = q1q2 + D, using Equations A1, and assuming that the population is close to equilibrium, the selection terms in Equation A4 give:
| (A5a) |
Provided that the fitness of A1A2 is greater than that of a1a2 (so that selection is purely directional), the multiplicand of D is negative.
The recombination term is ΔDr = −rD. This leaves the migration term to be evaluated, which is given by:
| (A5b) |
The net change in D near equilibrium is thus approximately:
| (A6) |
Equating this to zero gives Equation 1 of the main text.
Two-Locus Results: Exact Solution with Additive Selection
Here, we present the exact solution for the case when all processes are slow, with m, rD, s << 1, treating the population as evolving in continuous time, as in Bürger and Akerman (2011), whose treatment of the diploid model with semi-dominance is equivalent to this: their Eqs. 2.5 are equivalent to our A7 below. This analysis shows how the advantage of an inversion changes as migration increases, toward a critical value at which it overwhelms selection. In this case, the rates of change of allele frequencies and LD, and the selective advantage of the inversion, are given by:
| (A7) |
Provided that migration is not too high, both locally favored alleles can be maintained in the focal deme, giving the following equilibrium results:
| (A8) |
The critical migration rate, above which one or both alleles are swamped, is:
| (A9) |
The maximum advantage to the inversion is when migration is just below this critical value:
| (A10) |
With loose linkage, such that m, s << r, the advantage to the inversion can be written as:
| (A11) |
Footnotes
Communicating editor: G. Coop
Literature Cited
- Barton N. H., 1983. Multilocus clines. Evolution 37: 454–471. [DOI] [PubMed] [Google Scholar]
- Bürger R., Akerman A., 2011. The effects of linkage and gene flow on local selection in a continent-island model. Theor. Popul. Biol. 80: 272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1973. Selection of new inversions in multi-locus genetic systems. Genet. Res. 21: 167–183. [Google Scholar]
- Charlesworth B., Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts and Company, Greenwood Village, CO. [Google Scholar]
- Feldman M. W., 1972. Selection for linkage modification: 1. Random mating populations. Theor. Popul. Biol. 3: 324–346. [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. Part V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-R., Wang B., Mojica J. P., Mandakova T., Prasad K. V. S. K., et al. , 2017. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat. Ecol. Evol. 1: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P. A. P., 1962. The Statistical Processes of Evolutionary Theory. Oxford University Press, Oxford. [Google Scholar]
- Nordborg M., 1997. Structured coalescent processes on different time scales. Genetics 146: 1501–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B.-H., Clauss M. J., Pepper A., Mitchell-Olds T., 2006. Geographic patterns of microsatellite variation in Boechnera stricta, a close relative of Arabidopsis. Mol. Ecol. 15: 357–369. [DOI] [PubMed] [Google Scholar]