Barrington et al. examined the effect of four human diets (American, Mediterranean, Japanese, and Maasai/ketogenic) on metabolic health across four mouse...
Keywords: diet, mouse, metabolic syndrome
Abstract
The incidence of diet-induced metabolic disease has soared over the last half-century, despite national efforts to improve health through universal dietary recommendations. Studies comparing dietary patterns of populations with health outcomes have historically provided the basis for healthy diet recommendations. However, evidence that population-level diet responses are reliable indicators of responses across individuals is lacking. This study investigated how genetic differences influence health responses to several popular diets in mice, which are similar to humans in genetic composition and the propensity to develop metabolic disease, but enable precise genetic and environmental control. We designed four human-comparable mouse diets that are representative of those eaten by historical human populations. Across four genetically distinct inbred mouse strains, we compared the American diet’s impact on metabolic health to three alternative diets (Mediterranean, Japanese, and Maasai/ketogenic). Furthermore, we investigated metabolomic and epigenetic alterations associated with diet response. Health effects of the diets were highly dependent on genetic background, demonstrating that individualized diet strategies improve health outcomes in mice. If similar genetic-dependent diet responses exist in humans, then a personalized, or “precision dietetics,” approach to dietary recommendations may yield better health outcomes than the traditional one-size-fits-all approach.
OVER the last half-century, national dietary guidelines have failed to improve metabolic health in the United States, exemplified by the dramatic increase in metabolic syndrome (Cook et al. 2003). National dietary guidelines have largely been built upon epidemiological studies, which show that dietary patterns across populations are strongly correlated with the spectra of diseases (Knox 1977), and as dietary patterns change, so do disease spectra (Kagan et al. 1974; O’Dea 1992). However, a major limitation of population-level dietary studies is the absence of information on the relationship between individual and population-level responses.
There are clear examples in which genetically related subgroups within a population experience more severe health effects than the population as a whole, exemplified by Westernized indigenous populations that have disproportionately high incidences of type II diabetes (O’Dea 1992; Schulz et al. 2006). The importance of genetic background in diet response is supported by the strong similarity in weight gain within monozygotic twin pairs during long-term overfeeding (Bouchard et al. 1990). Clinical studies find wide variation across genetically diverse people in the health effects of diet, including weight gain and risk for heart disease (Liu et al. 1978; Dansinger et al. 2005; Hession et al. 2009). Some of the variation has been attributed to dietary adherence (Toubro and Astrup 1997). Yet, tightly controlled studies find extensive heterogeneity in physiological responses to identical diets (Levine et al. 1999; Zeevi et al. 2015), demonstrating that innate differences between people contribute to the heterogeneous effects of diets.
Mouse models provide a powerful resource for studying the interaction of genetics with diet. Similar to humans, genetically diverse mice vary in their susceptibility to diet-induced metabolic disease, but enable greater control of genetic and environmental factors. Studies in mice demonstrate that obesity has a strong genetic component and that identical diets affect weight gain differently across strains (West et al. 1992; Petro et al. 2004; Parks et al. 2013). While there are differences in metabolism and metabolic disease between mice and humans (Kennedy et al. 2010; Wong et al. 2016), a number of research groups have demonstrated the value of mouse studies in unraveling the genetic architecture underlying metabolic responses (Paigen 1995; Almind and Kahn 2004; Cheverud et al. 2004; Biddinger et al. 2005; Svenson et al. 2007; Hill-Baskin et al. 2009; Shockley et al. 2009; Ussar et al. 2015; Sinasac et al. 2016).
To explore the impact of the American diet on metabolic health across genetically diverse individuals, we designed a mouse version of the contemporary American diet and compared its metabolic health effects to that of a more typically fed control mouse diet across four inbred strains (A/J, C57BL/6J, FVB/NJ, and NOD/ShiLtJ, denoted as A, B6, FVB, and NOD, respectively). The clinical traits assayed were indicative of metabolic syndrome, a cluster of conditions that increase the risk of heart disease, stroke, and diabetes. We then compared each strain’s metabolic health when fed alternative human-relevant diets, including a Mediterranean diet, a Japanese diet, and a ketogenic diet analogous to that consumed by the Maasai, a tribal group in Kenya. In addition, we evaluated the liver metabolome and epigenetic changes underlying differential diet response. The diet (or diets) that was healthiest relative to the American diet was genetic-dependent, demonstrating that health outcomes in mice are improved through individualized dietary strategies, and raising the question of whether the development and implementation of personalized diet recommendations could also lead to better outcomes in people.
Materials and Methods
Animals and husbandry
Four-week-old A, B6, FVB, and NOD mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and acclimated for 2 weeks. Mice were randomized into diet groups. Two equally-sized and identical cohorts of five mice per diet, sex, and strain (480 in total) were studied in two locations, North Carolina (NC cohort) and Texas A&M University (TAMU cohort) over 6 months. The NC cohort was housed at the University of North Carolina during the first 4 months for analysis of body composition, metabolic rate, and physical activity. Mice were then transferred to North Carolina State University for the final 2 months for glucose tolerance testing (GTT), necropsy, and tissue collection. Mice were housed five per cage and maintained at 22° under a 12-hr light cycle; they were maintained and protocols followed in accordance with the University of North Carolina, North Carolina State University, and Texas A&M University Institution Animal Care and Use Committee guidelines. Mice were killed with carbon dioxide, and tissues were flash frozen in liquid nitrogen or fixed in formalin.
Diets
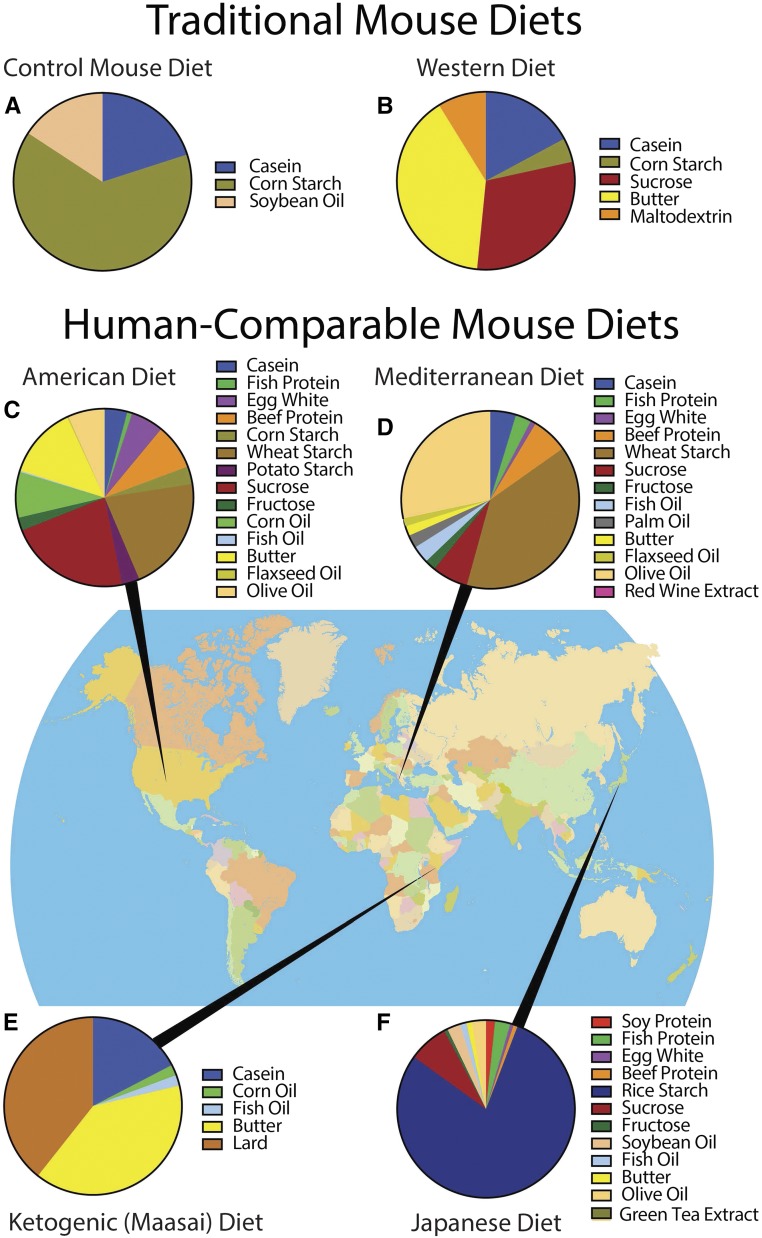
Powdered diets were designed in collaboration with Research Diets (New Brunswick, NJ). Traditional Mediterranean (D12052702) and Japanese (D12052703) diets were based on the Food and Agriculture Organization’s Food Balance Sheets from Greece and Japan in 1961 (Food and Agriculture Organization of the United Nations 2016). The American diet (D12052705) was based on the US Department of Agriculture’s 2008 Dietary Assessment of Major Food Trends (National Cancer Institute 2015). The Maasai/ketogenic diet (D12052706) was designed to allow mice to remain in ketosis, formulated with dairy sources as consumed by the Masaai (Mann et al. 1965), and with menhaden oil and corn oil added to ensure mice had essential lipids. Purified control mouse diet (D12052701; Research Diets) was used as a control diet for comparison to the American diet. Diets were designed to recapitulate human diets as closely as possible, matching macronutrient ratio, fiber content, types of ingredients, and fatty acid ratios to the human diets (Supplemental Material, Table S1, Table S2, Table S3, and Table S4 in File S1). Protein sources used included beef protein to match red meat intake, casein to match dairy intake, soy protein to match soy intake, egg white protein to match egg and white meat intake, and fish protein to match seafood intake. Cornstarch, wheat starch, rice starch, potato starch, sucrose, and fructose were matched accordingly to the types and amounts of starches and sugars in the diets. Soybean oil, corn oil, menhaden oil, sunflower oil, butter, lard, safflower oil, flaxseed oil, and olive oil were used to reconstruct lipid profiles.
Body composition and weight
Body composition (lean and fat mass) was assessed in both cohorts at 12 weeks (EchoMRI-130 Body Composition Analyzer). Body composition measurements were verified by comparing lean and fat mass measurements to scale weight.
Food consumption
Food consumption was measured in the TAMU cohort at 14 weeks by singly housing in wire bottom cages over a paper filter to collect spilled food. Starting, ending, and spilled-food weights were recorded. Two 24-hr acclimation periods were followed by two 24-hr testing periods. Each period was separated by 3 days. Diet-by-strain groups had n = 4–10 mice of both males and females, except female NOD mice fed the Maasai/ketogenic diet, which were omitted due to distress during the acclimation period.
Fasting glucose and GTT
Fasting glucose concentrations were measured following a 6-hr fast in both cohorts after 16 weeks on the diet. Glucose (2 g/kg) was administered by oral gavage. Blood glucose levels were measured with a Bionome GM100 glucose monitor (Bionome USA) at 0, 30, 60, 90, and 120 min. Area under the curve (from a baseline of 0) was calculated. All diet-strain groups had n = 9–20 mice. One American FVB male and one American B6 male did not show a change in blood glucose after gavaging and were omitted. Two NOD ketogenic males had glucose > 600 mg/dl (glucometer maximum) while fasting or at the first time point and were omitted.
Liver triglycerides
Following necropsy, liver triglyceride concentration was quantified in both cohorts as previously described (Folch et al. 1957). Briefly, 50 mg pieces of liver were homogenized in a 2:1 chloroform–methanol solution. After 30 min of incubation, a sodium chloride solution was added to the solution and vortexed. The lower phase was decanted and evaporated under nitrogen steam. Each sample was resuspended in a 0.5% Triton X-100/PBS solution. After sonication, samples were incubated at 55° for 5 min. Infinity Triglyceride reagent (Thermo Scientific) was added and samples were incubated for 5 min at 37°. Absorbance at 500 nm was measured and compared to a standard curve to quantify triglyceride concentration.
Metabolic rate and activity
Mice in the NC cohort were singly housed in Phenomaster Metabolic Chambers (TSE Systems) at 12 weeks. After an 8-hr acclimation period, data collection included two 12-hr night cycles and one 12-hr day cycle. Heat expenditure, oxygen consumption, and drinking volume were calculated per hour and normalized to lean mass, which was assessed prior to testing. Mice that failed to drink > 0.5 ml were omitted from water intake analysis (n < 2 per diet-strain group). Activity was determined by number of laser beam breaks in both vertical and horizontal axes and calculated per hour. Hyperactive mice (defined by activity > 100% strain mean) were omitted from activity analysis (n ≤ 1 per group). Respiratory exchange rate (RER) was calculated as an average per hour.
Liver histology
Formalin-fixed, paraffin-embedded right lobe liver samples were sectioned at 5 μm and stained with hematoxylin and eosin. The extent of steatosis was assessed in a blinded fashion by a board-certified veterinarian pathologist using a previously reported scoring system for nonalcoholic fatty liver disease (Liang et al. 2014). Briefly, the scoring system for macrovesicular steatosis, microvesicular steatosis, and cellular hypertrophy was based on the percentage of hepatocytes within the stained section. These parameters utilized the following categories: 0 (< 5% of hepatocytes), 1 (5–33%), 2 (34–66%), and 3 (> 66%). Inflammation was evaluated by counting the number of inflammatory foci per field, averaged across five fields of view at 100× magnification. The level of inflammation was assigned using the following categories: 0 (normal, < 0.5), 1 (slight, 0.5–1.0), 2 (moderate, 1.0–2.0), and 3 (severe, > 2).
Blood lipids and biochemistry
Fasted insulin was measured following a 6-hr fast at 18 weeks in both cohorts. Fasted blood samples were not collected from NOD mice in cohort 2, as they showed distress during fasting. Blood was collected via submandibular bleed, placed on ice for at least 30 min to allow clotting, then centrifuged at 10 × g for 5 min in 1.1 ml Z-Gel microtubes (Sarstedt). Insulin concentrations were quantified using a Mouse Serum Adipokine Immunoassay ELISA kit (Millipore, Bedford, MA) on a Bio-Plex 200 System (Bio-Rad, Hercules, CA).
Alanine aminotransferase (ALT) and cholesterol analysis was performed on serum samples taken at necropsy in both cohorts. Blood was collected via cardiac puncture, chilled for at least 30 min, then centrifuged at 10 × g for 5 min in 1.1 ml Z-Gel microtubes (Sarstedt). ALT activity was quantified in duplicate using a fluorometric ALT Activity Assay Kit per the manufacturer’s instructions (Sigma [Sigma Chemical], St. Louis, MO). Total, low-density lipoprotein (LDL), and high-density lipoprotein (HDL) cholesterol concentrations were measured in duplicate using a colorimetric Cholesterol Quantification Kit per the manufacturer’s instructions (Sigma).
Whole-genome bisulfite sequencing
DNA extraction and whole-genome bisulfite sequencing (WGBS) were performed on liver samples from 32 males spanning four diet-strain combinations: B6 control mouse diet, B6 American diet, A strain control mouse diet, and A strain American diet (n = 8 per group). Males were used because they had the most divergent diet responses in the B6 strain. Genomic DNA was isolated from liver using the DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA) and a modified protocol as follows: after tissue lysis and prior to spin column application, 50 μg of RNase A (Thermo Scientific) was added to each sample and samples were incubated at room temperature for 60 min. Samples were eluted in two cycles, in 100 and 60 μl of elution buffer.
WGBS single indexed libraries were generated using NEBNext Ultra DNA library Prep kit for Illumina (New England BioLabs, Beverly, MA), according to the manufacturer’s instructions with modifications. Next, 500 ng gDNA was quantified by Qubid dsDNA BR assay (Invitrogen, Carlsbad, CA) and 1% unmethylated λ DNA (Promega, Madison, WI) was spiked in to monitor bisulfite conversion efficiency. Samples were fragmented with a Covaris S2 or LE220 sonicator to an average insert size of 350 bp. Size selection was performed using AMPure XP beads and insert sizes of 300–400 bp were isolated (0.4× and 0.2× ratios). Samples were bisulfite converted after size selection using EZ DNA Methylation-Gold Kit (Zymo), following the manufacturer’s instructions. Amplification was performed after the bisulfite conversion using Kapa Hifi Uracil+ (Kapa Biosystems) polymerase using the following cycling conditions: 98° 45 sec/8 cycles, 98° 15 sec, 65° 30 sec, and 72° 30 sec/72° 1 min.
Final libraries were run on a 2100 Bioanalyzer (Agilent) High-Sensitivity DNA assay. Libraries were quantified by qPCR using the Library Quantification Kit for Illumina sequencing platforms (Kapa Biosystems), using the 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA). Libraries from 12 samples (three per group) were sequenced on Illumina a HiSeq2000 (100 bp), with the remainder sequenced on a HiSeq2500 (125 bp) paired-end single indexed run and 10% PhiX spike-in.
WGBS alignment and methylation analysis
Sequencing reads were aligned using the BSmooth (Hansen et al. 2012) bisulfite alignment pipeline (version 0.7.1) and Bowtie 2 version 2.1.0 (Langmead and Salzberg 2012). Samples from B6 and A strains were aligned to their respective genome builds, obtained from the Collaborative Cross page at the University of Northern Carolina Systems Genetics website (http://csbio.unc.edu/CCstatus/index.py?run=Pseudo), combined with the genome for λ phage. BSmooth was used to extract read-level measurements of methylation. To compare CG methylation across strains, the MODtools package (46), which functions similarly to the University of California, Santa Cruz Genome Browser’s liftOver, was used to convert A genomic coordinates to the B6 build. Following conversion to a common coordinate system, we smoothed the methylation data as previously described (Hansen et al. 2012).
Real-time PCR
Hepatic Avpr1a transcript abundance was analyzed in males in both cohorts, and in males from the 2-week follow-up study in which five A and B6 mice were fed ad libitum an American or control mouse diet for 2 weeks at TAMU. RNA was isolated from liver using a Maxwell 16 LEV simplyRNA kit (Promega). cDNA was generated using a Transcriptor First Strand cDNA Sythesis Kit (Roche). Primers were targeted to Avpr1a (5′-CATGTAGATCCACGGGTTGC-3′ and 5′-ACACCTTTCTTCATCGTCCAG-3′) and Rplp0 (5′-CGCTTGTACCCATTHATHATH-3′ and 5′-TTATAACCCTGAAGTGCTCGAC-3′) (Integrated DNA Technologies). Analysis was performed on a LightCycler 96 Thermocycler (Roche) using LightCycler 480 Sybr Green I Master reaction mix. All samples were run in duplicate and prepared on an EpMotion 5075 automated liquid-handling system. Cycling conditions were 95° for 5 min, followed by 35 cycles of 95° for 30 sec, 55° for 15 sec, and 72° for 60 sec. A high-resolution melting curve was produced by heating to 95° for 10 sec, cooling to 65° for 60 sec, and heating to 97° for 1 sec, followed by a cooling step of 37° for 30 sec. ∆CQ expression values were determined by normalizing to Rplp0 expression.
Isocaloric mouse experiments
Four-week-old C57BL/6J males were purchased from The Jackson Laboratory and acclimated for 2 weeks. Mice were isocalorically fed a control mouse or American diet for 90 days at TAMU. Ten males were used per group, as males had a more pronounced difference in adiposity between the control and American diets. Mice were housed five per cage and fed 11.5 kcal/mouse daily. Prior experiments optimized food quantity to ensure all food was eaten. Mice were weighed and body composition analyzed (EchoMRI-130 Body Composition Analyzer).
Metabolomics
Flash-frozen liver samples from the NC cohort were pulverized, and 0.025 g was added to 1.3 ml of prechilled methanol and incubated at −80° for 15 min. Samples were centrifuged for 5 min at 13.2 rpm at 4° and supernatant removed. Next, 200 ml of 80% methanol:20% water (solvent B) was added and incubated for 15 min at −80°. The samples were centrifuged for 5 min at 13.2 rpm at 4°, and the supernatants added to the same glass vial. Next, 200 ml of solvent B was added and incubated for 15 min at −80°. Samples were centrifuged for 5 min at 13.2 rpm at 4° and supernatants were added to the glass vial. The contents of the glass vial were dried using a nitrogen drying apparatus, then 160 μl of sterile water was added; 60 μl of 13C-labeled Escherichia coli was also added to the dried glass vial as an internal standard. Samples, kept at 4°, were placed in an autosampler tray. Next, 10μl from each sample was injected through a Synergi 2.5 μm Hydro-RP 100, 100 × 2.00 mm LC column (Phenomex) at 25°. The mass spectrometer was run in full-scan mode and negative-ionization mode, adapting a previous protocol (Clasquin et al. 2012). Samples were analyzed with a resolution of 140,000. A scan window of 85–800 m/z (mass-to-charge) was used from 0 to 9 min, and a window of 110–1000 m/z from 9 to 25 min. Solvent A consisted of 97:3 water:methanol, 10 mM tributylamine, and 15 mM acetic acid. Solvent B was methanol. The gradient from 0 to 5 min was 0% Solvent B, from 5 to 13 min was 20% Solvent B, from 13 to 15.5 min was 55% Solvent B, from 15.5 to 19 min was 95% Solvent B, and from 19 to 25 min was 0% Solvent B, with a flow rate of 200 µl/min. Raw files generated by Xcalibur were converted to mzML via msconvert (Chambers et al. 2012). MAVEN (Melamud et al. 2010; Clasquin et al. 2012) was used to correct total ion chromatograms based on retention time for each sample. Identified metabolites were manually chosen and peak abundance was integrated by mass (± 5 ppm) and retention time. Unidentified metabolites were chosen using an algorithm with the following settings: minimum peak width, 5; minimum signal/blank ratio, ≥ 3; minimum peak intensity, 10,000; and minimum peak/baseline, 3. Unidentified peaks were filtered manually to remove those that did not meet the above criteria.
Statistical analysis
Factors contributing to phenotypic variance:
To determine the relative contribution of each factor underlying phenotypic variance, multi-factor ANOVA was performed for each phenotype with the factors strain, diet, strain-by-diet interaction, sex, and cohort (where applicable). Log transformation was used for data that was not normally distributed (activity, ALT, GTT, insulin, LDL cholesterol, and liver triglycerides).
Comparison of metabolic effects within strains across diet:
To compare the effects of the American diet relative to the control diet, two-way or multi-factor ANOVA was performed independently for each strain using the factors diet and sex, and cohort where applicable. Cohen’s d effect sizes were calculated using the mean response on American diet minus the mean response on control mouse diet, divided by the pooled SD. The same methods were used to compare the effects of the Mediterranean, Japanese, and Maasai/ketogenic diets to the American diet, with P-values calculated using the American diet as a control. Dunnett’s Test correction was performed, which corrects for testing multiple comparisons to a control, given that we examined metabolic changes within a strain through diet modification. A P-value of 0.05 was used for the significance threshold.
Methylation analysis:
Methylation analysis was conducted in R via the bsseq package. The Avpr1a result was obtained by searching for differentially methylated regions genome-wide with a t-statistic cutoff of 4.6 and only considering CpGs, which had a coverage of ≥ 2 in all 31 samples. One sample from the B6 control mouse diet group was excluded from analysis because we observed that its source liver tissue had an abnormal tumor growth. This was confirmed by global hypomethylation, as previously described in human colon cancers (Reikvam et al. 2011).
Metabolomics analysis:
Metabolites missing ≥ 70% sample measurements were removed from analysis. Missing values in the remaining metabolites were imputed using k-nearest numbers. Data were assessed for normality using Q-Q plots, residuals, and the Shapiro–Wilks test after each step of the normalization process. Tissue weight and internal standard were treated as covariates. Tissue amount used in the extraction was weighed for each sample. Metabolites measured from the 13C E. coli internal standard were matched with their corresponding metabolite; otherwise, metabolites measured were matched with a 13C metabolite of the same class type. Class types were identified using the Human Metabolome Database (Wishart et al. 2013). Once metabolites were adjusted for tissue weight and internal standard, each metabolite was pareto-scaled across all mice using the package “MetabolAnalyze” (Nyamundanda et al. 2010). Each mouse was median normalized across all metabolites and metabolites transformed using cube root. The model assessed in each strain was:
Statistical analyses of metabolites were performed using R version 3.1.0 and 3.2.2. The α for all statistical tests was determined to be 0.05. P-values associated with metabolites were adjusted using the Benjamini–Hochberg correction factor.
Calculation of health category scores and metabolic health index score:
Health category scores were calculated by multiplying the effect size (Cohen’s d) of each alternative diet compared to the American diet for each phenotype in each strain by its confidence level, thereby weighting for both effect size and significance. Phenotypes within each category were designated as positive or negative (below) depending on their association with beneficial or detrimental health effects, respectively, as shown in the model below. To allow for comparison across categories, the category scores were standardized between −1 and +1 across all strain-diet groups within each category.
To calculate the metabolic health index score, a measure of the cumulative metabolic health effects of a given diet relative to the American diet, we calculated the mean of the health category scores, either with or without inclusion of the body composition score.
Data availability
File S1 contains supplemental data to support the conclusions in this article.
Results
Design of human-comparable mouse diets
We formulated diets based on historical dietary patterns to examine the metabolic effects of diets that span the spectrum of human dietary patterns and are associated with negative (American) or positive (Mediterranean, Japanese, and Maasai/ketogenic) epidemiological health outcomes in people. Many studies have compared effects of a control mouse diet (Figure 1a) to a high-fat or Western diet (Figure 1b). Many different high-fat diets have been formulated, each of which contains varying concentrations of fat and carbohydrate, and from different sources than shown in the figure. It is common to find one or two representative ingredients for each individual nutrient (i.e., casein provides protein, corn starch provides carbohydrate, and soybean oil provides fat). To better recapitulate the diversity of ingredients in human diets, we designed diets that match not only macronutrient ratios (i.e., proportions of protein, carbohydrate, and fat on a kcal basis), but also ingredients, including bioactive compounds (e.g., red wine and green tea extracts) (Figure 1, d and f), lipid profiles, and fiber content (Table S1, Table S2, Table S3, and Table S4 in File S1). The American diet is representative of contemporary dietary patterns in the United States (Figure 1c) (National Cancer Institute 2015). The Mediterranean and Japanese diets are representative of traditional eating patterns during the early 1960s, when these populations had among the longest life expectancies and lowest rates of chronic disease (Gordon 1957; Marmot et al. 1975; Trichopoulou and Vasilopoulou 2000; Knoops et al. 2004) (Figure 1, d and f). The ketogenic diet is analogous to that of the Maasai tribe, who do not develop heart disease despite eating high levels of fat and cholesterol (Mann et al. 1965) (Figure 1e). The ketogenic diet induces ketosis, a physiological state in which the body shifts metabolism to utilize ketones and preserve glucose.
Figure 1.
Diet ingredient profiles and geographic origins. (a) The purified control mouse diet, used for comparison to the American diet in our study, is typical of those used in mouse research. (b) Previous studies have evaluated metabolic effects using Western or high-fat diets. Instead, we designed human-comparable diets representative of the dietary patterns in human populations including: (c) a contemporary American diet, (d) a traditional Mediterranean diet, (e) a ketogenic diet analogous to the Maasai diet, and (f) a traditional Japanese diet.
Sources of phenotypic variation
Four human-comparable diets (American, Mediterranean, Japanese, and ketogenic) and the control mouse diet were fed ad libitum to 10 male and 10 female mice from four inbred strains (A, B6, FVB, and NOD). These strains were chosen due to their genetic and phenotypic diversity, with the aim of surveying mice with varying behavioral and metabolic profiles (Kirby et al. 2010). The B6 strain is most commonly used in studies and is susceptible to diet-induced obesity on a high-fat, high-sugar diet (West et al. 1992). FVB is more resistant to diet-induced obesity and is highly active, whereas the A strain is resistant to diet-induced obesity despite low levels of activity (Black et al. 1998; Parks et al. 2013). NOD is metabolically unique from the other strains in its predisposition to develop diabetes (Leiter et al. 1987).
Two equally-sized cohorts of mice began diets at 6 weeks and were aged on diets for 12 weeks to allow the manifestation of physiological effects, before the analysis of metabolic traits over an additional 12 weeks (Figure S1 in File S1). Phenotypes were compared using ANOVA models with diet, strain, the interaction of diet with strain, sex, and cohort as factors. Genetic variation accounted for a considerable proportion of the total phenotypic variation for most phenotypes (Table S5 in File S1). Sex also contributed significantly to many phenotypes. For example, sex effects accounted for about half of the variation in body weight and lean weight, 33–42% of the variation in metabolic rate, and 22% of the variation in water intake. Diet effects contributed to the total phenotypic variation of most traits, including 84% of the variation in RER, and 20–36% of the variation in plasma triglycerides, cholesterol, and metabolic rate. Cohort effects explained a relatively minor proportion of the total phenotypic variance, although some differences in body composition were observed (Table S5 in File S1).
Importantly, the interaction of genetic background with diet influenced most metabolic traits, and in some cases accounted for more variation than genetic or dietary effects alone (Table S5 in File S1). For instance, the genetic interaction with diet accounted for 13% of variation in food intake, whereas the genetic and diet effects each accounted for 11%. Genetic-by-diet interactions were important for water intake and metabolic rate, accounting for 12–15% of the phenotypic variance. While blood lipids were strongly influenced by diet, genetic-by-diet interaction effects contributed considerably, accounting for 10–12% of the variation in plasma triglycerides, and HDL, LDL, and total cholesterol. The genetic interaction with diet, where significant, complicates the interpretation of the main effects of genetics and diet, as the effect of genotype can vary across each diet. Together, the results indicate that both genetic and dietary factors contribute in varying degrees to most metabolic traits, and that the genetic interaction with diet plays a key role in influencing metabolic traits.
Influence of the American diet on activity, metabolic rate, and food intake
To model the potential impact of a precision nutrition approach, we sought to use the American diet as a baseline for comparison to other human diets. However, because the vast majority of mouse studies in any field use a chow diet, we first put the novel American diet into the perspective of the control mouse diet (Comparison A), before comparing the American diet to alternative human-relevant diets (Comparison B) (Figure S1 in File S1).
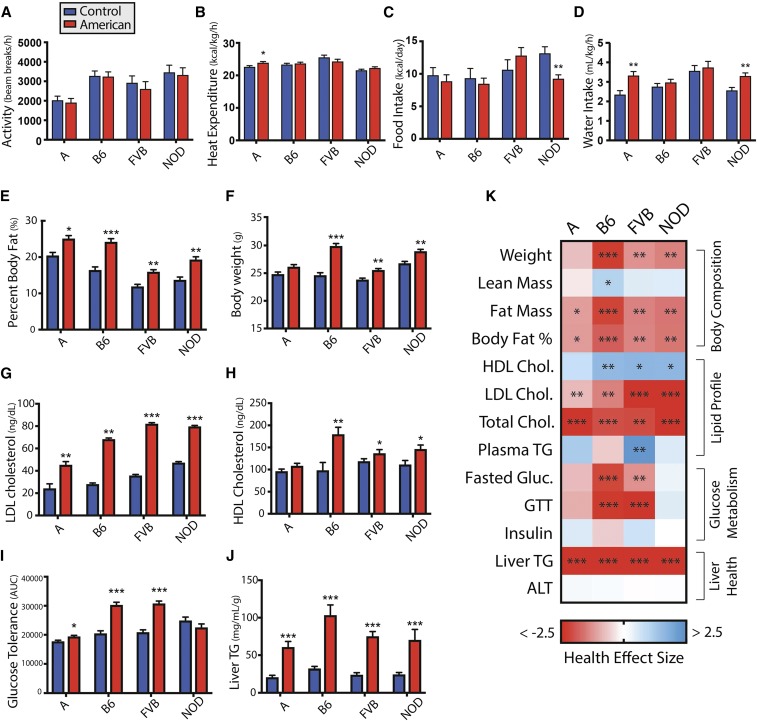
We examined basic physiological and behavioral parameters and quantified the changes in Cohen’s d effect size. Physical activity was not affected by diet (Figure 2a). Nonetheless, the American diet increased metabolic rate in the A strain, indicated by increase oxygen consumption (d = 0.98) and heat expenditure (d = 0.96) (Figure 2b and Figure S2a in File S1). Food intake decreased in the NOD strain (d = −2.20), but was not significantly altered in other strains (Figure 2c). Water intake increased significantly in A mice (d = 1.47) and NOD mice (d = 1.37) (Figure 2d). RER was reduced in B6 mice fed the American diet (d = −1.47), indicative of greater fat oxidation (Figure S2b in File S1).
Figure 2.
Comparison of metabolic phenotypes in each strain for mice fed the American diet relative to the control mouse diet. Effect of American diet relative to the control mouse diet in each strain for (a) activity (n = 9–10), (b) heat expenditure (n = 9–10), (c) food intake (n = 4–10), (d) water intake (n = 7–10), (e) body weight (n = 19–20), (f) percent body fat (n = 19–20), (g) HDL cholesterol (n = 4–10), (h) LDL cholesterol (n = 4–10), (i) GTT [n = 12–20, except NOD American (n = 4)], and (j) liver triglyceride concentration (n = 13–20). (k) Heatmap of health effect size (Cohen’s d, with higher value indicating improved health and lower value indicating diminished health) for metabolic phenotypes across strains. Data are mean ± SE. * P < 0.05, ** P < 0.01, and *** P < 0.001 by ANOVA between means. A, A/J strain mice; ALT, alanine aminotransferase; AUC, area under the curve; B6, C57BL/6J strain mice; FVB, FVB/NJ strain mice; GTT, glucose tolerance test; HDL high-density lipoprotein; LDL, low-density lipoprotein; NOD, NOD/ShiLtJ strain mice; TG, triglyceride.
The American diet induces varying degrees of fat gain across strains
We compared the body composition of mice fed the American diet relative to those fed the control mouse diet at 12 weeks. The American diet increased body fat to varying degrees across strains, with the largest effect in B6 mice (d = 2.18) compared to NOD (d = 1.57), FVB (d = 1.42), and A mice (d = 1.14) (Figure 2e and Figure S2c in File S1). This result is consistent with previous research feeding high-fat diets (West et al. 1992; Cheverud et al. 1999; Parks et al. 2013). Elevated body weight coincided with increased fat mass (Figure 2, f and k). Interestingly, food intake was poorly correlated with fat gain (Figure 2, c and e), but this observation is consistent with previous studies (Backhed et al. 2004; Petro et al. 2004; Hatori et al. 2012; Parks et al. 2013). To validate this finding, an independent cohort of B6 mice was isocalorically fed an American or control mouse diet for 14 weeks. Mice fed an American diet weighed 14% more with no change in lean mass, but a 74% increase in body fat (Figure S3 in File S1), emphasizing the importance of factors other than food intake in the accumulation of body fat.
The American diet negatively impacts blood lipid profiles
Across strains, mice fed the American diet had increased LDL cholesterol with variation in effect size (d = 0.82–3.09) (Figure 2g). Concomitantly, HDL cholesterol increased in B6, FVB, and NOD strains (d = 1.58–1.64) (Figure 2h). Total cholesterol was increased across strains (d = 2.01–3.31) (Figure S2e in File S1). Plasma triglyceride concentrations decreased in FVB mice (d = −2.33), but did not change in other strains (Figure S2f in File S1).
The impact of the American diet on glucose homeostasis differs by strain
The American diet’s effect on glucose homeostasis was evaluated by GTT, and measurement of fasting glucose and insulin. B6 mice, commonly susceptible to glucose intolerance on high-fat, high-sugar diets (Surwit et al. 1988), showed glucose intolerance when fed the American diet (d = 2.35), as demonstrated by increased area under the curve measurements (Figure 2i). Similar responses were observed in FVB (d = 2.69). Consistent with previous research, diet had minimal impact on glucose tolerance in the A strain (Surwit et al. 1988). Glucose tolerance was not significantly altered in the NOD strain; however, this may have been due to high interindividual variability in this strain, which is genetically predisposed to diabetes (Hattori et al. 1986).
Fasting glucose was increased by the American diet in B6 (d = 2.15), FVB (d = 1.30), and A (d = 0.80) strains (Figure S2g in File S1). We did not observe significant alterations to insulin levels (Figure S2h in File S1). Previous research has shown increased insulin levels in B6 mice fed a high-fat diet, albeit with high variability, requiring larger numbers of mice to detect a significant effect (Burcelin et al. 2002).
The American diet increases liver triglyceride concentrations
To determine how liver health is impacted by the American diet, we performed histological examinations, and measured liver triglycerides and serum ALT, a marker of liver damage. Liver triglyceride concentrations were consistently elevated across strains (d = 2.86–4.05) (Figure 2j). ALT concentrations did not significantly differ from mice fed the control diet (Figure S2i in File S1). Histological examination revealed increased macro- and microvesicular steatosis in the A, B6, and FVB strains, while only macrovesicular steatosis increased in NOD mice. NOD mice also exhibited increased hepatic inflammation (Table S6 in File S1).
Arginine vasopressin receptor 1A (Avrp1a) methylation status is associated with diet response in strains with divergent health effects
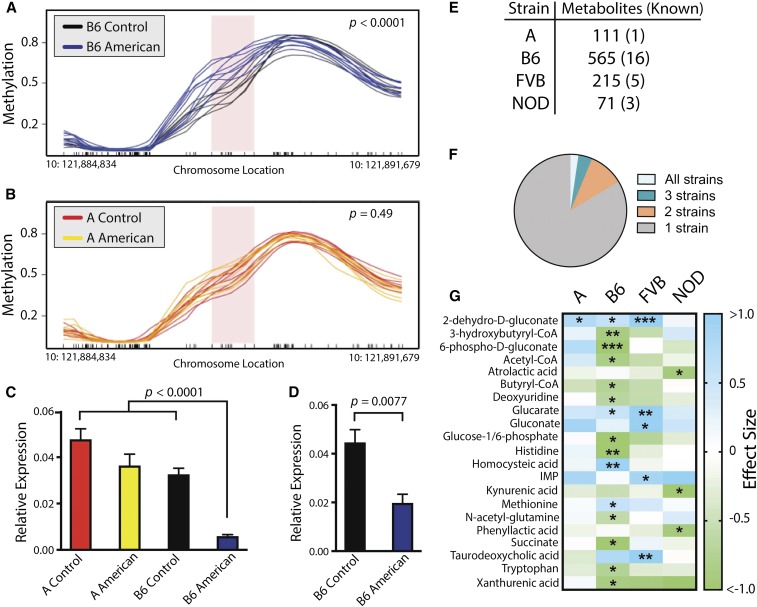
A mechanism by which individuals respond to or are protected from diet-associated changes in metabolic health phenotypes is through modification of the epigenome (Stover 2011; Janke et al. 2015; Barrett et al. 2016). To examine epigenetic modifications, we performed WGBS on livers from A and B6 mice fed control or American diets. These strains were chosen due to their divergent responses to the American diet, with B6 displaying greater increases in body weight, adiposity, and impaired glucose metabolism, while A was comparatively resistant to these phenotypic effects. Changes in methylation patterns at Avpr1a showed a significant contrast between A and B6 mice. B6 mice fed the American diet were hypermethylated at Avpr1a compared to their control diet counterparts (Figure 3a); in contrast, no significant difference was observed in A mice on the two diets, and A strain methylation was roughly equivalent to that of B6 mice fed the control diet (Figure 3b). Given the observed methylation differences and Avpr1a’s known association with metabolic disease in mice and humans (Aoyagi et al. 2007; Enhorning et al. 2009), we examined Avpr1a hepatic transcript abundance, which was greatly reduced in B6 mice fed the American diet (−84%) compared to the other three strain-diet groups (Figure 3c). In a follow-up study, we identified that repression of Avpr1a transcription occurs rapidly, as Avpr1a transcript abundance was reduced by 54% after 2 weeks in B6 mice fed the American diet (Figure 3d). Genetic variation within this region cannot account for methylation and expression differences, as Avpr1a sequences do not vary between the strains (Blake et al. 2017).
Figure 3.
Genetic-by-diet interactions in the methylation status of the Avpr1a locus and liver metabolome alterations. (a) B6 mice fed the American diet are hypermethylated relative to those fed the control mouse diet at the Avpr1a locus (n = 8), P < 0.0001 by Student’s t-test. (b) American diet feeding did not alter methylation status in the A strain (n = 7–8), P = 0.49 by Student’s t-test. (c) Transcript expression of Avpr1a was reduced by 84% in B6 mice fed the American diet relative to other strain-diet groups in mice fed diets for 6 months (n = 4–5), P < 0.0001 by ANOVA. (d) Transcript expression of Avpr1a is reduced by 54% in B6 mice fed the American diet for 2 weeks (n = 4), P = 0.0077 by Student’s t-test. (e) Number of liver metabolites, including both known and unknown, significantly altered by the American diet relative to the control mouse diet. (f) Proportion of metabolites significantly changed in all four strains (2%), three strains (4%), two strains (10%), or unique to one strain (84%). (g) Heatmap of effect sizes (Cohen’s d) for known metabolites significantly altered by the American diet relative to the control mouse diet across strains. * P < 0.05, ** P < 0.01, and *** P < 0.001 by ANOVA between means with Benjamin–Hochberg correction factor. A, A/J strain mice; B6, C57BL/6J strain mice; FVB, FVB/NJ strain mice; NOD, NOD/ShiLtJ strain mice.
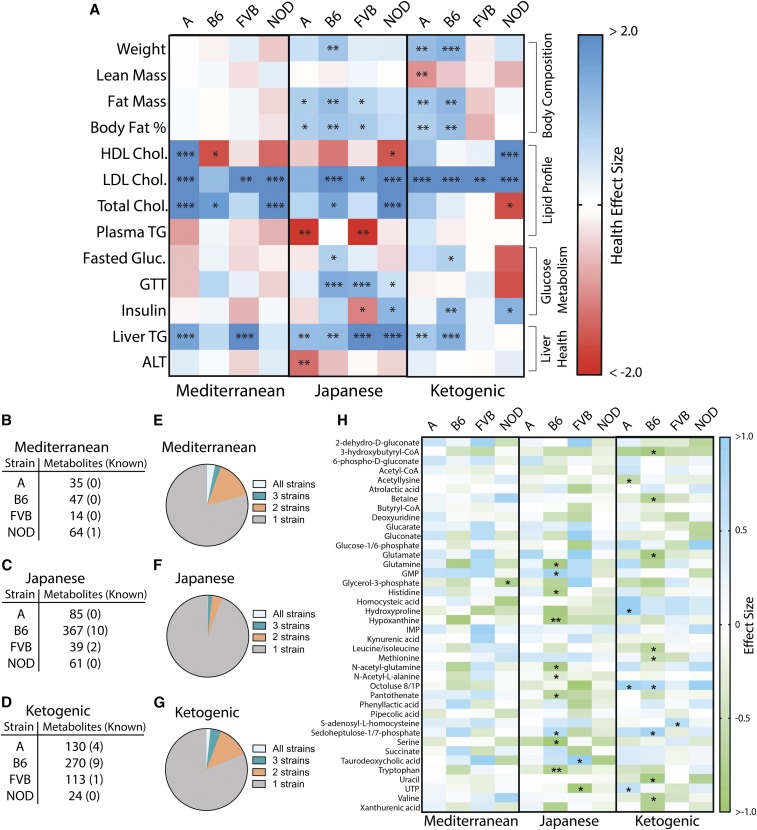
Liver metabolite changes vary by strain
Varying the composition of the diet causes corresponding metabolic shifts at the cellular level. Metabolite levels have been associated with increased risk of metabolic diseases like type 2 diabetes (C. F. Huang et al. 2013; T. Huang et al. 2013; Menni et al. 2013; Wang et al. 2013). Metabolomic profiling was used to determine the extent to which genetic variation impacted the tissue-level metabolic response to the American diet vs. the control diet. Profiling was performed in liver because of its primary role in nutrient allocation. Hepatic metabolomes of B6 mice were more affected by the American diet than other strains based on the number of metabolites that differed significantly in abundance between the two diet groups. A total of 16 known metabolites were affected by the American diet in B6 strain, compared to five (FVB), three (NOD) and one (A) in the other strains (Figure 3, e–g). Comparable diet-dependent differences across strains were also reflected in the numbers of unknown metabolites (i.e., spectral features that did not map to known compounds in our database) (Figure 3e). Diet-induced changes in metabolite levels were strain-dependent, with 84% of metabolites significantly altered by the American diet in only a single strain (Figure 3f).
A precision dietetics approach reveals strain-specific effects of diet on physiology and body composition
Dietary modification is a common initial intervention for patients with metabolic syndrome. While a number of studies have evaluated how obesigenic or atherogenic diets impact mice, studies on human-comparable diets are lacking. To test a precision dietetics approach by evaluating diet responses in the context of genetic background, we compared how physiology and health status differed in A, B6, FVB, and NOD mice fed Mediterranean, Japanese, and ketogenic diets relative to those fed an American diet.
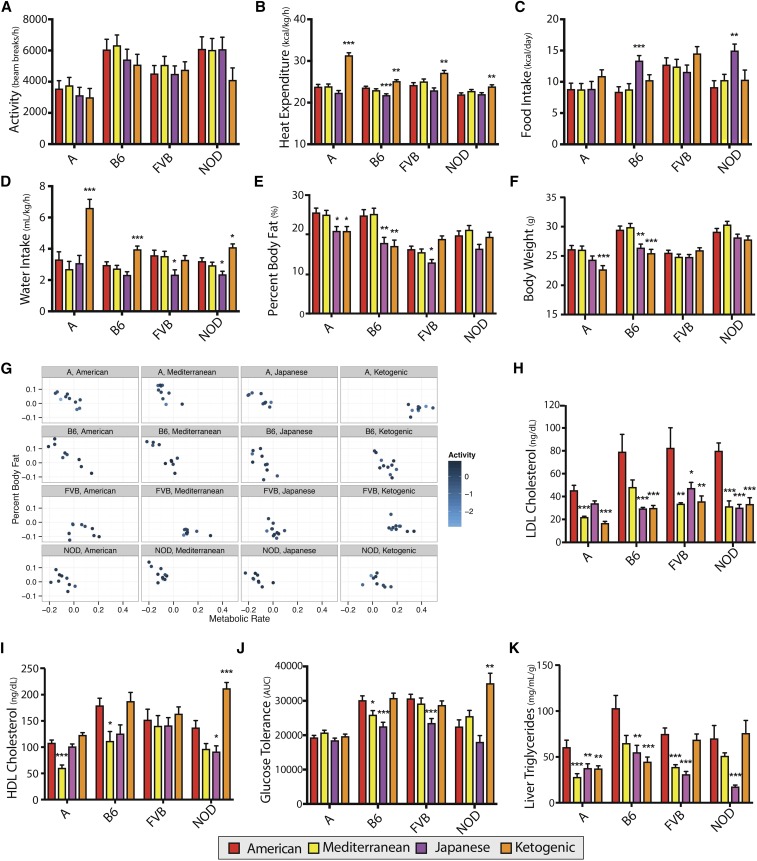
Physical activity was highly variable within strain-by-diet groups and was not significantly altered by diet (Figure 4a). Nonetheless, metabolic rate increased in all strains fed a ketogenic diet and the magnitude of increase was genetic-dependent (Figure 4b and Figure S4a in File S1). Food intake increased in B6 (d = 1.97) and NOD (d = 2.03) mice fed a Japanese diet (Figure 4c). All strains except FVB drank more water on the ketogenic diet (d = 1.42–2.13). RER decreased across strains fed the ketogenic diet and increased in all fed the Japanese diet (Figure S4b in File S1).
Figure 4.
Comparison of metabolic phenotypes in each strain for mice fed Mediterranean, Japanese, or ketogenic diets relative to the American diet. Effects of Mediterranean, Japanese, and ketogenic diets relative to the American diet in each strain are shown for (a) activity (n = 8–10), (b) heat expenditure (n = 8–10), (c) food intake (n = 4–10), (d) water intake (n = 7–10), (e) body weight (n = 17–20), and (f) percent body fat (n = 17–20). The influence of activity and metabolic rate on percent body fat varies by strain and diet, as shown for mice in which metabolic rate and activity was measured (g). Effects of Mediterranean, Japanese, and ketogenic diets relative to the American diet in each strain are shown for (h) HDL cholesterol (n = 4–10), (i) LDL cholesterol (n = 4–10), (j) glucose tolerance test (n = 9–20), and (k) liver triglyceride concentration (n = 11–20). Data are mean ± SE. * P < 0.05, ** P < 0.01, and *** P < 0.001 by ANOVA between means, with Dunnett’s correction to the American diet within each strain. A, A/J strain mice; B6, C57BL/6J strain mice; FVB, FVB/NJ strain mice; HDL high-density lipoprotein; LDL, low-density lipoprotein; NOD, NOD/ShiLtJ strain mice.
Body composition and weight did not significantly differ for any strain fed the Mediterranean diet relative to the American diet (Figure 4, e and f). Japanese and ketogenic diets yielded similar reductions in body fat in A and B6 mice (d = −0.93 to −1.17) (Figure 5a and Figure S4c in File S1), whereas FVB mice fed the Japanese diet had lower percent body fat (d = −0.94), but not those fed a ketogenic diet (Figure 4e). A reduction in lean weight was unique to A mice fed a ketogenic diet (d = −1.02) (Figure S4d in File S1). Across all diets, caloric intake was poorly correlated with body fat (Figure S5 in File S1).
Figure 5.
Comparison of metabolic phenotypes and liver metabolites in mice fed Mediterranean, Japanese, or ketogenic diets relative to the American diet. (a) Heatmap of health effect size (Cohen’s d, with higher value indicating improved health and lower value indicating diminished health) for metabolic phenotypes across strains. * P < 0.05, ** P < 0.01, and *** P < 0.001 by ANOVA between means, with Dunnett’s correction to the American diet within each strain. Number of known and unknown liver metabolites significantly altered compared to the American diet for (b) Mediterranean diet, (c) Japanese diet, and (d) ketogenic diet. Proportion of metabolites significantly changed in all four strains, three strains, two strains, or unique to one strain for (e) Mediterranean, (f) Japanese, and (g) ketogenic diets. (h) Heatmap of effect sizes (Cohen’s d) of known metabolites significantly altered relative to the American diet across strains, including metabolites that differed between American and control mouse diets (Figure 3g). * P < 0.05, ** P < 0.01, and *** P < 0.001 by ANOVA between means with Benjamin–Hochberg correction factor. A, A/J strain mice; ALT, alanine aminotransferase; B6, C57BL/6J strain mice; FVB, FVB/NJ strain mice; GTT, glucose tolerance test; HDL high-density lipoprotein; LDL, low-density lipoprotein; NOD, NOD/ShiLtJ strain mice; TG, triglyceride.
To better understand the relationship between percent body fat, metabolic rate, and activity, we plotted the residuals for each phenotype after accounting for sex differences in a subset of mice housed in metabolic cages for 3 days (Figure 4g). Metabolic rate and percent body fat varied by strain and diet, while activity level did not strongly contribute to shifts in metabolic rate or body fat. The A strain, which is considered to be resistant to the effects of diet (Black et al. 1998; Parks et al. 2013), had the greatest physiological shift of all strains on the ketogenic diet. Their metabolic rate and percent body fat were similar when fed American, Mediterranean or Japanese diets, but their metabolic rate greatly increased and body fat decreased when fed the ketogenic diet.
Variation persisted in some diet-by-strain groups even while accounting for sex differences. This is most evident in the percent body fat of B6 mice. Heterogeneity of diet response has been previously observed in B6 mice (Burcelin et al. 2002; Koza et al. 2006), and indicates that while genetic information can improve prediction of diet responses, other factors are also influential.
Effects of Mediterranean, Japanese, and ketogenic diets on blood lipid profiles
The response of blood lipids to diet is one of the most researched topics in biomedical sciences, with thousands of studies having been performed with varying results (Shekelle et al. 1981; Kris-Etherton et al. 1988; Mente et al. 2009). Utilizing inbred strains of mice clearly demonstrates that dietary effects are dependent on the underlying genetic architecture (West et al. 1992; Cheverud et al. 2004; Parks et al. 2013). In our study, the Mediterranean diet markedly reduced LDL cholesterol for A (d = −3.29), FVB (d = −2.56), and NOD mice (d = −2.97) (Figure 4h), and also decreased HDL cholesterol in A (d = −3.48) and B6 (d = −1.59) mice (Figure 4i). The Japanese diet decreased LDL cholesterol in NOD (d = −3.04), B6 (d = −2.21), and FVB (d = −1.69) strains (Figure 4h), and decreased HDL cholesterol only in NOD mice (d = −1.54) (Figure 4i). Plasma triglycerides were elevated in FVB (d = 2.20) and A (d = 1.87) mice fed the Japanese diet (Figure S3f in File S1).
The impact of low-carbohydrate diets on blood lipids is controversial. Studies have reported positive or negative effects depending on the age of the participants, obesity status, and duration of the diet (Sharman et al. 2002; Kwiterovich et al. 2003; Dashti et al. 2006). Our study found beneficial impacts of the ketogenic diet on cholesterol profiles, with decreased LDL cholesterol across strains (d = −2.21 to −4.72) and increased HDL cholesterol in NOD mice (d = 2.49) (Figure 4, h and i).
Influence of Mediterranean, Japanese, and ketogenic diets on glucose metabolism
Impaired glucose homeostasis is common in patients with metabolic syndrome. Glucose tolerance improved in B6 (d = −1.47) and FVB (d = −1.43) strains fed the Japanese diet (Figure 4j). In addition, fasted glucose concentration decreased in B6 mice (d = −0.88) (Figure S4g in File S1). Fasted insulin concentrations decreased in NOD (d = −1.30) mice but increased in FVB mice fed the Mediterranean diet (d = 1.14) (Figure 5a and Figure S4h in File S1). The ketogenic diet did not improve glucose tolerance in any strain (Figure 4j), but fasted glucose (d = −0.89) and insulin (d = −1.23) were reduced in B6 mice (Figure 5a, Figure S4g, and Figure S4h in File S1). In contrast, fasted glucose increased in NOD mice fed a ketogenic diet (d = 1.46), while fasted insulin was reduced (d = −1.39).
The effect of diet on liver health differs by strain
In addition to the effects of diet on the clinical plasma biomarkers of disease, we now appreciate the effects of lipid deposition in the liver on metabolic disease (Kotronen and Yki-Jarvinen 2008; Cohen et al. 2011).
We found that genome-by-diet interactions influenced liver phenotypes in mice fed the Mediterranean, Japanese, and ketogenic diets. The Mediterranean diet reduced liver triglyceride concentrations in the A (d = −1.20) and FVB (d = −2.25) strains (Figure 4k). The ketogenic diet reduced liver triglycerides in A (d = −1.02) and B6 (d = −1.43) mice. The Japanese diet lowered liver triglyceride concentrations across all strains (d = −1.21 to −2.75). Histological examination of A mice fed a ketogenic diet revealed that, while microvesicular steatosis was reduced (d = −1.57, P = 0.0008), macrovesicular steatosis was not (P = 0.97) (Table S7 in File S1). Importantly, serum ALT, a marker of liver damage, increased in A mice fed a Japanese diet (d = 1.31) (Figure S4i in File S1). While each diet benefitted some strains, no single diet was universally beneficial for liver health across strains (Figure 5a).
Diet alters the liver metabolome in a strain-specific manner
We compared the liver metabolomes of mice fed Mediterranean, Japanese, and ketogenic diets relative to those fed an American diet. The Mediterranean diet had little effect on the tissue metabolome across strains (Figure 5b), with glycerol-3-phosphate in NOD mice being the only known metabolite that was altered (d = −0.78) (Figure 5h). B6 mice were the most sensitive to dietary changes on the Japanese and ketogenic diets (Figure 5, b–d). Similar to the comparison between American and control mouse diets (Figure 3f), diet-induced changes were highly strain-dependent, with 79% of metabolite changes being unique to one strain for the Mediterranean diet, 81% for the ketogenic diet, and 94% for the Japanese diet (Figure 5, e–g).
Categorical health effects of Mediterranean, Japanese, and ketogenic diets
To provide a more comprehensive view of health effects, we analyzed four major categories of metabolic health (body composition, lipid profile, glucose metabolism, and liver health) by calculating health scores (HSs), by combining phenotypes within each category across all strain-diet groups and standardizing the effects on a scale from −1 to +1, with negative numbers indicating worsened health and positive numbers indicating improved health relative to mice fed the American diet.
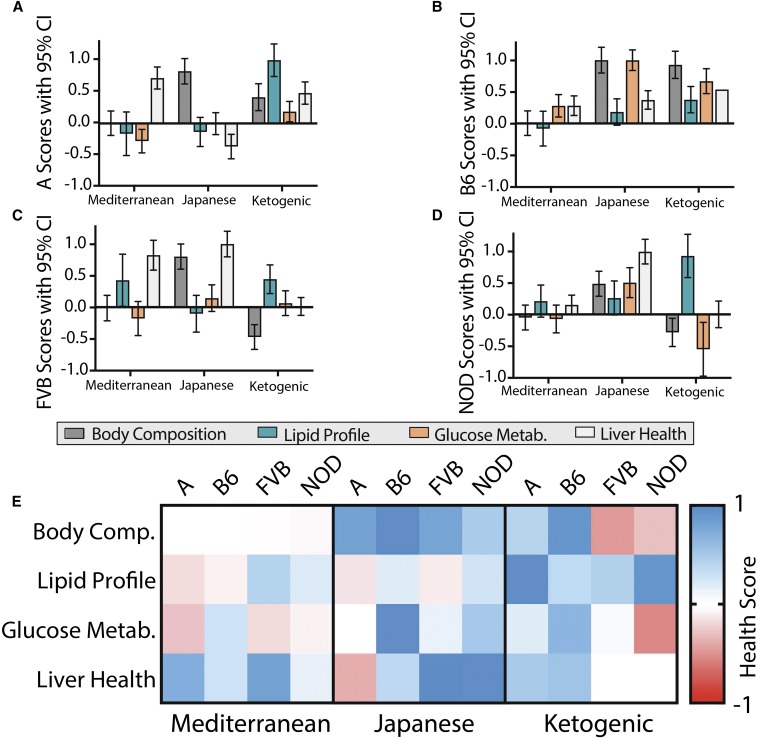
The Mediterranean diet did not improve body composition, but was beneficial for liver health in A (HS = 0.71) and FVB (HS = 0.82) mice (Figure 6). Despite high variability, the FVB Mediterranean group had an improved lipid profile (HS = 0.44). The Mediterranean diet improved glucose metabolism in B6 mice (HS = 0.27) but was detrimental for A mice (HS = −0.28).
Figure 6.
Scores of four health categories for alternative diets relative to the American diet in each strain. Health scores indicate cumulative health effects of alternative diets relative to the American diet for four categories: body composition (lean mass, fat mass, and percent body fat), lipid profile (HDL, LDL, and plasma triglycerides), glucose metabolism (fasted glucose, fasted insulin, and GTT), and liver health (liver triglycerides, ALT). A positive score represents improved health and a negative score represents diminished health. Scores and 95% C.I.s are shown for (a) A, (b) B6, (c) FVB, and (d) NOD . (e) The data are also represented in a heat map for comparison, with red showing improved health scores relative to the American diet. A, A/J strain mice; ALT, alanine aminotransferase; B6, C57BL/6J strain mice; FVB, FVB/NJ strain mice; GTT, glucose tolerance test; HDL high-density lipoprotein; LDL, low-density lipoprotein; NOD, NOD/ShiLtJ strain mice.
The Japanese diet improved body composition across strains (HS = 0.48–1.00). It did not consistently improve lipid profiles, but did provide the greatest improvement in glucose metabolism of all diets for B6 (HS = 1.00) and NOD mice (HS = 0.86). It also maximally improved liver health in the FVB (HS = 1.00) and NOD (HS = 1.00) strains, but had detrimental effects in the A strain (HS = −0.36).
The ketogenic diet improved body composition for B6 (HS = 0.93) and A (HS = 0.42) mice but was detrimental for NOD (HS = −0.28) and FVB (HS = −0.47) mice (Figure 7). It also improved blood lipid profiles across strains (HS = 0.37–1.00). Impacts on glucose metabolism varied, with a strong benefit in B6 mice (HS = 0.62), a mild benefit in A mice (HS = 0.18), no improvement in FVB mice, and a detrimental effect in NOD mice (HS = −0.55). Effects on liver health also varied by strain, with benefits in B6 (HS = 0.54) and A (HS = 0.48), but no benefit in FVB or NOD mice (Figure 6e).
Figure 7.
Mean Health Scores for comparison of overall metabolic health of mice fed alternative diets relative to the American diet in each strain. The four health category scores (Figure 6) were averaged to provide a measure of overall metabolic health for each alternative diet relative to the American diet. A positive score represents improved health and a negative score represents diminished health. (a) Scores were calculated with body composition included and are shown with 95% C.I. or (b) are represented by a heat map. (c) Scores were also calculated without the body composition parameter and are shown with 95% C.I. or (d) are represented by a heat map. A, A/J strain mice; B6, C57BL/6J strain mice; FVB, FVB/NJ strain mice; NOD, NOD/ShiLtJ strain mice.
The healthiest alternative to the American diet depends on genetic background
The mean of the four HSs (MHS) was quantified to provide a measure of collective metabolic health (Figure 7, a and b). Only the ketogenic diet improved health for the A strain (MHS = 0.52); both the Japanese (MHS = 0.64) and ketogenic (MHS = 0.61) diets improved health in the B6 strain; and the Japanese diet improved health in FVB (MHS = 0.47) and NOD mice (MHS = 0.65) (Figure 7, a and b).
There is an ongoing debate over the influence of adiposity on metabolic health, with some suggesting that it is not the accumulation of adipose tissue, but the dysfunction of adipose tissue that causes negative metabolic consequences (Goossens and Blaak 2015). The argument is strengthened by the observed lack of metabolic abnormalities in some obese individuals (Bluher 2013). To determine the metabolic effects of these diets without consideration of adiposity, we calculated MHSs without the body composition parameter (Figure 7, c and d). MHS rankings remained relatively consistent, with a notable exception that the FVB Mediterranean diet group showed a significant benefit (MHS = 0.36), similar to that of the Japanese diet (MHS = 0.35) (Figure 7d).
Discussion
In an effort to increase the relevance of rodent findings to people and provide a wider survey of responses to diets, we constructed mouse diets based on dietary patterns of historic populations that better recapitulate human dietary profiles than previous studies. We showed that the American diet caused negative health effects across strains relative to the control diet. However, as in humans (O’Dea 1992; Schulz et al. 2006), the severity of the effects varied across genetic backgrounds. Mice gained fat on the American diet, even though caloric intake did not significantly increase, which is in agreement with previous studies in mice and humans, and emphasizes the importance of factors other than caloric intake alone on fat gain and body weight (Keen et al. 1979; Wack and Rodin 1982; Baecke et al. 1983; Kromhout 1983; Braitman et al. 1985; Romieu et al. 1988; Nicklas et al. 1993; Prentice and Jebb 1995; Heini and Weinsier 1997; Jarvandi et al. 2011; Ford and Dietz 2013; Ladabaum et al. 2014). Metabolic rate increased in A mice fed the American diet, which may in part explain the strain’s resistance to weight gain, although additional research is needed to demonstrate causality.
This study revealed strain-specific, diet-induced epigenetic modification of the vasopressin receptor Avpr1a. The phenotypic changes observed in B6 mice fed the American diet are consistent with those of Avpr1a knockout mice (Aoyagi et al. 2007). Furthermore, people carrying a single-nucleotide polymorphism in the AVPR1A gene exhibit phenotypes consistent with both Avpr1a knockout mice and B6 mice fed a Western diet, including increased incidence of diabetes in those eating a Western-style diet (Enhorning et al. 2009). These data suggest that AVPR1A is influential in diet responsiveness.
After comparing the impact of the American diet to a control mouse diet, we investigated how health status differed in mice fed the American diet relative to those fed other human-comparable diets and identified that genome-by-diet interactions were influential for most metabolic phenotypes. Overall, while each strain had a diet or diets that improved health relative to the American diet, no single diet improved health across all genetic backgrounds.
The FVB strain showed beneficial health effects when fed the Mediterranean diet, even though body composition was not improved. This is consistent with human studies that find beneficial metabolic impacts of a Mediterranean diet in some individuals, without reducing food intake or body weight (Salas-Salvado et al. 2011; Estruch et al. 2013). The Japanese diet yielded metabolic benefits in all strains except A, in agreement with the epidemiological evidence showing generally positive health effects of a Japanese diet in people (Gordon 1957; Marmot et al. 1975; Marmot and Syme 1976; Kagawa 1978).
The ketogenic diet increased metabolic rate, in agreement with human research suggestive of a “metabolic advantage” of low-carbohydrate diets (Feinman and Fine 2003). Lipid profiles of all strains improved on the ketogenic diet, consistent with the healthy cardiovascular status of the Maasai population (Mann et al. 1964, 1965). Effects of high-fat, low-carbohydrate diets on glucose response in people are controversial, with studies having conflicting results (Accurso et al. 2008; Delahanty et al. 2009). Extrapolating from mouse data, our results suggest that genetic variation may underlie the heterogeneity of diet response, as A and B6 mice fed the ketogenic diet experienced benefits to glucose homeostasis, while FVB and NOD mice did not. The ketogenic diet improved overall health in the A and B6 strains, but not the FVB or NOD strains.
Our findings have potential limitations. While great care was taken to accurately recreate mouse versions of human diets, there are differences including a lack of fresh ingredients and spices that may contain additional bioactive compounds. The diets had a baseline vitamin and mineral content, whereas some human diets may lack certain vitamins or minerals. In addition, fiber was limited to one source of insoluble fiber (cellulose) and one source of soluble fiber (inulin) to easily manipulate the levels of these fiber classes, but a complete diet would contain several sources of each fiber class. While our study was not designed to detect the interaction of diet with sex, there is a growing appreciation that these interactions are influential in metabolic health (Bolnick et al. 2014; Arnold et al. 2017; Link et al. 2017; Reue 2017). There are differences in metabolism between humans and mice that could affect outcomes across species, although previous research has demonstrated the utility of mouse models in studying metabolic effects in humans (Rohner-Jeanrenaud and Jeanrenaud 1996; von Scheidt et al. 2017). This study demonstrates the utility of mouse models to dissect genetic-by-diet interactions; however, studies on diet responsiveness in humans are needed to extend findings to people.
Evaluating diet response in the context of genetic background enabled a much clearer understanding of dietary effects. Nonetheless, some variation persisted within diet-by-strain groups. Additional factors yet to be identified must play a role in diet response. One possibility is that the gut microbiome impacted phenotypes in this study (Backhed et al. 2007). Individual-specific epigenetic differences may also have played a role.
This study in mice demonstrates that the health effects of several popular human dietary patterns are dependent on genetic background, adding to a growing appreciation for individual variation in dietary considerations (West et al. 1992; Parks et al. 2013; Konstantinidou et al. 2014; Zeevi et al. 2015; Korem et al. 2017). Determining the extent to which genetic factors influence diet response in humans is difficult given complex genetic variability and environmental confounders. Even so, the disparate health consequences of a Western-style diet across genetically distinct human populations and the strong concordance of dietary response in monozygotic compared to dizygotic twins suggest that genetics plays an important role. If genetics impacts diet response similarly in people as in mice, then the implementation of personalized dietary recommendations will be important for the mitigation of metabolic disease.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300536/-/DC1.
Acknowledgments
We thank members of all laboratories for insightful discussion and suggestions; Andrew Hillhouse, Rachel Lynch, Liyang Zhao, Kuo-Chen Jung, Michael Yarboro, David Thornberg, Aaron Van Wettering, Ahmed Elsaadi, and Lora Moore for assistance with mouse experiments; Suzette Palmer for assistance with histology sample preparation; Jody Albright for assistance with hepatic triglyceride analysis; Stephen Dearth for assistance with mass spectrometry; Arnold Saxton for assistance with metabolome statistics; the Texas A&M Institute for Genome Sciences and Society Molecular Genomics Core for adipokine analysis; and the University of Tennessee Biological and Small Molecule Mass Spectrometry Core for metabolomic analysis. This work was supported by National Institutes of Health (NIH) grant CA-105417 to D.W.T., NIH grant ES-022579 to A.P.F., NIH grant HG-008529 to A.P.F. and D.W.T., and University of Tennessee AgResearch funding to B.H.V. A.P. was supported by NIH training grant OD-011083. Phenotypes were collected using the Texas A&M Institute for Genome Sciences and Society Rodent Phenotyping Core, and the Animal Metabolism Phenotyping core facility within the University of Northern Carolina’s Nutrition Obesity Research Center (NORC) (NIH grant DK-056350). File S1 contains additional data. The authors declare no competing financial interests.
Author contributions: W.T.B., D.W.T., D.P., and A.P.F. conceived and designed the study. W.T.B. and M.A.P. formulated diets. W.T.B., D.W.T., K.H., and D.P. performed mouse experiments. A.E.W. and B.H.V. performed metabolomic analysis. C.M.R. performed cholesterol analysis. S.Y.F.H. performed adipokine analysis. B.J.B. performed triglyceride analysis. A.P. performed histological analysis. P.W., K.D.H., and A.P.F. performed library preparation, sequencing, and epigenetic analysis; W.T.B., A.E.W., K.D.H., and P.W. performed statistical analysis. W.T.B. and D.W.T. designed the figures and wrote the manuscript. All authors revised the manuscript.
Footnotes
Communicating editor: E. Eskin
Literature Cited
- Accurso A., Bernstein R. K., Dahlqvist A., Draznin B., Feinman R. D., et al. , 2008. Dietary carbohydrate restriction in type 2 diabetes mellitus and metabolic syndrome: time for a critical appraisal. Nutr. Metab. (Lond.) 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almind K., Kahn C. R., 2004. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes 53: 3274–3285. [DOI] [PubMed] [Google Scholar]
- Aoyagi T., Birumachi J., Hiroyama M., Fujiwara Y., Sanbe A., et al. , 2007. Alteration of glucose homeostasis in V1a vasopressin receptor-deficient mice. Endocrinology 148: 2075–2084. [DOI] [PubMed] [Google Scholar]
- Arnold A. P., Cassis L. A., Eghbali M., Reue K., Sandberg K., 2017. Sex hormones and sex chromosomes cause sex differences in the development of cardiovascular diseases. Arterioscler. Thromb. Vasc. Biol. 37: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., et al. , 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 101: 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Manchester J. K., Semenkovich C. F., Gordon J. I., 2007. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 104: 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baecke J. A., van Staveren W. A., Burema J., 1983. Food consumption, habitual physical activity, and body fatness in young Dutch adults. Am. J. Clin. Nutr. 37: 278–286. [DOI] [PubMed] [Google Scholar]
- Barrett P., Mercer J. G., Morgan P. J., 2016. Preclinical models for obesity research. Dis. Model. Mech. 9: 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddinger S. B., Almind K., Miyazaki M., Kokkotou E., Ntambi J. M., et al. , 2005. Effects of diet and genetic background on sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes 54: 1314–1323. [DOI] [PubMed] [Google Scholar]
- Black B. L., Croom J., Eisen E. J., Petro A. E., Edwards C. L., et al. , 1998. Differential effects of fat and sucrose on body composition in A/J and C57BL/6 mice. Metabolism 47: 1354–1359. [DOI] [PubMed] [Google Scholar]
- Blake J. A., Eppig J. T., Kadin J. A., Richardson J. E., Smith C. L., et al. , 2017. Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 45: D723–D729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher M., 2013. Adipose tissue dysfunction contributes to obesity related metabolic diseases. Best Pract. Res. Clin. Endocrinol. Metab. 27: 163–177. [DOI] [PubMed] [Google Scholar]
- Bolnick D. I., Snowberg L. K., Hirsch P. E., Lauber C. L., Org E., et al. , 2014. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat. Commun. 5: 4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C., Tremblay A., Despres J. P., Nadeau A., Lupien P. J., et al. , 1990. The response to long-term overfeeding in identical twins. N. Engl. J. Med. 322: 1477–1482. [DOI] [PubMed] [Google Scholar]
- Braitman L. E., Adlin E. V., Stanton J. L., Jr, 1985. Obesity and caloric intake: the National Health and Nutrition Examination Survey of 1971–1975 (HANES I). J. Chronic Dis. 38: 727–732. [DOI] [PubMed] [Google Scholar]
- Burcelin R., Crivelli V., Dacosta A., Roy-Tirelli A., Thorens B., 2002. Heterogeneous metabolic adaptation of C57BL/6J mice to high-fat diet. Am. J. Physiol. Endocrinol. Metab. 282: E834–E842. [DOI] [PubMed] [Google Scholar]
- Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., et al. , 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30: 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., Pletscher L. S., Vaughn T. T., Marshall B., 1999. Differential response to dietary fat in large (LG/J) and small (SM/J) inbred mouse strains. Physiol. Genomics 1: 33–39. [DOI] [PubMed] [Google Scholar]
- Cheverud J. M., Ehrich T. H., Hrbek T., Kenney J. P., Pletscher L. S., et al. , 2004. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes 53: 3328–3336. [DOI] [PubMed] [Google Scholar]
- Clasquin M. F., Melamud E., Rabinowitz J. D., 2012. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics Chapter 14: Unit14.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Horton J. D., Hobbs H. H., 2011. Human fatty liver disease: old questions and new insights. Science 332: 1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S., Weitzman M., Auinger P., Nguyen M., Dietz W. H., 2003. Prevalence of a metabolic syndrome phenotype in adolescents: findings from the third National Health and Nutrition Examination Survey, 1988–1994. Arch. Pediatr. Adolesc. Med. 157: 821–827. [DOI] [PubMed] [Google Scholar]
- Dansinger M. L., Gleason J. A., Griffith J. L., Selker H. P., Schaefer E. J., 2005. Comparison of the Atkins, Ornish, Weight Watchers, and Zone diets for weight loss and heart disease risk reduction: a randomized trial. JAMA 293: 43–53. [DOI] [PubMed] [Google Scholar]
- Dashti H. M., Al-Zaid N. S., Mathew T. C., Al-Mousawi M., Talib H., et al. , 2006. Long term effects of ketogenic diet in obese subjects with high cholesterol level. Mol. Cell. Biochem. 286: 1–9. [DOI] [PubMed] [Google Scholar]
- Delahanty L. M., Nathan D. M., Lachin J. M., Hu F. B., Cleary P. A., et al. , 2009. Association of diet with glycated hemoglobin during intensive treatment of type 1 diabetes in the diabetes control and complications trial. Am. J. Clin. Nutr. 89: 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enhorning S., Leosdottir M., Wallstrom P., Gullberg B., Berglund G., et al. , 2009. Relation between human vasopressin 1a gene variance, fat intake, and diabetes. Am. J. Clin. Nutr. 89: 400–406. [DOI] [PubMed] [Google Scholar]
- Estruch R., Ros E., Salas-Salvado J., Covas M. I., Corella D., et al. , 2013. Primary prevention of cardiovascular disease with a Mediterranean diet. N. Engl. J. Med. 368: 1279–1290.23432189 [Google Scholar]
- Feinman R. D., Fine E. J., 2003. Thermodynamics and metabolic advantage of weight loss diets. Metab. Syndr. Relat. Disord. 1: 209–219. [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane Stanley G. H., 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- Ford E. S., Dietz W. H., 2013. Trends in energy intake among adults in the United States: findings from NHANES. Am. J. Clin. Nutr. 97: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens G. H., Blaak E. E., 2015. Adipose tissue dysfunction and impaired metabolic health in human obesity: a matter of oxygen? Front. Endocrinol. (Lausanne) 6: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon T., 1957. Mortality experience among the Japanese in the United States, Hawaii, and Japan. Public Health Rep. 72: 543–553. [PMC free article] [PubMed] [Google Scholar]
- Hansen K. D., Langmead B., Irizarry R. A., 2012. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E. A., et al. , 2012. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15: 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Buse J. B., Jackson R. A., Glimcher L., Dorf M. E., et al. , 1986. The NOD mouse: recessive diabetogenic gene in the major histocompatibility complex. Science 231: 733–735. [DOI] [PubMed] [Google Scholar]
- Heini A. F., Weinsier R. L., 1997. Divergent trends in obesity and fat intake patterns: the American paradox. Am. J. Med. 102: 259–264. [DOI] [PubMed] [Google Scholar]
- Hession M., Rolland C., Kulkarni U., Wise A., Broom J., 2009. Systematic review of randomized controlled trials of low-carbohydrate vs. low-fat/low-calorie diets in the management of obesity and its comorbidities. Obes. Rev. 10: 36–50. [DOI] [PubMed] [Google Scholar]
- Hill-Baskin A. E., Markiewski M. M., Buchner D. A., Shao H., DeSantis D., et al. , 2009. Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum. Mol. Genet. 18: 2975–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. F., Cheng M. L., Fan C. M., Hong C. Y., Shiao M. S., 2013. Nicotinuric acid: a potential marker of metabolic syndrome through a metabolomics-based approach. Diabetes Care 36: 1729–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Ren J., Huang J., Li D., 2013. Association of homocysteine with type 2 diabetes: a meta-analysis implementing Mendelian randomization approach. BMC Genomics 14: 867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke R., Dodson A. E., Rine J., 2015. Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 31: 473–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvandi S., Gougeon R., Bader A., Dasgupta K., 2011. Differences in food intake among obese and nonobese women and men with type 2 diabetes. J. Am. Coll. Nutr. 30: 225–232. [DOI] [PubMed] [Google Scholar]
- Kagan A., Harris B. R., Winkelstein W., Jr, Johnson K. G., Kato H., et al. , 1974. Epidemiologic studies of coronary heart disease and stroke in Japanese men living in Japan, Hawaii and California: demographic, physical, dietary and biochemical characteristics. J. Chronic Dis. 27: 345–364. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., 1978. Impact of Westernization on the nutrition of Japanese: changes in physique, cancer, longevity and centenarians. Prev. Med. 7: 205–217. [DOI] [PubMed] [Google Scholar]
- Keen H., Thomas B. J., Jarrett R. J., Fuller J. H., 1979. Nutrient intake, adiposity, and diabetes. BMJ 1: 655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. J., Ellacott K. L., King V. L., Hasty A. H., 2010. Mouse models of the metabolic syndrome. Dis. Model. Mech. 3: 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A., Kang H. M., Wade C. M., Cotsapas C., Kostem E., et al. , 2010. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics 185: 1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K. T., de Groot L. C., Kromhout D., Perrin A. E., Moreiras-Varela O., et al. , 2004. Mediterranean diet, lifestyle factors, and 10-year mortality in elderly European men and women: the HALE project. JAMA 292: 1433–1439. [DOI] [PubMed] [Google Scholar]
- Knox E. G., 1977. Foods and diseases. Br. J. Prev. Soc. Med. 31: 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidou V., Daimiel L., Ordovas J. M., 2014. Personalized nutrition and cardiovascular disease prevention: from Framingham to PREDIMED. Adv. Nutr. 5: 368S–371S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korem T., Zeevi D., Zmora N., Weissbrod O., Bar N., et al. , 2017. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 25: 1243–1253.e5. [DOI] [PubMed] [Google Scholar]
- Kotronen A., Yki-Jarvinen H., 2008. Fatty liver: a novel component of the metabolic syndrome. Arterioscler. Thromb. Vasc. Biol. 28: 27–38. [DOI] [PubMed] [Google Scholar]
- Koza R. A., Nikonova L., Hogan J., Rim J. S., Mendoza T., et al. , 2006. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kris-Etherton P. M., Krummel D., Russell M. E., Dreon D., Mackey S., et al. , 1988. The effect of diet on plasma lipids, lipoproteins, and coronary heart disease. J. Am. Diet. Assoc. 88: 1373–1400. [PubMed] [Google Scholar]
- Kromhout D., 1983. Energy and macronutrient intake in lean and obese middle-aged men (the Zutphen study). Am. J. Clin. Nutr. 37: 295–299. [DOI] [PubMed] [Google Scholar]
- Kwiterovich P. O., Jr, Vining E. P., Pyzik P., Skolasky R., Jr, Freeman J. M., 2003. Effect of a high-fat ketogenic diet on plasma levels of lipids, lipoproteins, and apolipoproteins in children. JAMA 290: 912–920. [DOI] [PubMed] [Google Scholar]
- Ladabaum U., Mannalithara A., Myer P. A., Singh G., 2014. Obesity, abdominal obesity, physical activity, and caloric intake in US adults: 1988 to 2010. Am. J. Med. 127: 717–727.e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter E. H., Prochazka M., Coleman D. L., 1987. The non-obese diabetic (NOD) mouse. Am. J. Pathol. 128: 380–383. [PMC free article] [PubMed] [Google Scholar]
- Levine J. A., Eberhardt N. L., Jensen M. D., 1999. Role of nonexercise activity thermogenesis in resistance to fat gain in humans. Science 283: 212–214. [DOI] [PubMed] [Google Scholar]
- Liang W., Menke A. L., Driessen A., Koek G. H., Lindeman J. H., et al. , 2014. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One 9: e115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link J. C., Hasin-Brumshtein Y., Cantor R. M., Chen X., Arnold A. P., et al. , 2017. Diet, gonadal sex, and sex chromosome complement influence white adipose tissue miRNA expression. BMC Genomics 18: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Stamler J., Dyer A., McKeever J., McKeever P., 1978. Statistical methods to assess and minimize the role of intra-individual variability in obscuring the relationship between dietary lipids and serum cholesterol. J. Chronic Dis. 31: 399–418. [DOI] [PubMed] [Google Scholar]
- Mann G. V., Shaffer R. D., Anderson R. S., Sandstead H. H., 1964. Cardiovascular disease in the Masai. J. Atheroscler. Res. 4: 289–312. [DOI] [PubMed] [Google Scholar]
- Mann G. V., Shaffer R. D., Rich A., 1965. Physical fitness and immunity to heart-disease in Masai. Lancet 2: 1308–1310. [DOI] [PubMed] [Google Scholar]
- Marmot M. G., Syme S. L., 1976. Acculturation and coronary heart disease in Japanese-Americans. Am. J. Epidemiol. 104: 225–247. [DOI] [PubMed] [Google Scholar]
- Marmot M. G., Syme S. L., Kagan A., Kato H., Cohen J. B., et al. , 1975. Epidemiologic studies of coronary heart disease and stroke in Japanese men living in Japan, Hawaii and California: prevalence of coronary and hypertensive heart disease and associated risk factors. Am. J. Epidemiol. 102: 514–525. [DOI] [PubMed] [Google Scholar]
- Melamud E., Vastag L., Rabinowitz J. D., 2010. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82: 9818–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni C., Fauman E., Erte I., Perry J. R., Kastenmuller G., et al. , 2013. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 62: 4270–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mente A., de Koning L., Shannon H. S., Anand S. S., 2009. A systematic review of the evidence supporting a causal link between dietary factors and coronary heart disease. Arch. Intern. Med. 169: 659–669. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations, 2016 FAOSTAT database. Available at: http://www.fao.org/faostat/en/#home. Accessed: December, 2016.
- National Cancer Institute, 2015 Usual dietary intakes: food intakes, U.S. population, 2007–10. Available at: https://epi.grants.cancer.gov/diet/usualintakes/pop/2007-10/.
- Nicklas T. A., Webber L. S., Srinivasan S. R., Berenson G. S., 1993. Secular trends in dietary intakes and cardiovascular risk factors of 10-y-old children: the Bogalusa Heart Study (1973–1988). Am. J. Clin. Nutr. 57: 930–937. [DOI] [PubMed] [Google Scholar]
- Nyamundanda, G., L. Brennan, and I. C. Gormley, 2010 Probabilistic principal component analysis for metabolomic data. BMC Bioinformatics 11: 571. [DOI] [PMC free article] [PubMed]
- O’Dea K., 1992. Diabetes in Australian aborigines: impact of the western diet and life style. J. Intern. Med. 232: 103–117. [DOI] [PubMed] [Google Scholar]
- Paigen B., 1995. Genetics of responsiveness to high-fat and high-cholesterol diets in the mouse. Am. J. Clin. Nutr. 62: 458S–462S. [DOI] [PubMed] [Google Scholar]
- Parks B. W., Nam E., Org E., Kostem E., Norheim F., et al. , 2013. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petro A. E., Cotter J., Cooper D. A., Peters J. C., Surwit S. J., et al. , 2004. Fat, carbohydrate, and calories in the development of diabetes and obesity in the C57BL/6J mouse. Metabolism 53: 454–457. [DOI] [PubMed] [Google Scholar]
- Prentice A. M., Jebb S. A., 1995. Obesity in Britain: gluttony or sloth? BMJ 311: 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikvam D. H., Erofeev A., Sandvik A., Grcic V., Jahnsen F. L., et al. , 2011. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 6: e17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue K., 2017. Sex differences in obesity: X chromosome dosage as a risk factor for increased food intake, adiposity and co-morbidities. Physiol. Behav. 176: 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner-Jeanrenaud F., Jeanrenaud B., 1996. The discovery of leptin and its impact in the understanding of obesity. Eur. J. Endocrinol. 135: 649–650. [DOI] [PubMed] [Google Scholar]
- Romieu I., Willett W. C., Stampfer M. J., Colditz G. A., Sampson L., et al. , 1988. Energy intake and other determinants of relative weight. Am. J. Clin. Nutr. 47: 406–412. [DOI] [PubMed] [Google Scholar]
- Salas-Salvado J., Bullo M., Babio N., Martinez-Gonzalez M. A., Ibarrola-Jurado N., et al. , 2011. Reduction in the incidence of type 2 diabetes with the Mediterranean diet: results of the PREDIMED-Reus nutrition intervention randomized trial. Diabetes Care 34: 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz L. O., Bennett P. H., Ravussin E., Kidd J. R., Kidd K. K., et al. , 2006. Effects of traditional and western environments on prevalence of type 2 diabetes in Pima Indians in Mexico and the U.S. Diabetes Care 29: 1866–1871. [DOI] [PubMed] [Google Scholar]
- Sharman M. J., Kraemer W. J., Love D. M., Avery N. G., Gomez A. L., et al. , 2002. A ketogenic diet favorably affects serum biomarkers for cardiovascular disease in normal-weight men. J. Nutr. 132: 1879–1885. [DOI] [PubMed] [Google Scholar]
- Shekelle R. B., Shryock A. M., Paul O., Lepper M., Stamler J., et al. , 1981. Diet, serum cholesterol, and death from coronary heart disease. The Western Electric study. N. Engl. J. Med. 304: 65–70. [DOI] [PubMed] [Google Scholar]
- Shockley K. R., Witmer D., Burgess-Herbert S. L., Paigen B., Churchill G. A., 2009. Effects of atherogenic diet on hepatic gene expression across mouse strains. Physiol. Genomics 39: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinasac D. S., Riordan J. D., Spiezio S. H., Yandell B. S., Croniger C. M., et al. , 2016. Genetic control of obesity, glucose homeostasis, dyslipidemia and fatty liver in a mouse model of diet-induced metabolic syndrome. Int. J. Obes. 40: 346–355. [DOI] [PubMed] [Google Scholar]
- Stover P. J., 2011. Polymorphisms in 1-carbon metabolism, epigenetics and folate-related pathologies. J. Nutrigenet. Nutrigenomics 4: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surwit R. S., Kuhn C. M., Cochrane C., McCubbin J. A., Feinglos M. N., 1988. Diet-induced type II diabetes in C57BL/6J mice. Diabetes 37: 1163–1167. [DOI] [PubMed] [Google Scholar]
- Svenson K. L., Von Smith R., Magnani P. A., Suetin H. R., Paigen B., et al. , 2007. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J. Appl. Physiol. (1985) 102: 2369–2378. [DOI] [PubMed] [Google Scholar]
- Toubro S., Astrup A., 1997. Randomised comparison of diets for maintaining obese subjects’ weight after major weight loss: ad lib, low fat, high carbohydrate diet v fixed energy intake. BMJ 314: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichopoulou A., Vasilopoulou E., 2000. Mediterranean diet and longevity. Br. J. Nutr. 84(Suppl. 2): S205–S209. [DOI] [PubMed] [Google Scholar]
- Ussar S., Griffin N. W., Bezy O., Fujisaka S., Vienberg S., et al. , 2015. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 22: 516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Scheidt M., Zhao Y., Kurt Z., Pan C., Zeng L., et al. , 2017. Applications and limitations of mouse models for understanding human atherosclerosis. Cell Metab. 25: 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wack J. T., Rodin J., 1982. Smoking and its effects on body weight and the systems of caloric regulation. Am. J. Clin. Nutr. 35: 366–380. [DOI] [PubMed] [Google Scholar]
- Wang T. J., Ngo D., Psychogios N., Dejam A., Larson M. G., et al. , 2013. 2-Aminoadipic acid is a biomarker for diabetes risk. J. Clin. Invest. 123: 4309–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West D. B., Boozer C. N., Moody D. L., Atkinson R. L., 1992. Dietary obesity in nine inbred mouse strains. Am. J. Physiol. 262: R1025–R1032. [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Jewison T., Guo A. C., Wilson M., Knox C., et al. , 2013. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 41: D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. K., Chin K. Y., Suhaimi F. H., Fairus A., Ima-Nirwana S., 2016. Animal models of metabolic syndrome: a review. Nutr. Metab. (Lond.) 13: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., et al. , 2015. Personalized nutrition by prediction of glycemic responses. Cell 163: 1079–1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 contains supplemental data to support the conclusions in this article.