Abstract
Highly regulated cell migration events are crucial during animal tissue formation and the trafficking of cells to sites of infection and injury. Misregulation of cell movement underlies numerous human diseases, including cancer. Although originally studied primarily in two-dimensional in vitro assays, most cell migrations in vivo occur in complex three-dimensional tissue environments that are difficult to recapitulate in cell culture or ex vivo. Further, it is now known that cells can mobilize a diverse repertoire of migration modes and subcellular structures to move through and around tissues. This review provides an overview of three distinct cellular movement events in Caenorhabditis elegans—cell invasion through basement membrane, leader cell migration during organ formation, and individual cell migration around tissues—which together illustrate powerful experimental models of diverse modes of movement in vivo. We discuss new insights into migration that are emerging from these in vivo studies and important future directions toward understanding the remarkable and assorted ways that cells move in animals.
Keywords: basement membrane, C. elegans, cell invasion, cell migration, cell signaling, F-actin, FGF pathway, integrin, netrin pathway, Wnt pathway, WormBook
THE ability of cells to move is crucial for many aspects of normal animal embryogenesis, organ formation, wound healing, tissue regeneration, and immune cell trafficking (Aman and Piotrowski 2010; Nourshargh and Alon 2014; Mayor and Etienne-Manneville 2016). Misregulation of cell movement also underlies human developmental disorders, immune dysfunction, and cancer (Kurosaka and Kashina 2008; Madsen and Sahai 2010; Friedl and Alexander 2011; Paul et al. 2017). Thus, understanding the mechanisms by which cells move in diverse cell and tissue environments has important basic and clinical relevance. Because of the challenge of examining dynamic cellular behaviors in native tissue settings, most studies of cell migration have been carried out in cell culture. While these in vitro studies have revealed mechanisms underlying key parameters of migration, such as cytoskeletal regulation, cell-cell and cell-extracellular matrix (ECM) adhesion, polarization machinery, and distinct modes of migration (Lammermann and Sixt 2009; Linder et al. 2011; Blanchoin et al. 2014; Te Boekhorst et al. 2016), in vitro conditions do not faithfully match the complexity of in vivo settings, and, therefore, their physiological significance often remains unclear.
The shortcomings of in vitro migration models are highlighted by the fact that cell-substrate adhesions and other cellular structures appear very different in cells plated on two-dimensional (2D) flat, rigid substrates as compared to more native three-dimensional (3D) cell and ECM environments, and often display different dynamics and biochemistry (Fraley et al. 2010; Geraldo et al. 2012; Petrie et al. 2012). Although 3D culture conditions are a step in the right direction, they do not reflect the richness of other physiologically relevant environmental factors that migrating cells encounter. These factors include diverse cell–cell interactions, diffusible cues, fluctuating nutrient conditions, changing oxygen levels, varying fluid dynamics, cell and tissue growth, and native mechanical properties of cells and extracellular matrices (Even-Ram and Yamada 2005; Friedl et al. 2012). Cells also have important intrinsic properties, such as unique transcriptional programs and chromatin states, that are likely not recapitulated in cell culture settings (Feil and Fraga 2012; Chen et al. 2013). Thus, in vivo models are essential, not only to verify or challenge mechanisms discovered in vitro, but also to discover new mechanisms of cell migration that are difficult, if not impossible, to recapitulate in vitro.
Studying cell movements in Caenorhabditis elegans provides a strong experimental model to examine cell motility in an in vivo setting. One of the advantages of studying cell migration in C. elegans is the simplicity of the gene families that encode cytoskeleton (Sawa et al. 2003; Schonichen and Geyer 2010; Mi-Mi et al. 2012; Abella et al. 2016; Pizarro-Cerda et al. 2017), ECM (Kramer 2005), and signaling proteins (Lai Wing Sun et al. 2011; Clevers and Nusse 2012; Sawa and Korswagen 2013) that guide cell migrations. This simplified genetic landscape reduces redundancy and makes gene perturbation studies easier to perform and interpret. Cell migration phenotypes are also straightforward to visualize, as the worm’s optical transparency allows for imaging of all cell migrations in real time. In addition, C. elegans anatomical simplicity (the adult has <1000 somatic cells) and its highly stereotyped development facilitate detailed analysis of even subtle phenotypes. C. elegans is also remarkably easy to manipulate genetically such that genes and proteins can be altered at the organismal and individual cell level using temporally controlled optogenetic, RNAi, CRISPR/Cas-9, and ubiquitin mediated methods (Hagedorn et al. 2009; Dickinson et al. 2013; Armenti et al. 2014; Shen et al. 2014; Corsi et al. 2015). Finally, the worm’s short life cycle and hermaphrodite mode of reproduction coupled with rapid whole-genome RNAi screening facilitate discovery of genes and pathways regulating cell migration that would not be found through candidate approaches (Jorgensen and Mango 2002; Kamath et al. 2003; Corsi et al. 2015). Together, these worm attributes permit exceptional experimental access to uncover the molecular and cell biological mechanisms that underlie migration in vivo.
C. elegans undergoes numerous cell migrations throughout embryonic and larval development (Hedgecock et al. 1987). Much information concerning mechanisms underlying cell migration in C. elegans has emerged from the study of a few major motile events. Some of these have recently been reviewed elsewhere, including ventral enclosure (Vuong-Brender et al. 2016), Q neuroblast migration (Rella et al. 2016) and axon guidance (Chisholm et al. 2016). Our review focuses on what has been learned and promising future studies on three distinct cellular movements that are common motility modes in animals: anchor cell (AC) invasion as a model for invasion through basement membrane (BM) barriers; distal tip cell (DTC) migration as a model for how a BM- encased leader cell directs organ formation; and sex myoblast (SM) migration as a model for how cells migrate between tissues.
AC Invasion: Breaching BM Barriers
BMs are thin, dense, highly cross-linked ECM composed of interlinked sheets of laminin and type IV collagen networks that surround and support most tissues (Yurchenco 2011; Jayadev and Sherwood 2017). Despite their barrier properties, BMs are breached and crossed by cells during development, blood vessel formation, and immune functioning (Yang and Weinberg 2008; Kelley et al. 2014; Seano et al. 2014). Inappropriate invasion also underlies numerous pathologies, most notably cancer cell metastasis (Valastyan and Weinberg 2011). Owing to the complexity of studying dynamic interactions between invasive cells, BMs, and the invaded tissue, cell invasion has been challenging to experimentally examine in native tissue environments (Beerling et al. 2011; Hagedorn and Sherwood 2011).
Most C. elegans tissues are enwrapped in BM, and the C. elegans genome harbors the major BM components laminin and type IV collagen, as well as the BM-associated proteins perlecan, nidogen, fibulin, agrin, hemicentin, SPARC, and collagen XVIII (Kramer 2005). Gene families encoding BM proteins in C. elegans have not undergone the extensive expansion observed in vertebrates (Kramer 2005), and many have been tagged with fluorophores allowing analysis of their localization and function (Kelley et al. 2014). Notably, C. elegans lacks interstitial matrix, and does not harbor genes encoding fibrillar collagens and other interstitial components (Hutter et al. 2000). Fibrillar collagens and interstitial matrix are thought to have originated near the time of metazoan emergence, but apparently were lost in the lineage that gave rise to C. elegans (Ozbek et al. 2010; Fidler et al. 2017).
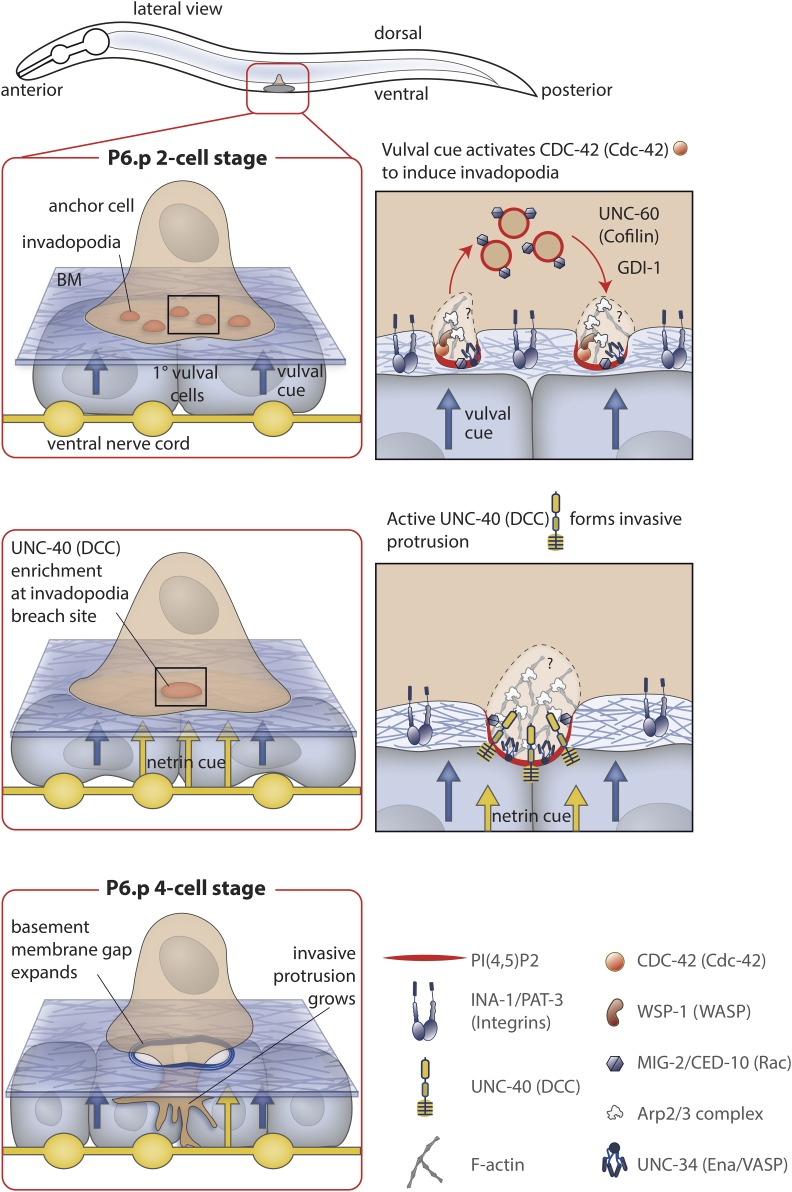
C. elegans AC invasion into the vulval epithelium is an in vivo model of invasive behavior that permits single cell and subcellular analysis of invasion through BM (Figure 1). The AC is a specialized uterine cell that invades through BM separating the uterine and vulval tissue to initiate uterine-vulval attachment—a connection required for mating and laying embryos. AC invasion occurs over a precise 90-min period during the L3 larval stage, and is coordinated with the divisions of the descendants of the P6.p epidermal cell. These cells are the 1° vulval precursor cells, and give rise to the centrally located vulval cells. The stereotyped nature of AC invasion, amenability to forward and reverse genetic screens, and visual accessibility to live-cell imaging have allowed mechanisms regulating invasion to be identified and characterized in vivo.
Figure 1.
AC invasion, a BM invasion event. Prior to invasion, the AC is positioned over the epidermal P6.p cell, which it induces to a 1° vulval precursor cell fate. The 1° fated P6.p divides three times. Invasion occurs at the time of the division from the P6.p 2-cell stage to P6.p 4-cell stage. Top panel: Prior to breaching the BM, AC invadopodia form along the AC’s invasive cell membrane, depress the BM, and then disassemble. Approximately 10 invadopodia are present at any one time. Invadopodia formation is stimulated by a cue from the 1° vulval cells that activates the Rho GTPase CDC-42. CDC-42 activates WSP-1, which presumably stimulates F-actin production through actin polymerization nucleators such as the Arp2/3 complex. UNC-34 (Ena/VASP) may also contribute to F-actin formation. Invadopodia generation is also dependent on an invadopodial membrane rich in PI(4,5)P2 and containing the lipid-anchored Rac GTPases, CED-10 and MIG-2. The invadopodial membrane is recycled through the endolysosome, and its trafficking is dependent on UNC-60 (cofilin) and the Rab GDP dissociation inhibitor (GDI-1). The integrin heterodimer INA-1/PAT-3 is required for trafficking of all known invadopodial components to the plasma membrane. Middle panel: when an invadopodium breaches the BM, the netrin receptor UNC-40 (DCC) traffics to the breach site, and is activated by its ligand UNC-6 (netrin) secreted by the underlying ventral nerve cord. UNC-40 recruits the actin regulators UNC-34 (Ena/VASP) and the Rac GTPases, which shuts down further invadopodia formation. Lower Panel: UNC-40 (DCC) directs the formation of a large invasive protrusion that expands the opening in the BM by degrading and physically displacing BM.
The AC breaches the BM with invadopodia
Studies on AC invasion have revealed that dynamic, ∼1.0 µm-diameter F-actin (filamentous actin) rich structures, termed invadopodia, form along the AC’s invasive cell membrane and breach the BM (Figure 1) (Hagedorn et al. 2013). Matrix degrading invadopodia were originally observed in transformed fibroblasts, human cancer cell lines, and primary tumor cells from human patients cultured on glass slides covered with simplified ECMs in vitro (Chen 1989; Linder et al. 2011; Genot and Gligorijevic 2014). Invadopodia have been extensively characterized in vitro, and numerous aspects of their composition, regulation, and formation have been elucidated in cell culture settings (Bergman et al. 2014). The identification of invadopodia in C. elegans has confirmed the in vivo existence of these structures, the importance of invadopodia in breaching BM, and regulation of invadopodia in native invasion events (Morrissey et al. 2013; Genot and Gligorijevic 2014; Lohmer et al. 2014).
AC invadopodia formation is stimulated ∼5 hr prior to invasion by an unidentified diffusible cue(s) from the 1° fated vulval precursor cells (Sherwood and Sternberg 2003; Lohmer et al. 2016). The vulval signal activates the Rho GTPase CDC-42 (vertebrate Cdc42) within the AC (Figure 1). CDC-42 seeds invadopodia in part through its effector WSP-1 (WASP), which activates the Arp2/3 complex, an actin polymerization nucleator (Shakir et al. 2008; Padrick and Rosen 2010; Lohmer et al. 2016). Approximately 10 invadopodia are present at any one time in the AC, and turn over rapidly with a median life time of 45 sec (Hagedorn et al. 2013). The rapid turnover of AC invadopodia is in stark contrast to invadopodia dynamics characterized in vitro in cancer cells, which have average lifetimes of 30 min or longer (Linder et al. 2011; Branch et al. 2012; Moshfegh et al. 2014). The rapid turnover of AC invadopodia might reflect the more physiologically relevant in vivo conditions (e.g., BM composition, microenvironmental signals, and intrinsic cell characteristics) or that AC invasion has evolved to be a quick invasion event, whereas cancer cells are less efficient in transmigrating BMs (Lohmer et al. 2014).
Other actin regulatory proteins are also associated with AC invadopodia, and have revealed important aspects of their formation and regulation. The actin filament severing protein UNC-60 (ADF/cofilin) is crucial to invadopodia turnover (Figure 1). In the absence of UNC-60, invadopodia formation ceases, and instead large aggregates of static F-actin form along the invasive cell membrane (Hagedorn et al. 2014). UNC-34 (Ena/VASP), another actin regulator associated with enhanced cell migration, is also localized to AC invadopodia, although its function in the AC has not been characterized (Chesarone and Goode 2009; Hagedorn et al. 2013). In addition to F-actin, invadopodia are constructed from a specialized invadopodial membrane containing the lipid phosphatidyl inositol 4,5 bisphosphate (PI(4,5)P2) and CED-10 and MIG-2, two members of the Rac subfamily of Rho GTPases that have lipid anchors (Lundquist et al. 2001; Hagedorn et al. 2014; Ridley 2015; Reiner and Lundquist 2016). The invadopodial membrane is actively recycled through the endolysosome during invadopodia formation and breakdown, and its delivery to the invasive cell membrane is dependent on the Rab GDP dissociation inhibitor GDI-1 as well as UNC-60 (cofilin) (Hagedorn et al. 2014; Lohmer et al. 2016). Cell culture studies with cancer cells have indicated that the membrane tethered matrix metalloproteinase MT1-MMP is delivered to invadopodia through the endolysosome, suggesting that active endolysosome recycling might be a shared feature of invadopodia formation (Castro-Castro et al. 2016; Hastie and Sherwood 2016).
Invadopodia formation also requires the ECM integrin receptor heterodimer INA-1/PAT-3 (Hagedorn et al. 2009). Integrin receptors are composed of an α and a β subunit. Unlike vertebrates, however, which encode 18 α and 8 β subunits and construct 24 known αβ integrin heterodimers, C. elegans has only a single β and two α subunits and make only two integrins: αINA-1/βPAT-3, most similar to vertebrate laminin binding integrins, and αPAT-2/βPAT-3, an RGD containing integrin (Baum and Garriga 1997; Kramer 2005; Campbell and Humphries 2011). INA-1/PAT-3 is the only integrin expressed in the AC, and mediates the trafficking of F-actin regulators and invadopodial membrane to the invasive cell membrane (Hagedorn et al. 2009). INA-1/PAT-3 might polarize the secretory apparatus of the AC (Wickstrom and Fassler 2011).
Invadopodia breach the BM during a narrow 20-min time period in the mid-L3 larval stage. These observations suggest that invadopodia formed in the 5-hr window prior to breach might not be fully mature and able to penetrate BM. The mechanisms that mediate the maturation/precise timing of the invadopodia breach are not known, but are likely connected to cues generated by the vulval cells, as precocious vulval cell formation accelerates the timing of AC invasion (Sherwood and Sternberg 2003). The AC expresses three matrix metalloproteinases (zmp-1, zmp-3, and zmp-6), a class of proteolytic enzymes that have been implicated in vertebrates in breaking down BMs to promote invasion (Sherwood et al. 2005; Hotary et al. 2006; Page-McCaw et al. 2007; Itoh 2015; Matus et al. 2015). Invadopodia also physically deform the BM as they extend, suggesting that a combination of proteolysis and mechanical disruption mediates invadopodial breaching of the BM (Figure 1) (Hagedorn et al. 2013).
The similarities of invadopodia regulation in cancer cells and the AC are striking. In numerous cancer cell lines Cdc-42 stimulates invadopodia formation (Di Martino et al. 2014; Razidlo et al. 2014), cofilin regulates invadopodia turnover (Bravo-Cordero et al. 2013; Beaty and Condeelis 2014), and integrins are essential for invadopodia generation (Destaing et al. 2011; Murphy and Courtneidge 2011). These observations suggest that invadopodia are conserved subcellular structures that evolved early in animals to allow cells to pass through BM barriers during development and tissue remodeling (Medwig and Matus 2017). A recent sensitized whole genome RNAi screen has identified many additional genes implicated in invadopodia regulation, including genes involved in G-protein signaling, extracellular matrix remodeling and the Hippo pathway (Lohmer et al. 2016). The characterization of these genes will provide new insight into mechanisms mediating invadopodia formation, turnover, and BM breaching in vivo.
A large invasive protrusion clears an opening in the BM
Invadopodia formation, turnover, and matrix breaching can be reasonably well recapitulated in cell culture on flat 2D surfaces such as ECM coated glass slides (Even-Ram and Yamada 2005). Capturing later events in BM invasion in vitro is challenging, as this requires 3D assays that faithfully mimic native BM and cellular architecture to follow how cells cross BM barriers and enter new tissues (Schoumacher et al. 2010, 2011). C. elegans AC invasion has been particularly crucial in extending our understanding of BM invasion following invadopodia-mediated BM breaching events.
Live-cell imaging revealed that usually only one or two AC invadopodia breach the BM and only one of these transitions into a protrusion that clears a large opening in the BM and expands between the central vulval cells (Hagedorn et al. 2013). The netrin pathway plays a crucial role in directing this dramatic transition (Morrissey et al. 2013). The netrin receptor UNC-40 (vertebrate DCC) clusters at the invadopodial breach site and in response to UNC-6 (netrin) recruits F-actin regulators (Ena/VASP, Rho GTPases) that direct invasive protrusive formation (Figure 1) (Hagedorn et al. 2013). UNC-40 acts as a molecular sink and depletes F-actin regulators from other invadopodia, shutting them down and focusing F-actin generation and invasion through a single breach site. In the absence of UNC-40 (DCC), invasive protrusion formation fails and invadopodia continue to form and turn over, creating multiple small breaches in the BM that hinder the ability of the AC to contact the vulval cells. This state likely mimics 2D culture conditions, where numerous holes are generated by tumor cell invadopodia on matrix covered glass slides (Martin et al. 2012).
Studies on AC invasion have also uncovered new mechanisms of how UNC-40 (DCC) receptors polarize toward sources of netrin. Surprisingly, UNC-40 (DCC) is still active in animals lacking UNC-6 (netrin): UNC-40 receptors randomly cluster in the cell membrane, recruit F-actin effectors, and generate F-actin. These transient clusters then break down and reform in a new location in an oscillatory cycle (Wang et al. 2014b). UNC-6 (netrin), which is localized in the BM and below it (secreted by the ventral nerve cord and later by the vulval cells) stabilizes UNC-40 (DCC) clustering toward the source of UNC-6 (netrin), thus directing protrusion formation through the BM and between the vulval cells (Wang et al. 2014b). This oscillatory behavior is likely a mechanism that allows UNC-40 (DCC) receptors to rapidly and robustly polarize toward sources of UNC-6 (netrin) and is probably a universal feature of UNC-40 (DCC) receptor polarization that is shared with other polarity systems (Bendezu and Martin 2013; Dyer et al. 2013; Kulkarni et al. 2013; Wang et al. 2014b).
Optical highlighting of the BM components laminin and type IV collagen revealed that the AC’s invasive protrusion utilizes a combination of proteolysis and physical displacement to both degrade and push aside the BM (Figure 1) (Hagedorn et al. 2013). This was an unforeseen finding, as BM invasion was not known to involve physical forces and instead was thought to rely solely on proteases dissolving the BM to clear a path for invasion (Valastyan and Weinberg 2011). How the AC’s invasive protrusion generates forces to push aside the BM remains an important area of future investigation (see below).
Identification of the transcriptional networks that specify cell invasive behavior
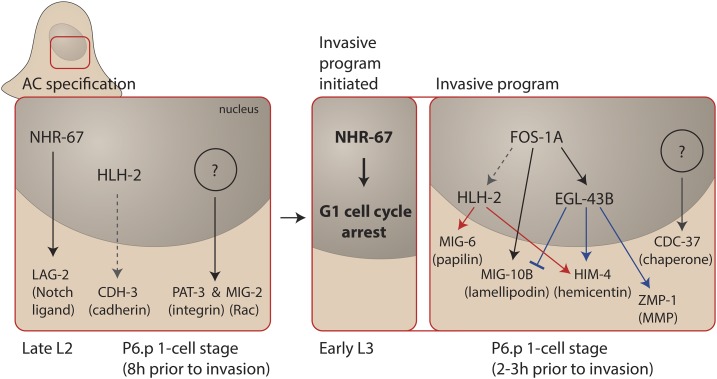
Study of AC invasion has allowed the transcriptional networks to be identified that endow cells with the specialized ability to breach BM (Figure 2). The AC is first specified during the late L2 larval stage (∼8 hr prior to invasion). Specification requires the helix-loop-helix transcription factor protein HLH-2 (Daughterless/vertebrate E proteins) and the nuclear hormone NHR-67 (vertebrate TLX) (Karp and Greenwald 2004; Schindler and Sherwood 2011; Verghese et al. 2011). These transcription factors are expressed prior to AC specification and throughout its differentiation, and appear to belong to a transcriptional network that regulates distinct transcriptional targets at different stages (Schindler and Sherwood 2011). During early specification, NHR-67 is required to express the gene encoding the Notch ligand LAG-2 and HLH-2 promotes expression of the FAT-like cadherin CDH-3 (Schindler and Sherwood 2011; Verghese et al. 2011), while unknown factors are responsible for upregulation of other invasive genes encoding the β-integrin subunit PAT-3, and the Rho GTPase MIG-2 (Sherwood and Sternberg 2003; Sherwood et al. 2005; Ziel et al. 2009).
Figure 2.
Transcription factors that specify invasive cell fate. Left: The AC is specified during the late L2 stage by the action of several transcription factors, including NHR-67 (Tailless) and HLH-2 (Daughterless/E proteins) that drive the expression of genes encoding LAG-2 (Notch ligand) and CDH-3 (cadherin), respectively, as well as other unknown transcription factors that promote the expression of genes encoding PAT-3 (β-integrin) and MIG-2 (Rac). Right: During the early L3 stage NHR-67 directs the AC into G1 cell cycle arrest, which allows full invasive fate differentiation. A central transcription factor that operates following G1 arrest is FOS-1A (Fos), which promotes the expression of the transcription factors HLH-2 and EGL-43B (EVI1). These transcription factors regulate the expression of several invasion effector genes that encode MIG-6 (papilin), MIG-10B (lamelipodin), HIM-4 (hemicentin), and ZMP-1 (MMP). Other transcription factors remain to be discovered as genes encoding proteins such as CDC-37 (Hsp90 cochaperone) are expressed in the AC and promote invasion, but are not regulated by the FOS-1A transcriptional network.
After the AC is specified, NHR-67 (TLX) transcription factor expression is further upregulated and induces G1 cell-cycle arrest (Matus et al. 2015). During development, G1 cell-cycle arrest is thought to allow cells to engage unique transcriptional programs to facilitate differentiation (Gonzales et al. 2015; Ruijtenberg and van den Heuvel 2015). This also appears to be the case in the AC as cell-cycle arrest is required for the AC to express genes associated with invasion and adopt the specialized features of an invasive cell. In the absence of G1 arrest, early aspects of AC specification occur normally, including expression of genes encoding PAT-3 (integrin) and CDH-3 (cadherin); however, later stages of invasive differentiation fail to occur. For example, the AC does not express genes encoding actin regulators such as the formin EXC-6 and the Ena/VASP ortholog UNC-34, the matrix metalloproteinases ZMP-1, -3 and -6, and the matrix component HIM-4 (hemicentin). In addition, invadopodia do not form (Matus et al. 2015). G1 cell cycle arrest might be a common feature of invasive cells, as invasive ability is correlated with G1 arrest in other developmental events involving BM transmigration and with invasive tumor cells (Kohrman and Matus 2017). This may be particularly important in treating metastatic cancers. As most chemotherapy drugs target dividing cells (Yano et al. 2014), such treatments would not be effective against nondividing invasive cells, and could select for more aggressive tumors by leaving invasive cells unscathed and later able to proliferate and seed new metastatic lesions.
G1 arrest is thought to permit cell-cycle dependent alterations in chromatin that allow the expression of differentiation genes (Ma et al. 2015). There is evidence in the AC that this might be mediated through the histone deacetylase HDA-1 and the zinc finger protein MEP-1, a component of the nucleosome remodeling NuRD complex. HDA-1 and MEP-1 are required for AC invasion and the expression of genes associated with later aspects of AC differentiation (Matus et al. 2010, 2015). An emerging transcription factor network is being identified that acts during G1 arrest (Figure 2). This network includes the conserved bZIP transcription factor FOS-1A (Fos), HLH-2 (Daughterless/vertebrate E proteins), and the zinc finger protein EGL-43B (vertebrate EVI1). These transcription factors regulate the expression of genes encoding invasion effectors such as matrix metalloproteinases (e.g., ZMP-1), extracellular matrix proteins [e.g., MIG-6 (papilin) and HIM-4 (hemicentin)], and actin cytoskeleton proteins [e.g., MIG-10B (lamellipodin); Sherwood et al. 2005; Hwang et al. 2007; Rimann and Hajnal 2007; Schindler and Sherwood 2011; L. Wang et al. 2014]. Interesting aspects of this network are beginning to emerge, such as an incoherent feedforward circuit where FOS-1A positively controls expression of the genes encoding MIG-10B and the transcription factor EGL-43B, while the EGL-43B protein negatively regulates mig-10b gene expression. Such networks likely provide fine control over the expression of key effector targets that promote invasion (L. Wang et al. 2014). The vertebrate Fos family of transcription factors are strongly implicated in promoting cell motility and invasion in normal development and multiple tumor types (Milde-Langosch 2005; Ozanne et al. 2007; Renaud et al. 2014). Further, vertebrate E proteins (HLH-2 orthologs) promote epithelial-mesenchymal transitions (EMTs) (Lamouille et al. 2014), which often involve breaching epithelial BMs. Thus, the transcriptional networks that program invasiveness might be conserved. Additional transcriptional mechanisms controlling AC invasion remain to be discovered, as many pro-invasive genes, such as those encoding MIG-2 (Rac), PAT-3 (integrin) and CDC-37 (Hsp 90 cochaperone), are upregulated in the AC, but are not controlled by any known transcriptional regulators (Sherwood et al. 2005; Matus et al. 2010; Schindler and Sherwood 2011).
The AC and vulval cells collaborate to further widen the BM opening
Following AC invasion, the BM opening enlarges past the edge of the AC. Widening the BM gap is crucial to allow direct attachment between the uterine and vulval cells that form the mature uterine-vulval connection. Optical highlighting of the BM components laminin and type IV collagen revealed that the BM moves over the growing vulval and uterine tissues to widen the BM gap through a newly described morphogenetic mechanism termed BM sliding (Ihara et al. 2011). It is thought that the rapid growth of the uterine and vulval tissue (approximately twofold growth during time of sliding) applies forces on the BM that drive its shifting (Ihara et al. 2011). In addition, the vulval cells invaginate, and move through the BM opening, which may further shift BM position (Ihara et al. 2011; Schindler and Sherwood 2013).
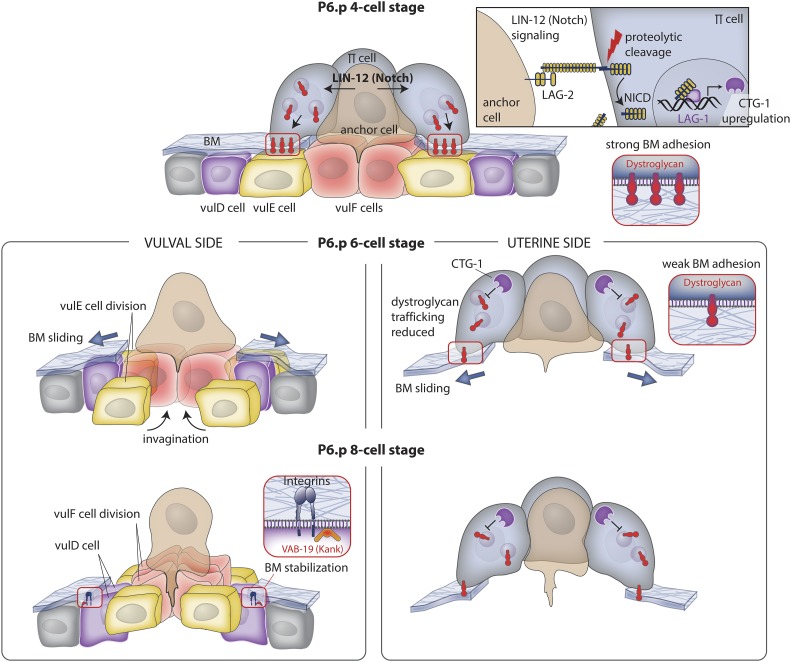
Both vulval cells and uterine cells act to control BM shifting to precisely enlarge the opening in the BM (Figure 3). On the vulval side, the centrally positioned vulE and vulF cells divide, and the rounding of the dividing cells causes these cells to lose contact with the BM, allowing the BM to slide over them to expand the BM gap (Matus et al. 2014). The BM position stabilizes on the nondividing vulD cell, where the integrin adhesion receptor INA-1/PAT-3 and VAB-19 (a cytosolic adaptor protein and ortholog of the vertebrate tumor suppressor Kank) localize at high levels to the vulD-BM interface, locking the position of the BM gap boundary (Ding et al. 2003; Ihara et al. 2011). The vulD cell is the only vulval cell that does not divide during the time of BM sliding in all rhabditid nematodes that have been examined (Kiontke et al. 2007), a group of nematodes that last shared a common ancestor 240–430 MYA (Dieterich et al. 2008). Examination of over 20 of these rhabditid species revealed that the BM gap always stabilizes over the vulD cell, suggesting that control of vulval cell division is a robust and evolutionary conserved mechanism to control BM gap enlargement.
Figure 3.
BM sliding following AC invasion widens the breach. Top panel (P6.p 4-cell stage): during invasion, the AC activates LIN-12 (Notch) signaling in neighboring uterine π cells via the ligand LAG-2 (Delta; see upper right box). Notch activation leads to proteolytic release of the LIN-12 (Notch) intracellular domain (NICD), which enters the nucleus, associates with LAG-1 (CSL), and upregulates expression of the gene encoding CTG-1 (Sec14-GOLD protein). Middle panel left (P6.p 6-cell stage): vulval and uterine tissue growth and vulval cell invagination apply forces on the BM that drive its shifting. VulF cells begin to invaginate and the vulE cells divide, lose contact with the BM, and allow the BM to slide over these cells, thus widening the BM gap. Middle panel right: CTG-1 activity in the uterine π cells inhibits the trafficking of the BM adhesion receptor dystroglycan to the cell-BM interface, weakening BM adhesion, and allowing the BM to move on the uterine side of the BM. Lower panel left (P6.p 8-cell stage): the vulF cells divide and further invaginate with the vulE cells. The BM stops shifting over the nondividing vulD cell, which sets the width of the opening of the BM gap. Integrin and VAB-19 (KANK) localize to the vulD-BM interface to stabilize BM adhesion. Lower panel right: dystroglycan levels continue to be reduced at the interface, allowing the BM to slide to a position determined by the underlying vulD cell.
The invading AC and neighboring uterine cells also have a role in promoting BM gap enlargement. During its invasion, the AC signals with the transmembrane Notch ligand LAG-2 to neighboring uterine π cells through the Notch receptor LIN-12 to upregulate the expression of the gene ctg-1, which encodes a Sec14 family phosphatidylinositol-transfer protein (Tripathi et al. 2014; McClatchey et al. 2016). During LIN-12 (Notch) activation, the intracellular domain of LIN-12 (Notch) is proteolytically cleaved (Notch intracellular domain, NICD) and enters the nucleus, where it forms a complex with the DNA binding protein LAG-1 (CSL), which promotes LIN-12 (Notch) effector gene expression (Greenwald 2005). The ctg-1 gene contains 19 putative LAG-1 binding sites, strongly suggesting it is a direct target of LIN-12 (Notch) signaling (Yoo et al. 2004). The CTG-1 protein limits the trafficking of the BM-adhesion receptor dystroglycan to the uterine cell-BM interface, allowing the BM to slide over the uterine cells to the position determined by the nondividing vulD on the other side of the BM (Figure 3) (Matus et al. 2014; McClatchey et al. 2016). The temporally coordinated mechanisms of tissue growth, movement, division, dystroglycan receptor downregulation in uterine cells, and integrin and VAB-19 upregulation in vulval cells at the BM gap edge act to precisely position the BM gap boundary to allow direct uterine-vulval tissue attachment.
The shifting of cell-BM interfaces has been observed in several important morphogenetic events in other organisms, including intestinal epithelial renewal, BM deposition, and branching morphogenesis (Haigo and Bilder 2011; Clevers and Batlle 2013; Harunaga et al. 2014). Thus, the mechanisms that slide the BM after AC invasion might be used in other important developmental processes to mediate BM remodeling. Further, as dystroglycan loss is a common occurrence in the progression of many epithelia cancers, such as breast and colon (Sgambato et al. 2003; Cross et al. 2008), it is possible that its loss in cancer allows BM openings to widen further, permitting more extensive tumor cell spread.
AC invasion: important unanswered questions
MMP expression is strongly associated with cell invasion in normal development and cancer (Overall and Kleifeld 2006; Srivastava et al. 2007; Page-McCaw 2008). Further, experimental work with in vitro and ex vivo models suggest MMPs are essential for BM transmigration (Rowe and Weiss 2008). As a result of their strong association with invasion, MMPs have been targeted in extensive clinical trials, which were unfortunately not effective for reasons that remain unclear (Zucker et al. 2000; Coussens et al. 2002; Dufour and Overall 2013), but could be due to changes in cancer cell invasion strategy (Te Boekhorst and Friedl 2016). There are over 20 encoded MMPs in vertebrate genomes, making genetic assessment of their necessity for invasion unfeasible (Rowe and Weiss 2008). In comparison, only six MMPs are encoded in the C. elegans genome (Altincicek et al. 2010), and it should be possible to determine if AC invasion can occur in their absence. Given the finding that the AC invades, in part, by displacing BM, it is possible that invasive cells may be capable of invading through solely physical means. An important future direction that should be addressed using the AC invasion model is to determine the necessity of MMPs during BM invasion.
It is also unclear how the invasive protrusion rapidly expands and generates forces to displace the encircling BM and vulval tissue. Actin-binding proteins are known to be important for cellular force production (Blanchoin et al. 2014), but it is unclear whether actin polymerization drives the expansion of the invasive protrusion in the AC, or what actin-binding proteins are involved in protrusion stabilization. The regulation of membrane dynamics during cell migration and invasion is also poorly understood (Lecuit and Pilot 2003; Fletcher and Rappoport 2010; Hastie and Sherwood 2016), and it is not known if invasive cells expand or alter their plasma membranes to breach BM barriers. The highly stereotyped nature of AC invasion should allow for the study of AC membrane addition and how it is regulated. Given the rapid enlargement of the protrusion, it will also be interesting to explore if aquaporins and ion channels play a role by generating hydrostatic forces that displace the BM (Schwab and Stock 2014).
It is unknown how the AC transitions rapidly from AC-BM adhesion prior to invasion to AC-vulval precursor cell (cell–cell) adhesion following invasion. Despite extensive screening, we have not found an adhesion system that mediates this connection (D. R. Sherwood, unpublished data). Interestingly, recent work has shown that when the AC adheres to the vulval precursor cells it stimulates the recruitment of the F-BAR-domain protein TOCA-1 to the vulval precursor cell-AC interface. TOCA-1 concentrates nonmuscle myosin NMY-2 at this cell surface and reorients contractile forces, which constricts the lateral membrane of the vulval precursor cells thus reshaping them (Yang et al. 2017). These observations indicate that invasive cells can also induce cell shape changes, which could possibly facilitate tissue invasion.
Finally, single-cell isolation procedures involving cell dissociation and fluorescence activated cell sorting (FACs) have been developed in C. elegans to determine the expression profile of individual cells (Spencer et al. 2014). Applied to the AC, this approach could reveal the first complete expression signature of an actively invading cell. By combining AC RNAseq studies with transcription factor mutants, it will be possible to carry out a comprehensive analysis of the transcriptional networks that direct invasive behavior, thus revealing how cells are programed to invade.
Distal Tip Cell Migration: a Leader Cell Migration that Shapes a Tissue
The distal tip cells (DTCs) are a pair of somatic gonad cells born during the L1 stage that act as leader cells in hermaphrodites, tracing out the symmetrical double lobed gonad through their migration trajectory (Kimble and Hirsh 1979; Kimble and White 1981). The ease of identifying defects in gonad shape with a dissecting microscope has led to the isolation of numerous genes regulating DTC migration through forward genetic (many termed mig, for Migration defective) as well as RNAi, screens (Hedgecock et al. 1987, 1990; Nishiwaki 1999; Cram et al. 2006; Suzuki et al. 2006). A similar leader cell function is mediated by the linker cell in the male C. elegans gonad, which establishes a single lobed J-shaped gonad (Kimble and Hirsh 1979). The migration of the linker cell has not been studied in as much detail as the DTC, but appears to be regulated by many, but not all, of the same genes controlling DTC movement (Kato and Sternberg 2009).
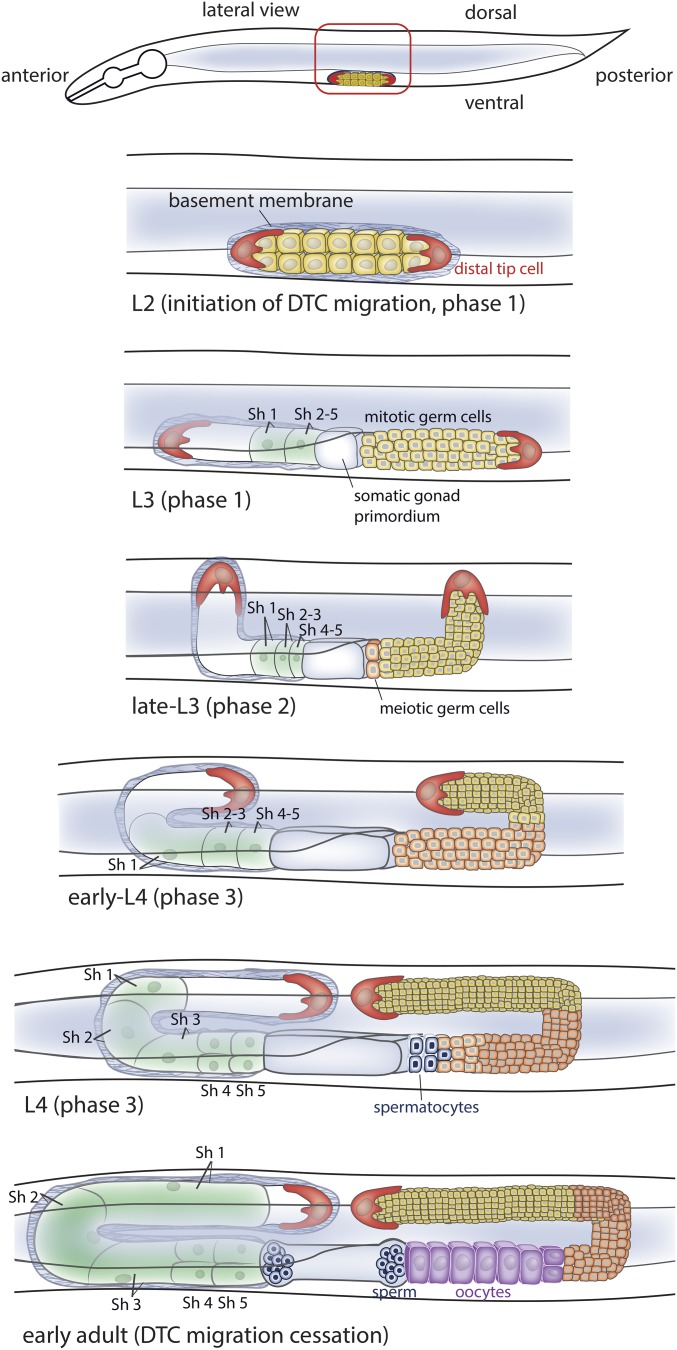
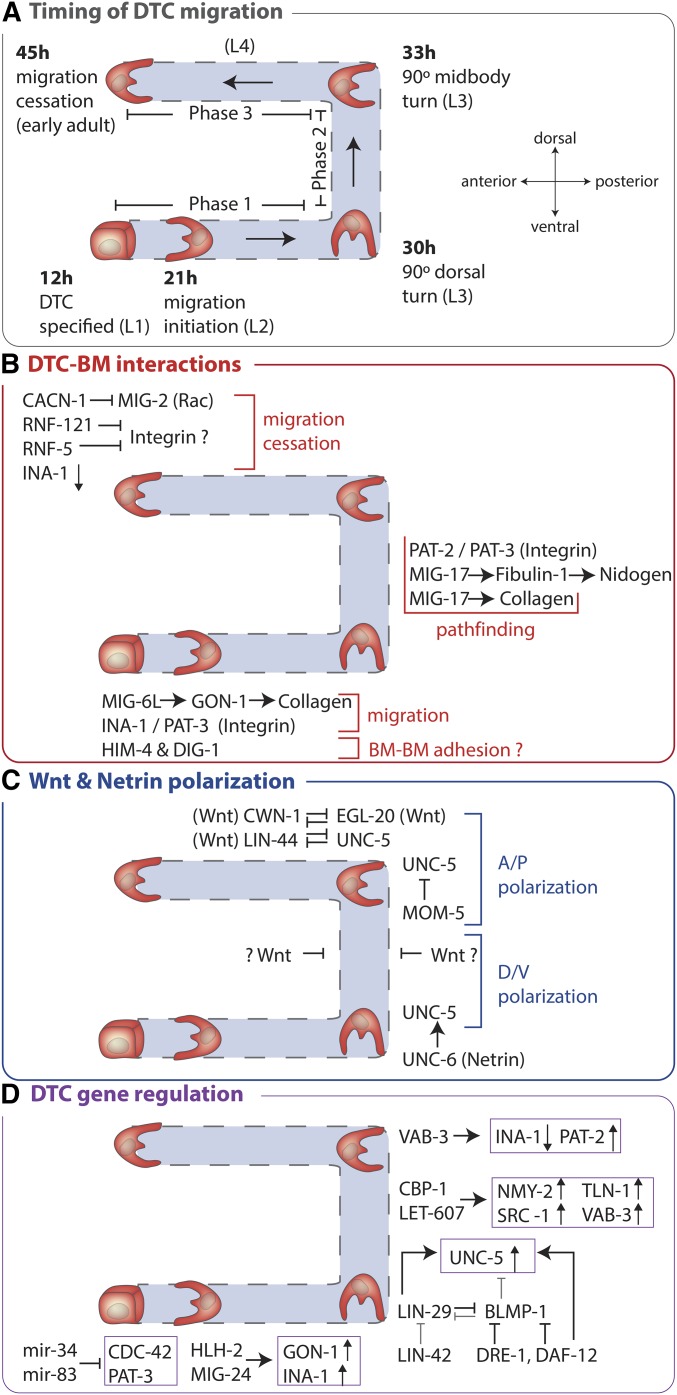
The DTCs initiate migration during the L2 stage, and continue moving at a variable rate of ∼6–10 µm/hr until the early adult stage, a duration of ∼25 hr (Lints and Hall 2017). The two DTCs, one in each gonad arm, are located at the anterior and posterior ends of the gonad, and their mirror imaged migration has been classified into three phases (Figure 4 and Figure 5A) (Hedgecock et al. 1987; Nishiwaki 1999): (phase 1) During the L2 and early-to-mid L3 stage, the DTCs move ventrally away from each other along the ventral body wall muscle BM toward the anterior (head) and posterior (tail) of the animal; (phase 2) during the late-L3 stage, both DTCs turn 90° and move from the ventral to dorsal surface, migrating along the BM that covers the lateral epidermis; (phase 3) during the L4 stage, the DTCs turn 90° and move back to the midsection along the BM covering the dorsal body wall muscles. Together, these different phases of migration build a gonad with two equivalent U shaped arms (Figure 4 and Figure 5A).
Figure 4.
DTC migration, a leading cell that shapes an organ. The pair of DTCs (one on the anterior the other on the posterior arm of the basement membrane enwrapped gonad) initiate migration at the L2 larval stage. During the L2 and L3 larval stages (phase 1 of migration), the DTCs move ventrally away from each other along the BMs of the ventral body wall muscles (data not shown) toward the anterior (head) and posterior (tail) of the animal. In the rest of the figure, the posterior gonad arm (right side) shows the germ cells from the L3 stage onward, while the anterior arm (left side) shows the basement membrane and five pairs of sheath cells (sh1-5) that cover the germ cells. The sheath cells follow the path of the DTCs. The digestive tube is shown in light gray and the anterior gonad arm passes underneath it to the other side of the animal. During the late-L3 stage, both DTCs turn 90° and move from the ventral to dorsal surface (phase 2 of migration), moving along the BM of the lateral epidermis. At the early L4 stage, the DTCs turn 90° and move back to the midsection along the BM of dorsal body wall muscles during the L4 stage (phase 3 of migration). DTC migration ceases in the early adult.
Figure 5.
DTC migration timing, BM interactions, polarization, and gene regulation. Only the posterior gonad arm is shown. (A) The timing of DTC migration shown is at 20°. (B–D) Details of proteins and interactions that regulate DTC migration are described in the text and outlined here for a global view. Note, the “Wnt ⊣” shown in (C) represents a hypothetical possible function for Wnt in inhibiting polarization along the anterior-posterior axis during phase 2 of DTC migration.
The DTC is shaped as a smooth cap that extends over the most distal three-to-five germ cells during its migration (Hall et al. 1999; Lints and Hall 2017). Five pairs of sheath cells encase the remaining germ cells and follow the path of the DTCs (Figure 4) (Killian and Hubbard 2005). The entire growing gonad, including the DTCs and sheath cells, are surrounded by a BM (Hall et al. 1999; C. C. Huang et al. 2003). The germ cells are thought to follow the DTC through extensive cell divisions, expanding from two cells at the beginning of larval development to ∼1000 at the young adult stage. The proliferation of the germ cells might help extend the distal gonad arms (Kimble and White 1981; Killian and Hubbard 2005). Although not quantified, loss of germline proliferation has not been reported to alter the DTC migration path (and thus the shape of the gonad), but the distance of DTC movement during the third phase of migration appears to be reduced (Austin and Kimble 1987). It is unclear how the sheath cells follow the DTC. They are thought to be either pulled along by the dividing germ cells or to actively migrate over the germ cells (Lints and Hall 2017). Interestingly, the DTC extends a single forward directed protrusion only during the initiation of the dorsal turn that begins the second phase of migration, and, otherwise, does not appear protrusive during its migration (Kim et al. 2011). In many respects, DTC migration has similarities to collective cell migration events in vertebrates that build branching tubular organs (Andrew and Ewald 2010). Very little is known about how leader cells in morphogenetic branching events migrate while enwrapped in tough, dense sheets of BM (Friedl and Wolf 2010). Such branching morphogenetic programs are thought to be misregulated in some epithelial tumors, allowing groups of cancer cells to collectively invade neighboring tissues (Friedl and Gilmour 2009; Gray et al. 2010). Thus, understanding DTC migration has significance to mechanisms underlying organogenesis and potentially some invasive cancers.
DTC modification of and interaction with BM during migration
The DTC expresses many BM components during its migration, including type IV collagen (C. elegans emb-9/collagen IV α1 and let-2/collagen IV α2), laminin (epi-1/laminin αB and lam-1/laminin β), nidogen (nid-1), and agrin (agr-1) (Graham et al. 1997; Kim and Wadsworth 2000; C. Huang et al. 2003; Hrus et al. 2007; Clay and Sherwood 2015), suggesting that the DTC secretes, deposits, and remodels the BM that surrounds it during its migration. Reduction of laminin and type IV collagen severely hampers DTC migration, indicating the important role of BM in DTC movement (C. Huang et al. 2003; Kao et al. 2006; Kubota et al. 2008; Kawano et al. 2009; Wong and Schwarzbauer 2012).
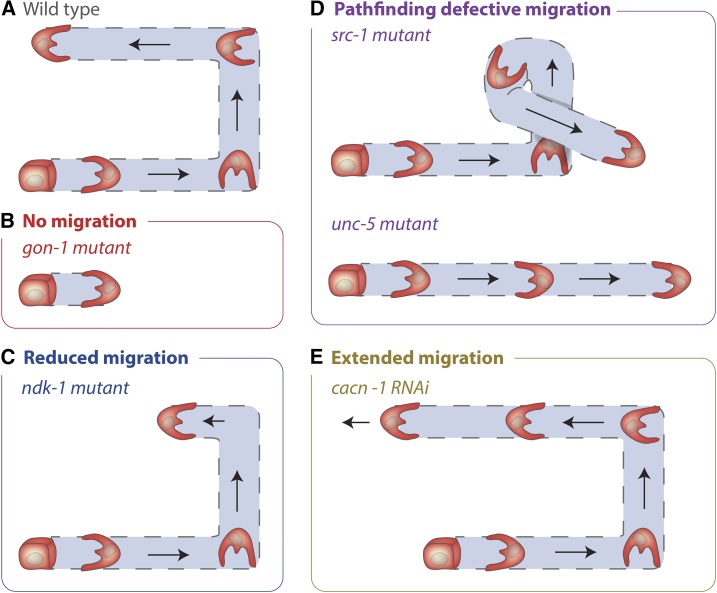
Throughout its migration the DTC expresses and secretes GON-1, an ortholog to the vertebrate proteases ADAMTS-9 and ADAMTS-20 (A Disintegrin And Metalloprotease with ThromboSpondin repeats protein) (Blelloch and Kimble 1999; Llamazares et al. 2003; Somerville et al. 2003). GON-1 is essential to facilitate DTC migration (Figure 5B) (Blelloch et al. 1999). In animals harboring null alleles of the gon-1 gene, or rescue constructs lacking the metalloprotease catalytic domain, the DTC fails to migrate (Figure 6, A and B) (Blelloch et al. 1999; Blelloch and Kimble 1999). The activity of GON-1 may be controlled by MIG-6L, a DTC expressed isoform of the conserved extracellular matrix protein papilin (Kawano et al. 2009). Genetic reduction of mig-6(l) gene activity leads to a similar defect in DTC migration as loss of the ADAMTS gon-1 (Cram et al. 2006; Kawano et al. 2009). In Drosophila, the ortholog of the MIG-6 protein, papilin, binds to and regulates the activity of a Drosophila ADAMTS collagenase (Kramerova et al. 2000). Genetic studies indicate that type IV collagen and the matrix protein fibulin-1 (C. elegans FBL-1) oppose GON-1 function (Hesselson et al. 2004; Kubota et al. 2012). As fibulin-1 maintains collagen in the BM (Kubota et al. 2012), an attractive model is that the ADAMTS GON-1 cleaves collagen to facilitate DTC movement and BM expansion during DTC migration (Figure 5B).
Figure 6.
Examples of DTC migration defects. Only the posterior gonad arm is shown. (A) Wild type migration, (B) a gon-1 mutant where no DTC migration occurs, (C) an ndk-1 mutant where DTC migration is incomplete, (D) a src-1 mutant and an unc-5 mutant where DTC migration shows pathfinding defects, (E) reduction of cacn-1 by RNAi leads to extended DTC migration (a cessation of migration defect).
Another ADAMTS protein, MIG-17, also regulates DTC migration (Nishiwaki et al. 2000). MIG-17 is secreted from the ventral and dorsal body wall muscle cells and accumulates in the gonadal BM during the L3 stage, where it regulates the directional migration of the DTC (Figure 5B) (Nishiwaki et al. 2000). In the absence of MIG-17, DTC migration occurs, but the DTC meanders and the gonad has an abnormal shape (Nishiwaki 1999). BM localization and the catalytic domain of MIG-17 are required for its function, suggesting that MIG-17 may cleave BM targets (Ihara and Nishiwaki 2007). MIG-17 recruits and activates (perhaps by proteolysis) an isoform of fibulin (fibulin-1C), which then recruits the BM protein nidogen to the gonad (Kubota et al. 2004, 2008). Based on genetic interactions, MIG-17 has also been proposed to modify type IV collagen to promote directional migration (Kubota et al. 2008). These observations imply that dynamic interactions of matrix proteins and proteases continually remodel the BM covering the DTC in a manner that both allows DTC migration and helps direct its path. ADAMTS proteins have complex functions in vertebrate tissue formation and maintenance, and mutations or misregulation of ADAMTS proteins are associated with numerous diseases, including cancer and arthritis (Cal and Lopez-Otin 2015; Kelwick et al. 2015). Understanding the functions of these proteases during DTC migration will likely provide insight into their roles in vertebrate tissue formation, and clues as to how their misregulation contributes to human diseases.
Several transmembrane proteins that act as BM receptors, or are strongly associated with BM, regulate DTC migration. These include the C. elegans orthologs of the proteins teneurin (C. elegans TEN-1), dystroglycan (DGN-1), and integrin (Baum and Garriga 1997; Lee et al. 2001; Drabikowski et al. 2005; Trzebiatowska et al. 2008; Topf and Chiquet-Ehrismann 2011). Of these, only the function of integrin has been carefully characterized during DTC migration. Both C. elegans integrins, αINA-1/βPAT-3 and αPAT-2/βPAT-3, are expressed in the DTC and regulate its migration (Baum and Garriga 1997; Lee et al. 2001; Meighan and Schwarzbauer 2007). The α-integrin INA-1 is expressed in the DTC prior to, and during, its migration, and promotes DTC motility, but does not appear to play a major role in DTC pathfinding (i.e., the direction of movement) (Baum and Garriga 1997). INA-1 expression is downregulated when DTC migration ceases and failure to turn off ina-1 gene expression results in DTCs that continue to migrate in adults (Meighan and Schwarzbauer 2007). Termination of migration is also regulated by the E3 ubiquitin ligases RNF-121 and RNF-5, which can target PAT-3 and a protein with similarity to the integrin effector paxillin (UNC-95) for degradation (Broday et al. 2004; Darom et al. 2010; Kovacevic et al. 2012). However, the specific targets of RNF-121 and RNF-5 that promote cessation of DTC migration are not yet clear. CACN-1, a conserved protein and component of the spliceosome, is also required to halt DTC migration (Figure 5B and Figure 6E). Genetic studies suggest CACN-1 inhibits the Rac GTPase MIG-2 (Tannoury et al. 2010; Doherty et al. 2014), although the precise mechanism of MIG-2 downregulation is not known.
The α-integrin PAT-2 is first expressed in the DTC during phase 2 of migration after the DTC turns dorsally, and is required for correct pathfinding along the dorsal body wall muscle BM surface during phase 3 of DTC migration. Knockdown of key components of the retrograde vesicular trafficking pathway has suggested that the polarized trafficking of integrin to the leading edge of the DTC is important for DTC pathfinding (Shafaq-Zadah et al. 2016); however, the effects of these knockdowns on the localization of the pathfinding integrin PAT-2 have not been reported. The requirement for both INA-1/PAT-3 and PAT-2/PAT-3 integrin heterodimers during phase 3 of DTC migration suggests that each simultaneously promotes distinct activities within the DTC, either via separate extracellular ligands or through different intracellular signaling partners (Meighan and Schwarzbauer 2007). Interestingly, there appears to be feedback between the amount of PAT-3 (β-integrin) in the DTC and the levels of type IV collagen in the BM, as lower concentrations of collagen in the BM decrease the levels of a PAT-3::GFP translational reporter (Kubota et al. 2012). These observations suggest that dynamic interactions between the composition of the BM and the receptors that bind to it regulate DTC migration.
Many known integrin downstream effectors mediate DTC migration and may function downstream of INA-1/PAT-3 and PAT-2/PAT-3. These include C. elegans orthologs of talin, kindlin, ILK, the tyrosine-protein kinase Src, the Rho GTPases Rac (C. elegans MIG-2 and CED-10), Cdc42, associated Rho GTPase regulators and effectors such as the nucleoside-diphosphate kinase NDK-1, and a GIT/PIX/PAK signaling pathway (Reddien and Horvitz 2000; Lundquist et al. 2001; Cram et al. 2003; Itoh et al. 2005; Xu et al. 2006; Meighan and Schwarzbauer 2007; Lucanic and Cheng 2008; Wong and Schwarzbauer 2012; Fancsalszky et al. 2014). Loss of most integrin effectors perturbs both DTC motility and pathfinding; however, some effectors predominantly alter motility (e.g., NDK-1; Figure 6C), while others pathfinding (e.g., SRC-1, Figure 6D) (Itoh et al. 2005; Fancsalszky et al. 2014). Thus, the INA-1 and PAT-2 integrins might achieve at least some of their respective roles in DTC motility and pathfinding through engagement with distinct effectors. Despite the characterization of many effectors, it remains unclear if integrin activity promotes DTC motility and directional migration through known roles in polarization, vesicular trafficking, BM deposition, adhesion strength, or cytoskeletal dynamics (Bokel and Brown 2002; Harburger and Calderwood 2009; Vicente-Manzanares et al. 2009; Huttenlocher and Horwitz 2011; Wickstrom and Fassler 2011; Yurchenco 2011) or if INA-1 and PAT-2 integrins act through novel mechanisms.
Stabilizing the DTC path: the matrix proteins HIM-4 (hemicentin) and DIG-1
Study of DTC migration and gonad formation has also revealed possible mechanisms that stabilize tissue positioning—a poorly understood aspect of morphogenesis. The DTC and gonad are encased in BM, and migrate along the BMs of the body wall muscle and epidermis, thus forming a BM–BM interface where the gonad must likely adhere to maintain its position within the body cavity. A potential molecule that stabilizes gonad positioning is HIM-4 (hemicentin), a large (>5000 amino acids) extracellular matrix protein of the immunoglobulin superfamily that is expressed by the DTC from the L1 stage and throughout its migration (Vogel and Hedgecock 2001). HIM-4 localizes between BMs of neighboring tissues in C. elegans and vertebrates (Vogel and Hedgecock 2001; Xu et al. 2007), and has recently been shown to connect neighboring BMs (Morrissey et al. 2014). Consistent with a possible role in stabilizing the position of the gonad along the neighboring BMs, loss of HIM-4 (hemicentin) can result in the ventral gonad detaching from the underlying body wall muscle BM (Vogel and Hedgecock 2001). Another secreted matrix associated molecule, DIG-1, which is a giant member of the immunoglobulin superfamily (>13,000 amino acids), might also mediate BM-BM interactions (Benard et al. 2006; Burket et al. 2006). In dig-1 mutants the entire gonad is often displaced from its normal position (most often relocated anteriorly). Strongly indicative of a possible role in BM–BM adhesion, the gonads of dig-1 mutants can be shifted within the body cavity by mechanical manipulation (Thomas et al. 1990; Burket et al. 2006). Reporter constructs indicate that the dig-1 gene is expressed in many muscles surrounding the gonad, but its expression has not been reported in the DTC (Benard et al. 2006).
Diffusible cues help orient DTC migration: netrin and Wnts
Studies on DTC migration have helped reveal a coordinated dorso-ventral (D/V) and anterior-posterior (A/P) positioning system within the worm involving the diffusible netrin and Wnt cues (Figure 5C). These navigation signals orient numerous migratory and cell outgrowth behaviors in vertebrates and invertebrates (Silhankova and Korswagen 2007; Lai Wing Sun et al. 2011; Hikasa and Sokol 2013), suggesting the navigational system revealed in C. elegans is conserved. Studies in worms have been simplified by the smaller gene families that encode these pathways. For example, vertebrates harbor five genes for netrin ligands, whereas C. elegans has only the UNC-6 protein (Lai Wing Sun et al. 2011). C. elegans netrin (UNC-6) is secreted from ventral cells and serves as an attractive source for cells expressing the transmembrane receptor UNC-40 (vertebrate DCC). UNC-6 (netrin) can also function as a repulsive cue for cells expressing the netrin receptor UNC-5 either alone or with UNC-40 (DCC) (Hedgecock et al. 1990; Ishii et al. 1992; Leung-Hagesteijn et al. 1992; Chan et al. 1996). C. elegans encodes four Wnt frizzled receptors (CFZ-2, LIN-17, MIG-1, and MOM-5), and five Wnt ligands (CWN-1, CWN-2, EGL-20, LIN-44, and MOM-5) (Eisenmann 2005), simplifying analysis as compared to mammalian genomes, which encode 19 Wnt proteins (Clevers and Nusse 2012; Sawa and Korswagen 2013). The C. elegans Wnt ligands have distinct graded distributions along the A/P axis, which can polarize cells (Whangbo and Kenyon 1999; Goldstein et al. 2006; Hilliard and Bargmann 2006; Pan et al. 2006; Levy-Strumpf 2016), thus providing a system for cells to assess positional information along the A/P axis.
Netrin and Wnts act as key directional signals for DTC migration along the body axes. For example, expression of the netrin receptor UNC-5 is activated in the DTC just prior to phase 2, and its expression helps initiate the 90° DTC dorsal turn and migration away from ventral UNC-6 (Figure 5C) (Su et al. 2000). In the absence of UNC-5, UNC-6 (netrin), or UNC-40 (DCC), both the anterior and posterior DTCs often fail to turn dorsally, and instead continue to migrate along the ventral body wall muscle (Figure 6D (Hedgecock et al. 1990)). The levels of the glycosaminoglycan chondroitin may regulate netrin signaling, as chondroitin synthase (SQV-5) and its cofactor chondroitin polymerizing factor (MIG-22) promote UNC-5 mediated dorsal DTC migration (Suzuki et al. 2006). In contrast to loss of netrin, mutations in Wnt pathway components lead to A/P migration defects during the third phase of DTC migration (Cabello et al. 2010; Levy-Strumpf and Culotti 2014). Genetic studies suggest that anterior and posterior localized Wnts (anterior CWN-1 and LIN-44 and posterior EGL-20) oppose each other’s activity to provide a precise positioning system to orient the posterior DTC along the A/P axis (Figure 5C). Similarly, the anterior expressed Wnt CWN-2 and Wnt inhibitor SFRP-1 and the posteriorly expressed Wnts LIN-44 and CWN-1 help direct A/P axis polarization of the anterior DTC during the third phase of migration (Levy-Strumpf and Culotti 2014). Whether Wnts act to polarize phase 1 of migration is unclear. Cross talk between the netrin and Wnt pathways is also important. The Wnt receptor MOM-5 is upregulated in both DTCs at the end of phase 2, and downregulates the UNC-5 receptor to ensure proper polarized migration of both the DTCs in phase 3 (Figure 5C) (Levy-Strumpf et al. 2015).
Notably, netrin and Wnt pathway mutants show only partially penetrant DTC polarity defects, suggesting that each cue acts with other signal(s) to direct DTC migration. Surprisingly, simultaneous impairment of both the netrin and Wnt pathways reveals that netrin and Wnt act redundantly (i.e., they function together) to polarize DTC migration (Levy-Strumpf and Culotti 2014). Thus, each cue provides polarity information along both axes (Levy-Strumpf and Culotti 2014). How these cues define the axis orthogonal to their graded distribution is not well understood, nor is it clear how they control polarization of both the anterior and the posterior DTCs such that they move in opposite directions during phase 3. There is evidence that Wnt ligands can exclude the polarization machinery along the A/P axis in the HSN neuron during axon outgrowth, thus helping direct polarity along its D/V axis (Kulkarni et al. 2013; Levy-Strumpf 2016). Wnt might act similarly during the dorsal migration of the DTC (Figure 5C). Further, opposing activities of Wnts, which are in graded distributions along the A/P axis, may help precisely position both anterior and posterior DTC migration along this axis (Levy-Strumpf and Culotti 2014). As integrin-BM interactions also impact polarity, and integrin, netrin, and Wnts share several downstream effectors, including Rac (CED-10) and Src kinase (SRC-1) (Gitai et al. 2003; Itoh et al. 2005; Meighan and Schwarzbauer 2007; Harburger and Calderwood 2009; Cabello et al. 2010; Lai Wing Sun et al. 2011; Levy-Strumpf and Culotti 2014; Wang et al. 2014a; Levy-Strumpf 2016), extensive collaboration and cross talk between Wnt, netrin, and integrin signaling likely exists during DTC polarization and migration.
Polarity of the nucleus and cytoskeleton during DTC migration
The nucleus is positioned at the leading front of the DTC throughout its migration. The VAB-10B1 protein, the C. elegans ortholog of the vertebrate cytoskeleton cross-linker spectraplakin, is required to move the nucleus to the leading edge during the dorsal turn of the DTC (phase 2 of migration) (Kim et al. 2011; Suozzi et al. 2012). The VAB-10B1 protein contains both F-actin and microtubule binding motifs, and both modules are required for nuclear translocation during the dorsal turn (Kim et al. 2011). Examination of microtubules and F-actin revealed that F-actin is present in filamentous structures that are loosely aligned along the axis of DTC migration, while microtubules are enriched at the trailing side of the DTC. Visualization of microtubule growth using a plus-end tracking protein indicated that microtubules grow dynamically toward the nucleus and leading edge during DTC migration, whereas microtubule growth in the rear of the DTC appears random. Notably, VAB-10B1 is largely dispensable for formation of polarized F-actin filaments, but is required for organization of the microtubule network within the DTC and polarized microtubule growth toward the nucleus and leading edge. Surprisingly, mutant analysis showed that VAB-10B1 is not required for DTC pathfinding, but is necessary for DTC migration: DTCs in vab-10 mutants migrate more slowly, and the animals have shortened gonad arms (Kim et al. 2011). These findings imply that VAB-10B1 (spectraplakin) and likely polarized microtubule dynamics are components of the engine that drives DTC migration, but they are not involved in the mechanism that orients DTC movement. Instead, the orientation of F-actin fibers along the axis of migration, which is not severely affected by loss of VAB-10B1, might be a component of the DTC orientation mechanism. It will be interesting to further explore if VAB-10B1 (spectraplakin) links mechanisms that orient DTC migration (Wnt, netrin, integrin regulation of F-actin polarity) to those that drive migration (microtubule polarity and polarized dynamic growth). Both C. elegans septin proteins, UNC-59 and UNC-61, are also required for robust DTC movement and pathfinding, although the specific role(s) this cytoskeletal system plays in DTC migration remains to be explored (Finger et al. 2003).
Transcriptional regulation of DTC migration
Transcriptional programing of the DTC plays a crucial role in all aspects of its migration (Figure 5D). As with the AC, an early regulator of the DTC transcriptional program is the basic helix-loop-helix (bHLH) transcription factor HLH-2 (vertebrate E proteins) (Karp and Greenwald 2004; Chesney et al. 2009). HLH-2 is upregulated in the DTC and controls the expression of key genes that help initiate DTC migration, including genes encoding GON-1 (ADAMTS), MIG-6 (papilin), and INA-1 (α-integrin) (Krause et al. 1997; Karp and Greenwald 2004; Cram et al. 2006; Tamai and Nishiwaki 2007; Meighan et al. 2015). bHLH factors function as heterodimers, and HLH-2 physically interacts with the Achaete-Scute bHLH transcription factor family member HLH-12 to control target gene expression in the DTC (Figure 5D). Importantly, gon-1 and ina-1 genes are still expressed, albeit weakly, in the DTC after loss of hlh-2 and hlh-12 (Tamai and Nishiwaki 2007; Meighan et al. 2015), suggesting that other transcription factors are also involved in the initiation of DTC migration. The role of HLH-2 in migration of the DTC, invasion of the C. elegans AC (Schindler and Sherwood 2011), and the role of the vertebrate HLH-2 ortholog E2A in epithelial-to-mesenchymal transition in mammalian cells (Sobrado et al. 2009), suggests a conserved role for HLH-2/E proteins in promoting cell migration and invasion.
The dorsal turn of the DTC depends on the UNC-5 protein, which is upregulated by a conserved circuit of heterochronic genes that controls developmental timing events in the worm (Figure 5D) (Su et al. 2000). This gene circuit involves the Blimp-1/PRDI-BF1 zinc finger transcriptional repressor BLMP-1, the zinc finger EGR (early growth response) family protein LIN-29, the steroid hormone receptor DAF-12, and the F-Box protein DRE-1, which is the key recognition subunit of the SCF ubiquitin ligase complex (Rougvie and Ambros 1995; Antebi et al. 1998, 2000; Su et al. 2000; Fielenbach et al. 2007; Huang et al. 2014). BLMP-1 is a transcriptional repressor that is expressed in the DTCs during the first phase of DTC migration. BLMP-1 binds to the upstream regulatory region of the unc-5 gene and is thought to repress unc-5 transcription. The repression of unc-5 is relieved at the beginning of the L3 stage by the global developmental timing cues of dafachronic acid release and lower levels of LIN-42 (Period protein), which help activate DAF-12 and LIN-29, respectively (Huang et al. 2014; Cecchetelli and Cram 2017). The levels of DRE-1 are also upregulated at the L3 stage (Fielenbach et al. 2007). LIN-29 and DAF-12 repress the transcription of the blmp-1 gene, while DRE-1 targets the BLMP-1 protein for degradation. Together, these activities remove BLMP-1 from the DTC and relieve the repression on unc-5 expression (Figure 5D). Interestingly, prior to its removal from the DTC, BLMP-1 also represses the expression of lin-29, suggesting that a double negative feedback loop between LIN-29 and BLMP-1 might act as a robust switch in the decision to turn on the expression of unc-5 (Huang et al. 2014). In addition, LIN-29 and DAF-12 act cooperatively to promote unc-5 expression (Huang et al. 2014). A consensus DAF-12 binding sequence in the unc-5 promoter suggests that DAF-12 may directly activate unc-5 transcription (Huang et al. 2014). Expression of unc-5 in the DTC triggers the dorsal turn, orienting the AC away from ventral sources of UNC-6 protein (Figure 5C) (Hedgecock et al. 1990; Su et al. 2000). Precocious expression of the unc-5 gene through ectopic expression or loss of blmp-1 function results in early DTCs dorsal turns, indicating the importance of the correct timing of unc-5 gene expression (Su et al. 2000; Huang et al. 2014). The vertebrate ortholog of DRE-1, Fbxo11, mediates the degradation of the pro-oncogene BLC6, which has sequence homology to BLMP-1. Overexpression of BLC6 is implicated in the pathogenesis of human B-cell lymphomas (Ci et al. 2008). Fbxo11 is deleted in diffuse large B-cell lymphomas, and, as a consequence, BLC6 expression is upregulated (Duan et al. 2012). BLC6 has recently been implicated in other cancers, including leukemia, breast cancer, and nonsmall-cell lung cancer (Cardenas et al. 2017). It will be interesting to determine if the DRE-1/BLMP-1 regulatory circuit that controls DTC migration might be a conserved switch that is misregulated in numerous cancers.
Following the dorsal turn, other transcription factors help guide DTC migration (Figure 5D). During the dorsal migration of the DTC (phase 2), VAB-3, a Pax6 transcription factor ortholog, turns on the expression of the α-integrin subunit gene encoding PAT-2, which regulates pathfinding during phase 3 of DTC movement (Meighan and Schwarzbauer 2007). In addition, VAB-3 downregulates the expression of the gene encoding INA-1 (integrin α-subunit), which is necessary to cease DTC migration in the early adult stage (Meighan and Schwarzbauer 2007). Several other transcriptional regulators also control pathfinding during phase 3. These include a CBP/p300 transcriptional coactivator CBP-1, and a CREBH transcription factor LET-607, which are both expressed in the DTC, and whose DTC-specific loss results in defects in the second turn of the DTC back toward the midsection (Wong et al. 2014). Functional DTC transcriptional targets of CBP-1 and LET-607 associated with integrin activity have been identified, including genes encoding SRC-1 (SRC kinase), TLN-1 (talin), NMY-2 (nonmuscle myosin heavy chain II), and PAT-2 (α-integrin subunit). Regulation of pat-2 gene expression is likely indirect, as CBP-1 and LET-607 promote vab-3 expression. Notably, expression of the gene encoding the matrix protein MIG-6 (papilin), and the bHLH transcription factor MIG-34, which promote the initiation of DTC migration, are not regulated by CBP-1 and CREBH (Wong et al. 2014). Together, these observations indicate that distinct sets of transcription factors and transcriptional regulators help direct the different steps of the DTC migration program. Thus, a combination of a dynamic transcriptional program within the DTC, and a complex extracellular environment of BM components, proteases, and diffusible signals (Wnt, netrin) directs and guides the specific path of DTC migration.
microRNAs confer robustness to DTC migration under temperature stress
Because of its stereotyped movement and ease of visual analysis, DTC migration can serve as a model to examine how developmental migration programs are buffered against environmental stresses. A pair of conserved microRNAs, mir-34 and mir-83 (orthologs of mammalian mir-34 and mir-29, respectively) act to ensure robust DTC migration, and appear to be particularly effective at maintaining the correct execution of DTC migration in the face of temperature stress (Figure 5D) (Burke et al. 2015). Loss of both mir-34 and mir-83 microRNAs results in a mild DTC migration defect, specifically affecting phase 1 and phase 3 of migration. Evidence suggests that both microRNAs function in the DTC and together directly suppress translation of the mRNA encoding the β-integrin subunit PAT-3 and one of its downstream effectors, the Rho GTPase CDC-42 (Figure 5D) (Burke et al. 2015). Temperature oscillations between 15 and 25° within a 2-hr window at the time of DTC birth in the L1 larva dramatically enhance the DTC migration defect of animals harboring mutations in mir-34 and mir-83. This suggests, somewhat perplexingly, that mir-34 and mir-83 may act at the birth of the DTC cells in the L1 stage, many hours (∼10–30) before DTC migration occurs in the L2, L3, and L4 stages. Alternatively, these microRNAs might function at the time of DTC migration to buffer gene expression changes set in motion by the earlier temperature oscillations. Misregulation of the vertebrate orthologs of mir-34 and mir-83 are associated with many cancers (Hermeking 2010; Jiang et al. 2014; Yan et al. 2015), and they also appear to function together to coregulate gene regulatory networks, such as a p53 network that promotes apoptosis (Burke et al. 2015). Thus, understanding how the DTC executes accurate migration in the face of stressful environmental conditions will reveal important mechanisms that maintain the fidelity and flexibility of gene regulatory networks in normal development, and the ways in which these networks go awry in human diseases.
DTC migration: key unanswered questions
An important unanswered question in DTC migration relates to how the DTC actually moves. The DTC migrates encased within a BM, a mode of cell migration that is widespread during branching morphogenesis, but poorly understood (Friedl and Wolf 2010). Germline proliferation may help propel DTC movement; however, the precise contribution of the dividing germ cells has not been determined. As mutants such as vab-3 lead to perpetual DTC migration in the absence of apparent germline hyperproliferation (Meighan and Schwarzbauer 2007), it strongly suggests that germline proliferation is not an essential driving force. Further, the male linker cell, which is functionally analogous to the DTC, can migrate when detached from the proliferating germline (Kato et al. 2014). Thus, it seems likely the DTC can move independently of germline proliferation.
It is not yet clear if the DTC employs the two primary modes of cell migration characterized to date—mesenchymal and bleb-based (Charras and Paluch 2008; Lammermann and Sixt 2009; Petrie and Yamada 2012, 2016; Te Boekhorst et al. 2016). During mesenchymal migration, cells extend protrusions through controlled F-actin formation. These protrusions adhere to the cell substrate, while the back end of the cell deadheres and retracts through actomyosin contractility, thus moving the cell or groups of cells forward (Friedl and Gilmour 2009). During bleb-based migration, cells use acto-myosin contractility to create rounded membrane protrusions that direct cell migration by wedging through and extending between spaces within the neighboring microenvironment (Friedl and Wolf 2010; Paluch and Raz 2013). Live cell imaging has indicated that the DTC only produces a single large protrusion during the dorsal turn at the initiation of the second migration phase, and this protrusion is not enriched with F-actin (Kim et al. 2011). Blebbing has also not been observed in the DTC during its movement (Kim et al. 2011). However, loss of GEX-3 a component of the WAVE complex that activates the actin nucleator the Arp2/3 complex (Soto et al. 2002; Shakir et al. 2008), and loss of components of the myosin machinery, cause mild DTC migration defects, consistent with roles in movement (Nishiwaki 1999; Cram et al. 2006; Wong et al. 2014). Notably, F-actin has only been observed via fusion of GFP to the actin binding domain from Moesin (Kim et al. 2011). As different populations of F-actin are bound by distinct F-actin probes (Washington and Knecht 2008), it will be important to examine F-actin using different actin binding probes that might label forms of F-actin that drive movement.
It is possible that the DTC uses a less well-established or novel mechanism to move. Given the importance of nuclear positioning in driving DTC movement, the role of the nucleus, which helps power movement of human fibroblasts and fibrosarcoma cells by acting as a piston to generate forward acting forces (Petrie et al. 2014, 2017), could be a contributing factor in DTC migration. In addition, the physically confining environment of the encasing BM might allow polarized water permeation or retrograde flow of actomyosin under the plasma membrane (a migration mode termed “chimneying”) to drive DTC movement, as it does for cancer cells in confining in vitro environments (Paluch and Raz 2013; Stroka et al. 2014; Bergert et al. 2015). The crucial role of integrins, secreted matrix proteases, and BM proteins in DTC migration further suggests the intriguing possibility that secretion or polarized assembly of BM that the DTC generates might help power DTC migration. Indeed, there is evidence that some bacteria power their movement via polysaccharide secretion (Jarrell and McBride 2008; Khayatan et al. 2015). It seems likely that the DTC might use multiple ways to propel movement and teasing out these mechanisms are important areas of future study.
Other unanswered questions center around DTC polarization. For example, how A/P localized Wnts oppose each other’s functions to precisely guide A/P migration is unclear. Further, how UNC-5 directs polarity away from UNC-6 (netrin) sources is poorly understood. It is also unclear what polarizes migration in the DTC. The polarized orientation of F-actin fibers within the DTC in the direction of migration suggests that these fibers might be associated with DTC polarity, and thus could help orient the mechanism(s) that generate movement. The convergence of polarizing signals from integrin, netrin, and Wnt pathways on small Rho GTPases strongly suggest their involvement in orienting DTC migration; however, their precise roles are uncertain.
Live cell imaging coupled with single cell molecular manipulation studies, which have helped elucidate AC invasion, will be crucial for advancing our understanding of DTC migration (Kelley et al. 2017). Dynamic methods to visualize and perturb Rho GTPases, F-actin, BM protein deposition, secretion, and removal, as well as trafficking, activity, and localization of integrins, the Wnt receptor MOM-5 and the netrin receptors UNC-40 and UNC-5 should help clarify how the DTC polarizes and migrates. Branching morphogenesis in the lung, mammary gland, kidney and salivary gland is driven by BM encased leader cells, which are not protrusive and have a smooth leading front like the DTC (Williams and Daniel 1983; Ewald et al. 2008; Andrew and Ewald 2010; Harunaga et al. 2014). Thus, a deeper understanding of the mechanisms underlying DTC migration will have significance to vertebrate organ formation. Further, as invasive tumor growth hijacks these morphogenetic mechanisms to spread into adjoining tissues (Gray et al. 2010), it is likely that examining DTC migration will reveal mechanisms that are misregulated in metastatic cancers.
Sex Myoblast Migration: Navigating Around Tissues To Position Muscles
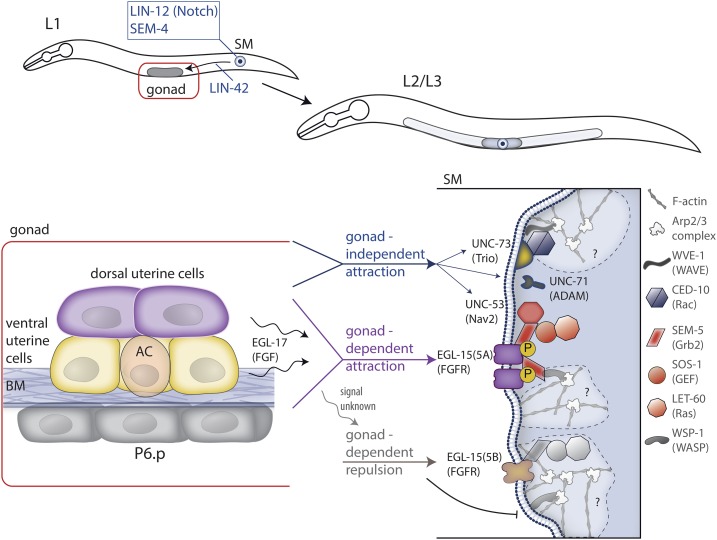
Unlike the DTC, which directs a collective cell migration event while encased in a BM, many cells also migrate individually through and around animal tissues unencumbered by a surrounding BM. Examples of this mode of migration include primordial germ cells, leukocytes, hematopoietic stem cells, and tumor cells (Aman and Piotrowski 2010; Friedl and Alexander 2011). The SMs are two bilaterally symmetric cells, born at the end of the first larval stage among the posterior ventral body muscles. Each SM undergoes an individual long-distance migration along the left and right sides of the worm body cavity. In the hermaphrodite, ∼2 hr after the SMs are formed and over the course of L2 and early L3 stages of development, the SMs migrate anteriorly to a final position at the exact center of the gonad, where the future uterine-vulval attachment will form (Figure 7). Following migration, the SM cells divide to form 16 vulval and uterine muscle cells (the sex muscles), which make attachments to the uterus, the vulva, and lateral epidermis (Sulston and Horvitz 1977; Lints and Hall 2017). The SMs migrate ∼65 μm in 4 hr (Branda and Stern 2000), and are not encased in BM (D. R. Sherwood and E. L. Hastie, unpublished data). SMs in male animals migrate toward the tail, and differentiate into the tail muscles used for mating (Sulston and Horvitz 1977). Little is known about male SM migration, except that it is cell-autonomous: in an otherwise hermaphrodite animal, mosaic loss of tra-1, a gene linked with sex determination, creates male SMs that migrate posteriorly and produce male-like sex muscles in the tail (Hunter and Wood 1990). In the following, we discuss SM migration in the hermaphrodite.
Figure 7.
SM migration, a cell navigation event. Top panels: two SMs are specified by LIN-12 (Notch) signaling and the action of the zinc finger transcriptional regulator SEM-4 in the tail of the L1 larva and migrate independently of each other through the body of the worm over the course of the L2 and L3 larval stages until reaching the central gonad (shown with darker shading). The transcriptional regulator LIN-42 (Period) prevents the SMs from dividing precociously during their migration. Only one SM is shown for simplicity. Bottom panel: SMs are directed to the proper location by an EGL-17 (FGF) signal emanating from the central gonad region and vulval precursor cells, and by an additional unknown signal originating from nongonadal tissue. In addition to these attractive signals, an unidentified cue, also originating in the gonad, repels the SMs. Both EGL-17 (FGF)-dependent attractive and gonad-dependent repulsive signals are sensed by the FGF receptor EGL-15 in the SM, but different isoforms (5A and 5B) respond to attractive and repulsive cues. Signals are transduced to LET-60 (Ras) via the adaptor molecule SEM-5 (Grb2), which may also communicate to Arp2/3 complex-driven actin assembly. EGL-17 (FGF)-independent attraction is less well-understood, but depends on various molecules with potential roles in cell adhesion and motility such as UNC-53 (Nav2), UNC-71 (ADAM), and UNC-73 (Trio).
Attraction and repulsion in SM migration
SM migration depends on a complex mix of both attractive and repulsive signals originating from somatic gonadal as well nongonadal tissues that precisely position the SM cells at the uterine-vulval connection [for review see Chen and Stern (1998); Figure 7]. During the L2 and early L3 stages, the central part of the gonad is composed of six somatic (nongermline) cells: three ventral uterine cells, two dorsal uterine cells and the AC. The AC marks the center of the gonad and aligns with the vulval precursor cell P6.p. During the late L2 stage the AC secretes the EGF-like ligand LIN-3, which induces the P6.p cell to take on the 1° vulval precursor fate (see AC invasion section) (Sternberg 2005). In dig-1 mutants, where the gonad is displaced anteriorly, and sometimes also dorsally, SMs still target the gonad center, navigating through new territory to precisely center on the displaced AC (Thomas et al. 1990). When the gonad is entirely deleted by laser ablation, SMs nevertheless still migrate, but their final positions cover a broad, centrally dispersed region (Thomas et al. 1990). SM migration therefore seems to be guided by a combination of gonad-dependent and gonad-independent attractive signals that direct the SM to the site of uterine-vulval connection.
The fibroblast growth factor EGL-17 (FGF) is a key attractive cue that helps guide SM migration (Burdine et al. 1997, 1998; Branda and Stern 2000). Vertebrates encode 23 fibroblast growth factor (FGF) ligands and five FGF receptors, while C. elegans has just two FGF ligands (EGL-17 and LET-756) and one receptor (EGL-15) (Borland et al. 2001). In egl-17 mutant animals, SMs display severe premature termination of migration, stopping ∼50 μm short of the gonad center (Stern and Horvitz 1991). Strikingly, gonad ablation in egl-17 mutant animals restores the broad positioning around P6.p as observed in wild-type animals with ablated gonads. These experiments suggest that EGL-17 (FGF) is an attractive cue for SM migration and that the gonad generates an unknown repulsive cue. Further, these results indicate that, in the absence of both the attractive cue (EGL-17) and the repulsive cue from the gonad, a separate nongonadal (and non-EGL-17) signal acts to help position the SM cells. It is unknown how the presence of EGL-17 (FGF) masks the repulsive effect of the gonad during normal development, or what the physiological role of repulsion might be.
The egl-17 gene is expressed in the P6.p vulval precursor cell after it is induced to a 1° vulval fate by the gonadal AC, thus providing a source of EGL-17 (FGF) protein that guides migration and coordinates SM migration with vulval induction during the early L3 stage (Burdine et al. 1998). Analysis of the promoter region of egl-17 revealed three enhancer regions that control egl-17 expression both temporally and spatially within the 1° fated P6.p cell (Cui and Han 2003; Sternberg 2005). When egl-17 is expressed in neighboring vulval precursor cells, the SM targets to the cell expressing egl-17 (Burdine et al. 1998). The vulval precursor cells can be ablated, however, and SMs still position precisely (Burdine et al. 1998). EGL-17 (FGF) is also expressed in the dorsal uterine cells, and there is evidence for its presence in the ventral uterine cells and the AC as well (Branda and Stern 2000). Thus egl-17 from the uterine cells within the gonad likely also guide SM migration (Branda and Stern 2000). Overall, these observations suggest that EGL-17 (FGF) from multiple sources near the uterine-vulval connection overcomes the negative SM migration signal. Together these EGL-17 (FGF) attractive cues have been referred to as the “gonad dependent signal,” as egl-17 expression in the 1° fated P6.p depends on AC-mediated vulval induction. In addition to directing SMs toward the gonad, EGL-17 (FGF) acts with the C. elegans Robo receptor SAX-3 to maintain SMs ventrally during their migration (Branda and Stern 2000).
The C. elegans FGF receptor EGL-15 (FGFR) is expressed in SMs and is necessary for correct targeting of the gonad by the SMs (DeVore et al. 1995; Sundaram et al. 1996; Branda and Stern 2000; Lo et al. 2008). It was originally reported that mutation of EGL-15 phenocopies what is observed with egl-17 mutant worms, with posteriorly displaced SMs indicating that egl-17 mediated attraction is turned off, while gonad repulsion is maintained (Stern and Horvitz 1991). However, later it was discovered that alternative splicing generates two EGL-15 (FGFR) isoforms, 5A and 5B, which each have a unique domain in their extracellular region after the first IG motif of EGL-15 (FGFR) (Burdine et al. 1997, 1998; Goodman et al. 2003; Huang and Stern 2004; Lo et al. 2008). The original egl-15 allele interferes with only the splicing of isoform 5A, leaving 5B intact (Goodman et al. 2003). Loss of both EGL-15 (FGFR) isoforms leads to SMs that are loosely centered on the AC—a phenotype observed with gonad ablation (i.e., when both gonad repulsion and attraction are lost) (Lo et al. 2008). Site of action studies similarly suggest that 5A is the form of the FGF receptor that mediates attraction to EGL-17 (FGF) sources, while 5B has a role in gonad-dependent repulsion. Expression and genetic evidence suggests that repulsion is mediated through an unidentified ligand that is not the other C. elegans FGF, LET-756 (Lo et al. 2008).
It is not uncommon for ligands and receptors associated with migration to play both attractive and repulsive roles. This occurs with semaphorins and ephrin signaling systems (Klein 2012; Gurrapu and Tamagnone 2016). As discussed earlier, this is also how the netrin signaling system functions, as the ligand UNC-6 (netrin) attracts cells that express the receptor UNC-40 (DCC) and repels cells expressing the receptor UNC-5 and UNC-40 together or UNC-5 alone (Lai Wing Sun et al. 2011). There is evidence that opposing signals might act by biasing self-organizing stochastic intracellular polarity systems (Tang and Wadsworth 2014; Chisholm et al. 2016). For example, UNC-6 (netrin) stabilizes where randomly directed self-organizing UNC-40 polarizes during AC invasion (Wang et al. 2014b). Whatever the mechanism, the FGF-dependent guidance system for SMs seems to be a conserved: Ppa-egl-17 phenotypes and ablation experiments indicate that gonad-dependent and independent attraction and gonad-dependent repulsion mechanisms are operational in the evolutionarily distant Pristionchus pacificus (Photos et al. 2006).
Ras in SM positioning
LET-60 (Ras) is a small GTPase that is part of the core C. elegans RTK (Receptor Tyrosine Kinase)/Ras/MAPK (Mitogen-Activated Protein Kinase) signaling pathway, which converts growth factor signals to cellular transformations via LET-60 (Ras)-dependent intracellular phosphorylation events (Sundaram 2013). LET-60 is expressed in the SMs and the somatic gonad, as well as in the vulval precursor cells, where EGL-17 (FGF) expression is dependent on induction by LET-60 (Burdine et al. 1998; Dent and Han 1998). The LET-60 (Ras) signaling cascade begins with EGL-17 (FGF) binding to EGL-15 (FGFR) on the cell surface, which triggers dimerization and autophosphorylation of the intracellular domain of EGL-15 (FGFR), promoting docking of the adaptor protein SEM-5 (Grb2) ((Sundaram 2013) and Figure 7). In C. elegans, EGL-15 (FGFR) binds directly to SEM-5 (Grb2), unlike in vertebrates, which require an additional FRS2 molecule to link FGFRs to Grb2 (Borland et al. 2001; Lo et al. 2010). SEM-5 associates with the Ras-GEF, SOS-1 (also known as LET-341), which activates LET-60, leading to a variety of downstream signaling events including phosphorylations via the MAP kinase cascade and nuclear translocations that change gene expression (Borland et al. 2001; Sundaram 2013).
Mutations in the sem-5 (Grb2) gene lead to posteriorly displaced SMs, but the defects are far weaker than egl-17 mutants, and SM distributions more closely resemble those of gonad-ablated animals, suggesting that both gonad-dependent repulsion and attraction are eliminated when SEM-5 is not functional (Clark et al. 1992; DeVore et al. 1995; Chen et al. 1997). Likewise, let-60 loss-of-function mutations lead to SMs that are loosely centered on the AC, indicating loss of EGL-15 (FGFR) mediated activity, both instructive and repulsive (Sundaram 2013). When LET-60 (Ras) is restored mosaically in one SM cell, but not the other, the genetically wild-type SM shows rescue of positioning and the mutant SM does not, indicating that LET-60 (Ras) is required cell-autonomously (Sundaram 2013). Mutations in Ras regulators, such as ksr-1, the C. elegans equivalent of the positive Ras regulator KSR, give phenotypes like those observed for let-60 deletion (Ohmachi et al. 2002; Oakes et al. 2012); similarly loss of RasGAPs that control Ras activity also affects SM migration (Stetak et al. 2008). Overall, LET-60 and its associated adaptors play a major role in guidance of SM migration by the gonad, via both attraction and repulsion.
FGF independent mechanisms of SM migration
The EGL-17/EGL-15-independent migration of SMs, revealed in the absence of the gonad, is not well understood, but mutations in the genes unc-53, unc-71, and unc-73 (corresponding to mammalian Nav2, ADAM, and Trio, respectively) abrogate this migration mode and SMs are posteriorly displaced in gonad-ablated animals instead of being loosely grouped around the AC (Chen et al. 1997). Mutations in these genes have no effect on EGL-15 (FGFR)-dependent SM migration. The posterior SM displacement of unc-53 gonad-ablated animals is worsened by mutations in let-60 and sem-5, indicating that LET-60 (Ras) and SEM-5 (Grb2) also contribute to gonad-independent SM migration, seemingly in addition to their participation in the FGFR signaling pathway (Chen et al. 1997; Chen and Stern 1998). Similarly, in gonad-ablated animals, expression of constitutively active LET-60 (Ras) produces an almost wild-type targeting of SMs, reducing the broad distribution usually observed in gonad-ablated animals (Sundaram 2013). Taken together, these results suggest that SM migration is controlled by EGL-15 (FGFR) signaling to SEM-5 (Grb2) and LET-60 (Ras), which also play FGFR-independent roles in SM movement by an undetermined signaling pathway.
Possible signaling to the actin cytoskeleton for SM migration
Due to the different compensatory and competing mechanisms driving SM movement, it is somewhat difficult to link genetic interactions to molecular mechanisms of SM migration, and little is known concerning membrane or cytoskeleton dynamics in the SM downstream of the different signaling pathways described above. However drawing analogies with what is known in mammalian systems, SEM-5 (Grb2) could directly link EGL-15 (FGFR) to the actin cytoskeleton, since Grb2 is an known activator of the actin polymerization factor N-WASP, enhancing its ability to activate actin polymerization nucleation via the Arp2/3 complex (Carlier et al. 2000, and Figure 7). Additionally the molecules involved in gonad-independent SM migration have connections to the actin cytoskeleton: UNC-53 (NAV-2) physically interacts with ABI-1, part of the WAVE complex, as well as SEM-5, thus potentially linking UNC-53 to WASP activity as well (Stringham et al. 2002; Schmidt et al. 2009). UNC-71 (ADAM) is an ADAM protein, lacking protease activity, and involved in cell adhesion during cell motility possibly via integrins (X. Huang et al. 2003) and UNC-73 (Trio) is a GEF that accelerates the activity of the Rac GTPases CED-10 and MIG-2. MIG-2 is expressed in SMs, and expression of a constitutively active MIG-2 protein perturbs SM migration (Zipkin et al. 1997). CED-10 and MIG-2 are the upstream activators of the WAVE complex and WASP, respectively, for actin polymerization via the Arp 2/3 complex (Shakir et al. 2008; Walck-Shannon et al. 2015). All of this points to a possible scenario in which signals arriving from the gonad, and other, as-yet-unidentified, sources signal to the actin cytoskeleton and adhesion systems of the SM to enable its migration and correct positioning at the center of the gonad.
Specification of migratory SM cells
Specification of the SM fate from surrounding mesodermal precursor cells initiates before movement begins (Figure 7). Genetic analysis has indicated that SM cells require LIN-12 (Notch) activity in the mid-to-late L1 stage for initial specification (Greenwald et al. 1983; Foehr and Liu 2008). In the absence of the lin-12 gene, the SM precursor cells adopt the coelomocyte fate (another mesoderm progenitor fate). LIN-12 (Notch) is activated by its transmembrane ligands LAG-2 and APX-1, which are expressed in adjacent epidermal cells in the mid-to-late L1 (Foehr and Liu 2008). A zinc finger-type transcription factor encoded by the gene sem-4, appears to function after LIN-12 (Notch) activity. In the absence of sem-4, cells resembling the SMs are generated, but they do not migrate toward the vulva or divide, and instead take on the appearance of body wall muscle cells (Basson and Horvitz 1996). SEM-4, which is an ortholog of Drosophila SPALT, is conserved in P. pacificus, and as in C. elegans, disruption of Ppa-sem-4 causes SM fate specification to fail (Photos et al. 2006).
Once SM migration halts, and they are positioned near the vulval precursor cells in the L3 stage, the SM cells divide and differentiate into four types of sex muscles [for mechanisms regulating postmigratory SM lineage differentiation, see Hale et al. (2014)]. Molecular mechanisms regulate the timing of SM division, thereby assuring that SMs arrive at their vulval destination before proceeding with their developmental program and dividing to create the sex muscles. Loss of LIN-42, the C. elegans ortholog of the Per family of circadian rhythm proteins, causes SMs to prematurely undergo division during the L2 migratory stage (Tennessen et al. 2006). LIN-42 (PER) is believed to interfere with transcriptional activators, thus repressing the expression of target genes. EGL-15 (FGFR), while necessary for EGL-17 (FGF)-dependent SM attraction and gonad-dependent repulsion, is a negative regulator of terminal differentiation of SMs into muscle cells. When hyperactivation of EGL-15 (FGFR) is induced via constitutive dimerization, the vulval muscles descendant from the SMs do not form properly; in particular actin cables and myosin are reduced (Sasson and Stern 2004). This phenotype is strongly dependent on the 5A isoform, and less so on 5B, mirroring the isoform specificities for SM positioning. Additionally, the function of EGL-15 (FGFR) in sex muscle differentiation is controlled by glycosylation. When a key glycosylation site in EGL-15 (FGFR) is mutated, an EGL-15 hyperactivation phenotype is observed, with incorrect formation of the vulval muscles. This result indicates that glycosylation negatively regulates the role of EGL-15 (FGFR) in SM differentiation to muscle. Glycosylation, however, has no effect on SM migration (Polanska et al. 2009), suggesting that glycosylation could be a means of inactivating EGL-15 (FGFR) specifically during differentiation.
Unanswered questions in SM migration
Many questions remain to be addressed concerning SM migration, including the identity of the gonad-dependent repellant signal, how the presence of EGL-17 (FGF) masks the repulsive effect, and what physiological function repulsion serves in SM focusing. Additionally, the signals and receptors involved in gonad-independent attraction are unknown. Furthermore, little is known about the molecular details regarding how signals are transduced intracellularly leading to gonad-dependent attraction/repulsion, and whether the motility mechanism is different in gonad-dependent vs. -independent movement. Although actin assembly is surely involved in SM movement, the actin cytoskeleton has not been imaged in moving SMs, nor has the cytoskeleton been examined under conditions that affect SM movement. Real-time imaging has not yet been performed on the migrating SM cells, and we do not know what migration mode—mesenchymal, bleb-based, or novel—they use to move. Finally, since male SMs placed in a hermaphrodite environment still migrate toward their normal posterior location, SM migration could serve as a powerful model to determine how the intrinsic programs of migrating cells control the manner in which they respond to extrinsic guidance cues.
Summary and outlook
Cell migration is crucial during tissue formation and homeostasis in animals, and cells have developed a rich toolkit to drive and control movement in different tissue environments (Aman and Piotrowski 2010; Mayor and Etienne-Manneville 2016). Our review of three types of cell movement in C. elegans has focused on distinct types of movements found in animals: AC invasion, a model for how cells breach BM barriers; DTC migration, an example of how a leader cell encased in a BM directs organ shape; and SM migration, a model for how individual cells migrate between tissues. The importance of studying cell movement in vivo is highlighted by many novel findings from these models. These include (1) in the case of the AC, the identification of invadopodia in a native tissue environment, the discovery that the netrin ligand stabilizes intrinsic oscillatory clustering of its receptor UNC-40 (DCC) to polarize cells, the finding that BM sliding widens BM openings, and that the AC induces cell shape changes when it contacts the underlying vulval precursor cells; (2) in the case of DTCs, the elucidation of a Netrin-Wnt global positioning system that orients migration along the A/V and D/V axes, and a role for conserved microRNAs in buffering cell migration to environmental fluctuations in temperature; and (3) in the case of SMs, the intricate roles played by FGF/FGFR-dependent and independent signaling. These in vivo studies have also revealed the key functions that dynamic transcriptional programs play in changing the ways in which migrating cells respond to their environment to alter movement. Finally, work in C. elegans has identified significant areas for future study of cell motility, such as the puzzling observation that the DTC is largely nonprotrusive and moves by means that are currently poorly understood.
One important implication from these studies in C. elegans is that a deep understanding of cell migration requires the ability to visualize migration, to view the subcellular structures and molecular components (as well as their activity) that regulate migration, and to simultaneously perturb this molecular machinery. The challenge of combining these approaches in vivo is a key reason why in vitro studies of cell migration have been so predominant in examination of cell movement. However, in vivo systems are needed to confirm and extend these in vitro findings. With its amenability to live cell imaging, simple tissue architecture, and temporally and spatially controlled methods for gene and protein manipulation (Chai et al. 2012; Wei et al. 2012; Armenti et al. 2014; Corsi et al. 2015; Zhang et al. 2015), C. elegans is emerging as more than just a model to identify genes involved in migration. The worm is developing into a powerful in vivo system that should remain at the forefront of discovering key conserved cellular and molecular mechanisms that allow cells to enact exquisite migratory programs to navigate through diverse tissue environments.
Acknowledgments
We thank Agnieszka Kawska of IlluScientia.com for her work on the figures, and thank Naomi Levy-Strumpf, Jane Hubbard, Andy Ewald, Peter Friedl, and David Matus for helpful discussions on the material covered. D.R.S. was supported by National Institute of General Medical Sciences (NIGMS) R01 GM079320, National Institute of Child Health and Human Development (NICHD) R21HD084290 and NIGMS R35 Maximizing Investors’ Research Award (MIRA) GM118049. J.P. was supported by Fondation Association pour la Recherche sur le Cancer (ARC) and the Idex ANR-10-IDEX-0001-02 PSL.
Footnotes
Communicating editor: M. Labouesse
Literature Cited
- Abella J. V., Galloni C., Pernier J., Barry D. J., Kjaer S., et al. , 2016. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat. Cell Biol. 18: 76–86. [DOI] [PubMed] [Google Scholar]
- Altincicek B., Fischer M., Luersen K., Boll M., Wenzel U., et al. , 2010. Role of matrix metalloproteinase ZMP-2 in pathogen resistance and development in Caenorhabditis elegans. Dev. Comp. Immunol. 34: 1160–1169. [DOI] [PubMed] [Google Scholar]
- Aman A., Piotrowski T., 2010. Cell migration during morphogenesis. Dev. Biol. 341: 20–33. [DOI] [PubMed] [Google Scholar]
- Andrew D. J., Ewald A. J., 2010. Morphogenesis of epithelial tubes: insights into tube formation, elongation, and elaboration. Dev. Biol. 341: 34–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A., Culotti J. G., Hedgecock E. M., 1998. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development 125: 1191–1205. [DOI] [PubMed] [Google Scholar]
- Antebi A., Yeh W. H., Tait D., Hedgecock E. M., Riddle D. L., 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14: 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Armenti S. T., Lohmer L. L., Sherwood D. R., Nance J., 2014. Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development 141: 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J., Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599. [DOI] [PubMed] [Google Scholar]
- Basson M., Horvitz H. R., 1996. The Caenorhabditis elegans gene sem-4 controls neuronal and mesodermal cell development and encodes a zinc finger protein. Genes Dev. 10: 1953–1965. [DOI] [PubMed] [Google Scholar]
- Baum P. D., Garriga G., 1997. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron 19: 51–62. [DOI] [PubMed] [Google Scholar]
- Beaty B. T., Condeelis J., 2014. Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 93: 438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerling E., Ritsma L., Vrisekoop N., Derksen P. W., van Rheenen J., 2011. Intravital microscopy: new insights into metastasis of tumors. J. Cell Sci. 124: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard C. Y., Boyanov A., Hall D. H., Hobert O., 2006. DIG-1, a novel giant protein, non-autonomously mediates maintenance of nervous system architecture. Development 133: 3329–3340. [DOI] [PubMed] [Google Scholar]
- Bendezu F. O., Martin S. G., 2013. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr. Biol. 23: 42–47. [DOI] [PubMed] [Google Scholar]
- Bergert M., Erzberger A., Desai R. A., Aspalter I. M., Oates A. C., et al. , 2015. Force transmission during adhesion-independent migration. Nat. Cell Biol. 17: 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A., Condeelis J. S., Gligorijevic B., 2014. Invadopodia in context. Cell Adhes. Migr. 8: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J., 2014. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94: 235–263. [DOI] [PubMed] [Google Scholar]
- Blelloch R., Kimble J., 1999. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature 399: 586–590. [DOI] [PubMed] [Google Scholar]
- Blelloch R., Anna-Arriola S. S., Gao D., Li Y., Hodgkin J., et al. , 1999. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev. Biol. 216: 382–393. [DOI] [PubMed] [Google Scholar]
- Bokel C., Brown N. H., 2002. Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev. Cell 3: 311–321. [DOI] [PubMed] [Google Scholar]
- Borland C. Z., Schutzman J. L., Stern M. J., 2001. Fibroblast growth factor signaling in Caenorhabditis elegans. Bioessays 23: 1120–1130. [DOI] [PubMed] [Google Scholar]
- Branch K. M., Hoshino D., Weaver A. M., 2012. Adhesion rings surround invadopodia and promote maturation. Biol. Open 1: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda C. S., Stern M. J., 2000. Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol. 226: 137–151. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero J. J., Magalhaes M. A., Eddy R. J., Hodgson L., Condeelis J., 2013. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 14: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L., Kolotuev I., Didier C., Bhoumik A., Podbilewicz B., et al. , 2004. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J. Cell Biol. 165: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine R. D., Chen E. B., Kwok S. F., Stern M. J., 1997. egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 94: 2433–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine R. D., Branda C. S., Stern M. J., 1998. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Burke S. L., Hammell M., Ambros V., 2015. Robust distal tip cell pathfinding in the face of temperature stress is ensured by two conserved microRNAS in Caenorhabditis elegans. Genetics 200: 1201–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burket C. T., Higgins C. E., Hull L. C., Berninsone P. M., Ryder E. F., 2006. The C. elegans gene dig-1 encodes a giant member of the immunoglobulin superfamily that promotes fasciculation of neuronal processes. Dev. Biol. 299: 193–205. [DOI] [PubMed] [Google Scholar]
- Cabello J., Neukomm L. J., Gunesdogan U., Burkart K., Charette S. J., et al. , 2010. The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 8: e1000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cal S., Lopez-Otin C., 2015. ADAMTS proteases and cancer. Matrix Biol. 44–46: 77–85. [DOI] [PubMed] [Google Scholar]
- Campbell I. D., Humphries M. J., 2011. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3: a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M. G., Oswald E., Yu W., Xue F., MacKerell A. D., Jr., et al. , 2017. The expanding role of the BCL6 oncoprotein as a cancer therapeutic target. Clin. Cancer Res. 23: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.-F., Nioche P., Broutin-L’Hermite I., Boujemaa R., Le Clainche C., et al. , 2000. GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich syndrome protein (N-WASP) with actin-related protein (Arp2/3) complex. J. Biol. Chem. 275: 21946–21952. [DOI] [PubMed] [Google Scholar]
- Castro-Castro A., Marchesin V., Monteiro P., Lodillinsky C., Rosse C., et al. , 2016. Cellular and molecular mechanisms of MT1-MMP-dependent cancer cell invasion. Annu. Rev. Cell Dev. Biol. 32: 555–576. [DOI] [PubMed] [Google Scholar]
- Cecchetelli A. D., Cram E. J., 2017. Regulating distal tip cell migration in space and time. Mech. Dev. pii: S0925–4773(16)30131–9. DOI: 10.1016/j.mod.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Li W., Feng G., Yang Y., Wang X., et al. , 2012. Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat. Protoc. 7: 2090–2102. [DOI] [PubMed] [Google Scholar]
- Chan S. S., Zheng H., Su M. W., Wilk R., Killeen M. T., et al. , 1996. UNC-40, a C. elegans homolog of DCC (deleted in colorectal cancer), is required in motile cells responding to UNC-6 netrin cues. Cell 87: 187–195. [DOI] [PubMed] [Google Scholar]
- Charras G., Paluch E., 2008. Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 9: 730–736. [DOI] [PubMed] [Google Scholar]
- Chen E. B., Stern M. J., 1998. Understanding cell migration guidance: lessons from sex myoblast migration in C. elegans. Trends Genet. 14: 322–327. [DOI] [PubMed] [Google Scholar]
- Chen E. B., Branda C. S., Stern M. J., 1997. Genetic enhancers of sem-5 define components of the gonad-independent guidance mechanism controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol. 182: 88–100. [DOI] [PubMed] [Google Scholar]
- Chen P., Edelman J. D., Gharib S. A., 2013. Comparative evaluation of miRNA expression between in vitro and in vivo airway epithelium demonstrates widespread differences. Am. J. Pathol. 183: 1405–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. T., 1989. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J. Exp. Zool. 251: 167–185. [DOI] [PubMed] [Google Scholar]
- Chesarone M. A., Goode B. L., 2009. Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney M. A., Lam N., Morgan D. E., Phillips B. T., Kimble J., 2009. C. elegans HLH-2/E/Daughterless controls key regulatory cells during gonadogenesis. Dev. Biol. 331: 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. D., Hutter H., Jin Y., Wadsworth W. G., 2016. The genetics of axon guidance and axon regeneration in Caenorhabditis elegans. Genetics 204: 849–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci W., Polo J. M., Melnick A., 2008. B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma. Curr. Opin. Hematol. 15: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. G., Stern M. J., Horvitz H. R., 1992. C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature 356: 340–344. [DOI] [PubMed] [Google Scholar]
- Clay M. R., Sherwood D. R., 2015. Basement membranes in the worm: a dynamic scaffolding that instructs cellular behaviors and shapes tissues. Curr. Top. Membr. 76: 337–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Batlle E., 2013. SnapShot: the intestinal crypt. Cell 152: 1198–1198.e2. [DOI] [PubMed] [Google Scholar]
- Clevers H., Nusse R., 2012. Wnt/beta-catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Corsi A. K., Wightman B., Chalfie M., 2015. A transparent window into biology: a primer on Caenorhabditis elegans (June 18, 2015) WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.177.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens L. M., Fingleton B., Matrisian L. M., 2002. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295: 2387–2392. [DOI] [PubMed] [Google Scholar]
- Cram E. J., Clark S. G., Schwarzbauer J. E., 2003. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J. Cell Sci. 116: 3871–3878. [DOI] [PubMed] [Google Scholar]
- Cram E. J., Shang H., Schwarzbauer J. E., 2006. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J. Cell Sci. 119: 4811–4818. [DOI] [PubMed] [Google Scholar]
- Cross S. S., Lippitt J., Mitchell A., Hollingsbury F., Balasubramanian S. P., et al. , 2008. Expression of beta-dystroglycan is reduced or absent in many human carcinomas. Histopathology 53: 561–566. [DOI] [PubMed] [Google Scholar]
- Cui M., Han M., 2003. Cis regulatory requirements for vulval cell-specific expression of the Caenorhabditis elegans fibroblast growth factor gene egl-17. Dev. Biol. 257: 104–116. [DOI] [PubMed] [Google Scholar]
- Darom A., Bening-Abu-Shach U., Broday L., 2010. RNF-121 is an endoplasmic reticulum-membrane E3 ubiquitin ligase involved in the regulation of beta-integrin. Mol. Biol. Cell 21: 1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J. A., Han M., 1998. Post-embryonic expression pattern of C. elegans let-60 ras reporter constructs. Mech. Dev. 72: 179–182. [DOI] [PubMed] [Google Scholar]
- Destaing O., Block M. R., Planus E., Albiges-Rizo C., 2011. Invadosome regulation by adhesion signaling. Curr. Opin. Cell Biol. 23: 597–606. [DOI] [PubMed] [Google Scholar]
- DeVore D. L., Horvitz H. R., Stern M. J., 1995. An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell 83: 611–620. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., et al. , 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino J., Paysan L., Gest C., Lagree V., Juin A., et al. , 2014. Cdc42 and Tks5: a minimal and universal molecular signature for functional invadosomes. Cell Adhes. Migr. 8: 280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M., Goncharov A., Jin Y., Chisholm A. D., 2003. C. elegans ankyrin repeat protein VAB-19 is a component of epidermal attachment structures and is essential for epidermal morphogenesis. Development 130: 5791–5801. [DOI] [PubMed] [Google Scholar]
- Doherty M. F., Adelmant G., Cecchetelli A. D., Marto J. A., Cram E. J., 2014. Proteomic analysis reveals CACN-1 is a component of the spliceosome in Caenorhabditis elegans. G3 Bethesda 4: 1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski K., Trzebiatowska A., Chiquet-Ehrismann R., 2005. ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev. Biol. 282: 27–38. [DOI] [PubMed] [Google Scholar]
- Duan S., Cermak L., Pagan J. K., Rossi M., Martinengo C., et al. , 2012. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 481: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A., Overall C. M., 2013. Missing the target: matrix metalloproteinase antitargets in inflammation and cancer. Trends Pharmacol. Sci. 34: 233–242. [DOI] [PubMed] [Google Scholar]
- Dyer J. M., Savage N. S., Jin M., Zyla T. R., Elston T. C., et al. , 2013. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 23: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., 2005. Wnt signaling (June 25, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.7.1, http://www.wormbook.org. [Google Scholar]
- Even-Ram S., Yamada K. M., 2005. Cell migration in 3D matrix. Curr. Opin. Cell Biol. 17: 524–532. [DOI] [PubMed] [Google Scholar]
- Ewald A. J., Brenot A., Duong M., Chan B. S., Werb Z., 2008. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell 14: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancsalszky L., Monostori E., Farkas Z., Pourkarimi E., Masoudi N., et al. , 2014. NDK-1, the homolog of NM23–H1/H2 regulates cell migration and apoptotic engulfment in C. elegans. PLoS One 9: e92687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Fraga M. F., 2012. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13: 97–109. [DOI] [PubMed] [Google Scholar]
- Fidler A. L., Darris C. E., Chetyrkin S. V., Pedchenko V. K., Boudko S. P., et al. , 2017. Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. Elife 6: e24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., Guardavaccaro D., Neubert K., Chan T., Li D., et al. , 2007. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev. Cell 12: 443–455. [DOI] [PubMed] [Google Scholar]
- Finger F. P., Kopish K. R., White J. G., 2003. A role for septins in cellular and axonal migration in C. elegans. Dev. Biol. 261: 220–234. [DOI] [PubMed] [Google Scholar]
- Fletcher S. J., Rappoport J. Z., 2010. Moving forward: polarised trafficking in cell migration. Trends Cell Biol. 20: 71–78. [DOI] [PubMed] [Google Scholar]
- Foehr M. L., Liu J., 2008. Dorsoventral patterning of the C. elegans postembryonic mesoderm requires both LIN-12/Notch and TGFbeta signaling. Dev. Biol. 313: 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley S. I., Feng Y., Krishnamurthy R., Kim D.-H., Celedon A., et al. , 2010. A distinctive role for focal adhesion proteins in threedimensional cell motility. Nat. Cell Biol. 12: 598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Alexander S., 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147: 992–1009. [DOI] [PubMed] [Google Scholar]
- Friedl P., Gilmour D., 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10: 445–457. [DOI] [PubMed] [Google Scholar]
- Friedl P., Wolf K., 2010. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Sahai E., Weiss S., Yamada K. M., 2012. New dimensions in cell migration. Nat. Rev. Mol. Cell Biol. 13: 743–747. [DOI] [PubMed] [Google Scholar]
- Genot E., Gligorijevic B., 2014. Invadosomes in their natural habitat. Eur. J. Cell Biol. 93: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S., Simon A., Elkhatib N., Louvard D., Fetler L., et al. , 2012. Do cancer cells have distinct adhesions in 3D collagen matrices and in vivo? Eur. J. Cell Biol. 91: 930–937. [DOI] [PubMed] [Google Scholar]
- Gitai Z., Yu T. W., Lundquist E. A., Tessier-Lavigne M., Bargmann C. I., 2003. The netrin receptor UNC-40/DCC stimulates axon attraction and outgrowth through enabled and, in parallel, Rac and UNC-115/AbLIM. Neuron 37: 53–65. [DOI] [PubMed] [Google Scholar]
- Goldstein B., Takeshita H., Mizumoto K., Sawa H., 2006. Wnt signals can function as positional cues in establishing cell polarity. Dev. Cell 10: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales K. A., Liang H., Lim Y. S., Chan Y. S., Yeo J. C., et al. , 2015. Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell 162: 564–579. [DOI] [PubMed] [Google Scholar]
- Goodman S. J., Branda C. S., Robinson M. K., Burdine R. D., Stern M. J., 2003. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development 130: 3757–3766. [DOI] [PubMed] [Google Scholar]
- Graham P. L., Johnson J. J., Wang S., Sibley M. H., Gupta M. C., et al. , 1997. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J. Cell Biol. 137: 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R. S., Cheung K. J., Ewald A. J., 2010. Cellular mechanisms regulating epithelial morphogenesis and cancer invasion. Curr. Opin. Cell Biol. 22: 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I., 2005. LIN-12/Notch signaling in C. elegans (August 4, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.10.1, http://www.wormbook.org. [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R., 1983. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444. [DOI] [PubMed] [Google Scholar]
- Gurrapu S., Tamagnone L., 2016. Transmembrane semaphorins: multimodal signaling cues in development and cancer. Cell Adhes. Migr. 10: 675–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn E. J., Sherwood D. R., 2011. Cell invasion through basement membrane: the anchor cell breaches the barrier. Curr. Opin. Cell Biol. 23: 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn E. J., Yashiro H., Ziel J. W., Ihara S., Wang Z., et al. , 2009. Integrin acts upstream of netrin signaling to regulate formation of the anchor cell’s invasive membrane in C. elegans. Dev. Cell 17: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn E. J., Ziel J. W., Morrissey M. A., Linden L. M., Wang Z., et al. , 2013. The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J. Cell Biol. 201: 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn E. J., Kelley L. C., Naegeli K. M., Wang Z., Chi Q., et al. , 2014. ADF/cofilin promotes invadopodial membrane recycling during cell invasion in vivo. J. Cell Biol. 204: 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo S. L., Bilder D., 2011. Global tissue revolutions in a morphogenetic movement controlling elongation. Science 331: 1071–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale J. J., Amin N. M., George C., Via Z., Shi H., et al. , 2014. A role of the LIN-12/Notch signaling pathway in diversifying the non-striated egg-laying muscles in C. elegans. Dev. Biol. 389: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., et al. , 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. [DOI] [PubMed] [Google Scholar]
- Harburger D. S., Calderwood D. A., 2009. Integrin signalling at a glance. J. Cell Sci. 122: 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harunaga J. S., Doyle A. D., Yamada K. M., 2014. Local and global dynamics of the basement membrane during branching morphogenesis require protease activity and actomyosin contractility. Dev. Biol. 394: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie E. L., Sherwood D. R., 2016. A new front in cell invasion: the invadopodial membrane. Eur. J. Cell Biol. 95: 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Hall D. H., Stern B. D., 1987. Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100: 365–382. [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Hall D. H., 1990. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron 4: 61–85. [DOI] [PubMed] [Google Scholar]
- Hermeking H., 2010. The miR-34 family in cancer and apoptosis. Cell Death Differ. 17: 193–199. [DOI] [PubMed] [Google Scholar]
- Hesselson D., Newman C., Kim K. W., Kimble J., 2004. GON-1 and fibulin have antagonistic roles in control of organ shape. Curr. Biol. 14: 2005–2010. [DOI] [PubMed] [Google Scholar]
- Hikasa H., Sokol S. Y., 2013. Wnt signaling in vertebrate axis specification. Cold Spring Harb. Perspect. Biol. 5: a007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard M. A., Bargmann C. I., 2006. Wnt signals and frizzled activity orient anterior-posterior axon outgrowth in C. elegans. Dev. Cell 10: 379–390. [DOI] [PubMed] [Google Scholar]
- Hotary K., Li X. Y., Allen E., Stevens S. L., Weiss S. J., 2006. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 20: 2673–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrus A., Lau G., Hutter H., Schenk S., Ferralli J., et al. , 2007. C. elegans agrin is expressed in pharynx, IL1 neurons and distal tip cells and does not genetically interact with genes involved in synaptogenesis or muscle function. PLoS One 2: e731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. C., Hall D. H., Hedgecock E. M., Kao G., Karantza V., et al. , 2003. Laminin alpha subunits and their role in C. elegans development. Development 130: 3343–3358. [DOI] [PubMed] [Google Scholar]
- Huang P., Stern M. J., 2004. FGF signaling functions in the hypodermis to regulate fluid balance in C. elegans. Development 131: 2595–2604. [DOI] [PubMed] [Google Scholar]
- Huang T. F., Cho C. Y., Cheng Y. T., Huang J. W., Wu Y. Z., et al. , 2014. BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 10: e1004428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Huang P., Robinson M. K., Stern M. J., Jin Y., 2003. UNC-71, a disintegrin and metalloprotease (ADAM) protein, regulates motor, axon guidance and sex myoblast migration in C. elegans. Development 130: 3147–3161. [DOI] [PubMed] [Google Scholar]
- Hunter C. P., Wood W. B., 1990. The tra-1 gene determines sexual phenotype cell-autonomously in C. elegans. Cell 63: 1193–1204. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A., Horwitz A. R., 2011. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 3: a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter H., Vogel B. E., Plenefisch J. D., Norris C. R., Proenca R. B., et al. , 2000. Conservation and novelty in the evolution of cell adhesion and extracellular matrix genes. Science 287: 989–994. [DOI] [PubMed] [Google Scholar]
- Hwang B. J., Meruelo A. D., Sternberg P. W., 2007. C. elegans EVI1 proto-oncogene, EGL-43, is necessary for Notch-mediated cell fate specification and regulates cell invasion. Development 134: 669–679. [DOI] [PubMed] [Google Scholar]
- Ihara S., Nishiwaki K., 2007. Prodomain-dependent tissue targeting of an ADAMTS protease controls cell migration in Caenorhabditis elegans. EMBO J. 26: 2607–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara S., Hagedorn E. J., Morrissey M. A., Chi Q., Motegi F., et al. , 2011. Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat. Cell Biol. 13: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N., Wadsworth W. G., Stern B. D., Culotti J. G., Hedgecock E. M., 1992. UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron 9: 873–881. [DOI] [PubMed] [Google Scholar]
- Itoh B., Hirose T., Takata N., Nishiwaki K., Koga M., et al. , 2005. SRC-1, a non-receptor type of protein tyrosine kinase, controls the direction of cell and growth cone migration in C. elegans. Development 132: 5161–5172. [DOI] [PubMed] [Google Scholar]
- Itoh Y., 2015. Membrane-type matrix metalloproteinases: their functions and regulations. Matrix Biol. 44–46: 207–223. [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., McBride M. J., 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6: 466–476. [DOI] [PubMed] [Google Scholar]
- Jayadev R., Sherwood D. R., 2017. Basement membranes. Curr. Biol. 27: R207–R211. [DOI] [PubMed] [Google Scholar]
- Jiang H., Zhang G., Wu J. H., Jiang C. P., 2014. Diverse roles of miR-29 in cancer (review). Oncol. Rep. 31: 1509–1516. [DOI] [PubMed] [Google Scholar]
- Jorgensen E. M., Mango S. E., 2002. The art and design of genetic screens: caenorhabditis elegans. Nat. Rev. Genet. 3: 356–369. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kao G., Huang C. C., Hedgecock E. M., Hall D. H., Wadsworth W. G., 2006. The role of the laminin beta subunit in laminin heterotrimer assembly and basement membrane function and development in C. elegans. Dev. Biol. 290: 211–219. [DOI] [PubMed] [Google Scholar]
- Karp X., Greenwald I., 2004. Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev. Biol. 272: 460–469. [DOI] [PubMed] [Google Scholar]
- Kato M., Sternberg P. W., 2009. The C. elegans tailless/Tlx homolog nhr-67 regulates a stage-specific program of linker cell migration in male gonadogenesis. Development 136: 3907–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Chou T. F., Yu C. Z., DeModena J., Sternberg P. W., 2014. LINKIN, a new transmembrane protein necessary for cell adhesion. Elife 3: e04449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T., Zheng H., Merz D. C., Kohara Y., Tamai K. K., et al. , 2009. C. elegans mig-6 encodes papilin isoforms that affect distinct aspects of DTC migration, and interacts genetically with mig-17 and collagen IV. Development 136: 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. C., Lohmer L. L., Hagedorn E. J., Sherwood D. R., 2014. Traversing the basement membrane in vivo: a diversity of strategies. J. Cell Biol. 204: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. C., Wang Z., Hagedorn E. J., Wang L., Shen W., et al. , 2017. Live-cell confocal microscopy and quantitative 4D image analysis of anchor-cell invasion through the basement membrane in Caenorhabditis elegans. Nat. Protoc. 12: 2081–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R., Desanlis I., Wheeler G. N., Edwards D. R., 2015. The ADAMTS (A disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 16: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayatan B., Meeks J. C., Risser D. D., 2015. Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria. Mol. Microbiol. 98: 1021–1036. [DOI] [PubMed] [Google Scholar]
- Killian D. J., Hubbard E. J., 2005. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev. Biol. 279: 322–335. [DOI] [PubMed] [Google Scholar]
- Kim H. S., Murakami R., Quintin S., Mori M., Ohkura K., et al. , 2011. VAB-10 spectraplakin acts in cell and nuclear migration in Caenorhabditis elegans. Development 138: 4013–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Wadsworth W. G., 2000. Positioning of longitudinal nerves in C. elegans by nidogen. Science 288: 150–154. [DOI] [PubMed] [Google Scholar]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. [DOI] [PubMed] [Google Scholar]
- Kimble J. E., White J. G., 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81: 208–219. [DOI] [PubMed] [Google Scholar]
- Kiontke K., Barriere A., Kolotuev I., Podbilewicz B., Sommer R., et al. , 2007. Trends, stasis, and drift in the evolution of nematode vulva development. Curr. Biol. 17: 1925–1937. [DOI] [PubMed] [Google Scholar]
- Klein R., 2012. Eph/ephrin signalling during development. Development 139: 4105–4109. [DOI] [PubMed] [Google Scholar]
- Kohrman A. Q., Matus D. Q., 2017. Divide or conquer: cell cycle regulation of invasive behavior. Trends Cell Biol. 27: 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic I., Ho R., Cram E. J., 2012. CCDC-55 is required for larval development and distal tip cell migration in Caenorhabditis elegans. Mech. Dev. 128: 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., 2005. Basement membranes. (September 1, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.16.1, http://www.wormbook.org. [Google Scholar]
- Kramerova I. A., Kawaguchi N., Fessler L. I., Nelson R. E., Chen Y., et al. , 2000. Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development 127: 5475–5485. [DOI] [PubMed] [Google Scholar]
- Krause M., Park M., Zhang J. M., Yuan J., Harfe B., et al. , 1997. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development 124: 2179–2189. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Kuroki R., Nishiwaki K., 2004. A fibulin-1 homolog interacts with an ADAM protease that controls cell migration in C. elegans. Curr. Biol. 14: 2011–2018. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Ohkura K., Tamai K. K., Nagata K., Nishiwaki K., 2008. MIG-17/ADAMTS controls cell migration by recruiting nidogen to the basement membrane in C. elegans. Proc. Natl. Acad. Sci. USA 105: 20804–20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Nagata K., Sugimoto A., Nishiwaki K., 2012. Tissue architecture in the Caenorhabditis elegans gonad depends on interactions among fibulin-1, type IV collagen and the ADAMTS extracellular protease. Genetics 190: 1379–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni G., Xu Z., Mohamed A. M., Li H., Tang X., et al. , 2013. Experimental evidence for UNC-6 (netrin) axon guidance by stochastic fluctuations of intracellular UNC-40 (DCC) outgrowth activity. Biol. Open 2: 1300–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka S., Kashina A., 2008. Cell biology of embryonic migration. Birth Defects Res. C Embryo Today 84: 102–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Wing Sun K., Correia J. P., Kennedy T. E., 2011. Netrins: versatile extracellular cues with diverse functions. Development 138: 2153–2169. [DOI] [PubMed] [Google Scholar]
- Lammermann T., Sixt M., 2009. Mechanical modes of ‘amoeboid’ cell migration. Curr. Opin. Cell Biol. 21: 636–644. [DOI] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R., 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., Pilot F., 2003. Developmental control of cell morphogenesis: a focus on membrane growth. Nat. Cell Biol. 5: 103–108. [DOI] [PubMed] [Google Scholar]
- Lee M., Cram E. J., Shen B., Schwarzbauer J. E., 2001. Roles for beta(pat-3) integrins in development and function of Caenorhabditis elegans muscles and gonads. J. Biol. Chem. 276: 36404–36410. [DOI] [PubMed] [Google Scholar]
- Leung-Hagesteijn C., Spence A. M., Stern B. D., Zhou Y., Su M. W., et al. , 1992. UNC-5, a transmembrane protein with immunoglobulin and thrombospondin type 1 domains, guides cell and pioneer axon migrations in C. elegans. Cell 71: 289–299. [DOI] [PubMed] [Google Scholar]
- Levy-Strumpf N., 2016. Orchestrating A/P and D/V guidance—a Wnt/Netrin tale. Worm 5: e1146857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Strumpf N., Culotti J. G., 2014. Netrins and Wnts function redundantly to regulate antero-posterior and dorso-ventral guidance in C. elegans. PLoS Genet. 10: e1004381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Strumpf N., Krizus M., Zheng H., Brown L., Culotti J. G., 2015. The Wnt frizzled receptor MOM-5 regulates the UNC-5 netrin receptor through small GTPase-dependent signaling to determine the polarity of migrating cells. PLoS Genet. 11: e1005446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., Wiesner C., Himmel M., 2011. Degrading devices: invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 27: 185–211. [DOI] [PubMed] [Google Scholar]
- Lints, R., and D. H. Hall, 2017 Reproductive System. Wormatlas. 10.3908/wormatlas.1.17. [Google Scholar]
- Llamazares M., Cal S., Quesada V., Lopez-Otin C., 2003. Identification and characterization of ADAMTS-20 defines a novel subfamily of metalloproteinases-disintegrins with multiple thrombospondin-1 repeats and a unique GON domain. J. Biol. Chem. 278: 13382–13389. [DOI] [PubMed] [Google Scholar]
- Lo T.-W., Branda C. S., Huang P., Sasson I. E., Goodman S. J., et al. , 2008. Different isoforms of the C. elegans FGF receptor are required for attraction and repulsion of the migrating sex myoblasts. Dev. Biol. 318: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T. W., Bennett D. C., Goodman S. J., Stern M. J., 2010. Caenorhabditis elegans fibroblast growth factor receptor signaling can occur independently of the multi-substrate adaptor FRS2. Genetics 185: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmer L. L., Kelley L. C., Hagedorn E. J., Sherwood D. R., 2014. Invadopodia and basement membrane invasion in vivo. Cell Adh. Migr. 8: 246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmer L. L., Clay M. R., Naegeli K. M., Chi Q., Ziel J. W., et al. , 2016. A sensitized screen for genes promoting invadopodia function in vivo: CDC-42 and Rab GDI-1 direct distinct aspects of invadopodia formation. PLoS Genet. 12: e1005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M., Cheng H. J., 2008. A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 4: e1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist E. A., Reddien P. W., Hartwieg E., Horvitz H. R., Bargmann C. I., 2001. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128: 4475–4488. [DOI] [PubMed] [Google Scholar]
- Ma Y., Kanakousaki K., Buttitta L., 2015. How the cell cycle impacts chromatin architecture and influences cell fate. Front. Genet. 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen C. D., Sahai E., 2010. Cancer dissemination–lessons from leukocytes. Dev. Cell 19: 13–26. [DOI] [PubMed] [Google Scholar]
- Martin K. H., Hayes K. E., Walk E. L., Ammer A. G., Markwell S. M., et al. , 2012. Quantitative measurement of invadopodia-mediated extracellular matrix proteolysis in single and multicellular contexts. J. Vis. Exp. 66: e4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus D. Q., Li X. Y., Durbin S., Agarwal D., Chi Q., et al. , 2010. In vivo identification of regulators of cell invasion across basement membranes. Sci. Signal. 3: ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus D. Q., Chang E., Makohon-Moore S. C., Hagedorn M. A., Chi Q., et al. , 2014. Cell division and targeted cell cycle arrest opens and stabilizes basement membrane gaps. Nat. Commun. 5: 4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus D. Q., Lohmer L. L., Kelley L. C., Schindler A. J., Kohrman A. Q., et al. , 2015. Invasive cell fate requires G1 cell-cycle arrest and histone deacetylase-mediated changes in gene expression. Dev. Cell 35: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R., Etienne-Manneville S., 2016. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17: 97–109. [DOI] [PubMed] [Google Scholar]
- McClatchey S. T., Wang Z., Linden L. M., Hastie E. L., Wang L., et al. , 2016. Boundary cells restrict dystroglycan trafficking to control basement membrane sliding during tissue remodeling. Elife 5: e17218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medwig T. N., Matus D. Q., 2017. Breaking down barriers: the evolution of cell invasion. Curr. Opin. Genet. Dev. 47: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighan C. M., Schwarzbauer J. E., 2007. Control of C. elegans hermaphrodite gonad size and shape by vab-3/Pax6-mediated regulation of integrin receptors. Genes Dev. 21: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighan C. M., Kann A. P., Egress E. R., 2015. Transcription factor hlh-2/E/Daughterless drives expression of alpha integrin ina-1 during DTC migration in C. elegans. Gene 568: 220–226. [DOI] [PubMed] [Google Scholar]
- Milde-Langosch K., 2005. The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer 41: 2449–2461. [DOI] [PubMed] [Google Scholar]
- Mi-Mi L., Votra S., Kemphues K., Bretscher A., Pruyne D., 2012. Z-line formins promote contractile lattice growth and maintenance in striated muscles of C. elegans. J. Cell Biol. 198: 87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey M. A., Hagedorn E. J., Sherwood D. R., 2013. Cell invasion through basement membrane: the netrin receptor DCC guides the way. Worm 2: e26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey M. A., Keeley D. P., Hagedorn E. J., McClatchey S. T., Chi Q., et al. , 2014. B-LINK: a hemicentin, plakin, and integrin-dependent adhesion system that links tissues by connecting adjacent basement membranes. Dev. Cell 31: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshfegh Y., Bravo-Cordero J. J., Miskolci V., Condeelis J., Hodgson L., 2014. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat. Cell Biol. 16: 574–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. A., Courtneidge S. A., 2011. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12: 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K., 1999. Mutations affecting symmetrical migration of distal tip cells in Caenorhabditis elegans. Genetics 152: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K., Hisamoto N., Matsumoto K., 2000. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science 288: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Nourshargh S., Alon R., 2014. Leukocyte migration into inflamed tissues. Immunity 41: 694–707. [DOI] [PubMed] [Google Scholar]
- Oakes P. W., Beckham Y., Stricker J., Gardel M. L., 2012. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196: 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmachi M., Rocheleau C. E., Church D., Lambie E., Schedl T., et al. , 2002. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr. Biol. 12: 427–433. [DOI] [PubMed] [Google Scholar]
- Overall C. M., Kleifeld O., 2006. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 6: 227–239. [DOI] [PubMed] [Google Scholar]
- Ozanne B. W., Spence H. J., McGarry L. C., Hennigan R. F., 2007. Transcription factors control invasion: AP-1 the first among equals. Oncogene 26: 1–10. [DOI] [PubMed] [Google Scholar]
- Ozbek S., Balasubramanian P. G., Chiquet-Ehrismann R., Tucker R. P., Adams J. C., 2010. The evolution of extracellular matrix. Mol. Biol. Cell 21: 4300–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick S. B., Rosen M. K., 2010. Physical mechanisms of signal integration by WASP family proteins. Annu. Rev. Biochem. 79: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A., 2008. Remodeling the model organism: matrix metalloproteinase functions in invertebrates. Semin. Cell Dev. Biol. 19: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A., Ewald A. J., Werb Z., 2007. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch E. K., Raz E., 2013. The role and regulation of blebs in cell migration. Curr. Opin. Cell Biol. 25: 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C. L., Howell J. E., Clark S. G., Hilliard M., Cordes S., et al. , 2006. Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10: 367–377. [DOI] [PubMed] [Google Scholar]
- Paul C. D., Mistriotis P., Konstantopoulos K., 2017. Cancer cell motility: lessons from migration in confined spaces. Nat. Rev. Cancer 17: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R. J., Yamada K. M., 2012. At the leading edge of three-dimensional cell migration. J. Cell Sci. 125: 5917–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R. J., Yamada K. M., 2016. Multiple mechanisms of 3D migration: the origins of plasticity. Curr. Opin. Cell Biol. 42: 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R. J., Gavara N., Chadwick R. S., Yamada K. M., 2012. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197: 439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R. J., Koo H., Yamada K. M., 2014. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science 345: 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R. J., Harlin H. M., Korsak L. I., Yamada K. M., 2017. Activating the nuclear piston mechanism of 3D migration in tumor cells. J. Cell Biol. 216: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Photos A., Gutierrez A., Sommer R. J., 2006. sem-4/spalt and egl-17/FGF have a conserved role in sex myoblast specification and migration in P. pacificus and C. elegans. Dev. Biol. 293: 142–153. [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerda J., Chorev D. S., Geiger B., Cossart P., 2017. The diverse family of Arp2/3 complexes. Trends Cell Biol. 27: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanska U. M., Duchesne L., Harries J. C., Fernig D. G., Kinnunen T. K., 2009. N-Glycosylation regulates fibroblast growth factor receptor/EGL-15 activity in Caenorhabditis elegans in vivo. J. Biol. Chem. 284: 33030–33039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razidlo G. L., Schroeder B., Chen J., Billadeau D. D., McNiven M. A., 2014. Vav1 as a central regulator of invadopodia assembly. Curr. Biol. 24: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Horvitz H. R., 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2: 131–136. [DOI] [PubMed] [Google Scholar]
- Reiner D. J., Lundquist E. A., 2016. Small GTPases (January 17, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.67.1, http://www.wormbook.org. [Google Scholar]
- Rella L., Fernandes Povoa E. E., Korswagen H. C., 2016. The Caenorhabditis elegans Q neuroblasts: a powerful system to study cell migration at single-cell resolution in vivo. Genesis 54: 198–211. [DOI] [PubMed] [Google Scholar]
- Renaud S. J., Kubota K., Rumi M. A., Soares M. J., 2014. The FOS transcription factor family differentially controls trophoblast migration and invasion. J. Biol. Chem. 289: 5025–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J., 2015. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 36: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimann I., Hajnal A., 2007. Regulation of anchor cell invasion and uterine cell fates by the egl-43 Evi-1 proto-oncogene in Caenorhabditis elegans. Dev. Biol. 308: 187–195. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Ambros V., 1995. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development 121: 2491–2500. [DOI] [PubMed] [Google Scholar]
- Rowe R. G., Weiss S. J., 2008. Breaching the basement membrane: who, when and how? Trends Cell Biol. 18: 560–574. [DOI] [PubMed] [Google Scholar]
- Ruijtenberg S., van den Heuvel S., 2015. G1/S inhibitors and the SWI/SNF complex control cell-cycle exit during muscle differentiation. Cell 162: 300–313. [DOI] [PubMed] [Google Scholar]
- Sasson I. E., Stern M. J., 2004. FGF and PI3 kinase signaling pathways antagonistically modulate sex muscle differentiation in C. elegans. Development 131: 5381–5392. [DOI] [PubMed] [Google Scholar]
- Sawa H., Korswagen H. C., 2013. Wnt signaling in C. elegans (December 9, 2013), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.7.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Suetsugu S., Sugimoto A., Miki H., Yamamoto M., et al. , 2003. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116: 1505–1518. [DOI] [PubMed] [Google Scholar]
- Schindler A. J., Sherwood D. R., 2011. The transcription factor HLH-2/E/Daughterless regulates anchor cell invasion across basement membrane in C. elegans. Dev. Biol. 357: 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler A. J., Sherwood D. R., 2013. Morphogenesis of the caenorhabditis elegans vulva. Wiley Interdiscip. Rev. Dev. Biol. 2: 75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K. L., Marcus-Gueret N., Adeleye A., Webber J., Baillie D., et al. , 2009. The cell migration molecule UNC-53/NAV2 is linked to the ARP2/3 complex by ABI-1. Development 136: 563–574. [DOI] [PubMed] [Google Scholar]
- Schonichen A., Geyer M., 2010. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim. Biophys. Acta 1803: 152–163. [DOI] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R. D., Louvard D., Vignjevic D. M., 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189: 541–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Louvard D., Vignjevic D., 2011. Cytoskeleton networks in basement membrane transmigration. Eur. J. Cell Biol. 90: 93–99. [DOI] [PubMed] [Google Scholar]
- Schwab A., Stock C., 2014. Ion channels and transporters in tumour cell migration and invasion. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seano G., Chiaverina G., Gagliardi P. A., di Blasio L., Puliafito A., et al. , 2014. Endothelial podosome rosettes regulate vascular branching in tumour angiogenesis. Nat Cell Biol. 16: 931–941, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgambato A., Migaldi M., Montanari M., Camerini A., Brancaccio A., et al. , 2003. Dystroglycan expression is frequently reduced in human breast and colon cancers and is associated with tumor progression. Am. J. Pathol. 162: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaq-Zadah M., Gomes-Santos C. S., Bardin S., Maiuri P., Maurin M., et al. , 2016. Persistent cell migration and adhesion rely on retrograde transport of beta(1) integrin. Nat. Cell Biol. 18: 54–64. [DOI] [PubMed] [Google Scholar]
- Shakir M. A., Jiang K., Struckhoff E. C., Demarco R. S., Patel F. B., et al. , 2008. The Arp2/3 activators WAVE and WASP have distinct genetic interactions with Rac GTPases in Caenorhabditis elegans axon guidance. Genetics 179: 1957–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., et al. , 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 30: 625–636. [DOI] [PubMed] [Google Scholar]
- Sherwood D. R., Sternberg P. W., 2003. Anchor cell invasion into the vulval epithelium in C. elegans. Dev. Cell 5: 21–31. [DOI] [PubMed] [Google Scholar]
- Sherwood D. R., Butler J. A., Kramer J. M., Sternberg P. W., 2005. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell 121: 951–962. [DOI] [PubMed] [Google Scholar]
- Silhankova M., Korswagen H. C., 2007. Migration of neuronal cells along the anterior-posterior body axis of C. elegans: Wnts are in control. Curr. Opin. Genet. Dev. 17: 320–325. [DOI] [PubMed] [Google Scholar]
- Sobrado V. R., Moreno-Bueno G., Cubillo E., Holt L. J., Nieto M. A., et al. , 2009. The class I bHLH factors E2–2A and E2–2B regulate EMT. J. Cell Sci. 122: 1014–1024. [DOI] [PubMed] [Google Scholar]
- Somerville R. P., Longpre J. M., Jungers K. A., Engle J. M., Ross M., et al. , 2003. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J. Biol. Chem. 278: 9503–9513. [DOI] [PubMed] [Google Scholar]
- Soto M. C., Qadota H., Kasuya K., Inoue M., Tsuboi D., et al. , 2002. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 16: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., McWhirter R., Miller T., Strasbourger P., Thompson O., et al. , 2014. Isolation of specific neurons from C. elegans larvae for gene expression profiling. PLoS One 9: e112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Pastor-Pareja J. C., Igaki T., Pagliarini R., Xu T., 2007. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc. Natl. Acad. Sci. USA 104: 2721–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. J., Horvitz H. R., 1991. A normally attractive cell interaction is repulsive in two C. elegans mesodermal cell migration mutants. Development 113: 797–803. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., 2005. Vulval development (June 25, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetak A., Gutierrez E., Hajnal A., 2008. Tissue-specific functions of the Caenorhabditis elegans p120 Ras GTPase activating protein GAP-3. Dev. Biol. 323: 166–176. [DOI] [PubMed] [Google Scholar]
- Stringham E., Pujol N., Vandekerckhove J., Bogaert T., 2002. unc-53 controls longitudinal migration in C. elegans. Development 129: 3367–3379. [DOI] [PubMed] [Google Scholar]
- Stroka K. M., Jiang H., Chen S. H., Tong Z., Wirtz D., et al. , 2014. Water permeation drives tumor cell migration in confined microenvironments. Cell 157: 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Merz D. C., Killeen M. T., Zhou Y., Zheng H., et al. , 2000. Regulation of the UNC-5 netrin receptor initiates the first reorientation of migrating distal tip cells in Caenorhabditis elegans. Development 127: 585–594. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sundaram M., Yochem J., Han M., 1996. A Ras-mediated signal transduction pathway is involved in the control of sex myoblast migration in Caenorhabditis elegans. Development 122: 2823–2833. [DOI] [PubMed] [Google Scholar]
- Sundaram M. V., 2013. Canonical RTK-Ras-ERK signaling and related alternative pathways (July 1, 2013), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.80.2, //www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suozzi K. C., Wu X., Fuchs E., 2012. Spectraplakins: master orchestrators of cytoskeletal dynamics. J. Cell Biol. 197: 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Toyoda H., Sano M., Nishiwaki K., 2006. Chondroitin acts in the guidance of gonadal distal tip cells in C. elegans. Dev. Biol. 300: 635–646. [DOI] [PubMed] [Google Scholar]
- Tamai K. K., Nishiwaki K., 2007. bHLH transcription factors regulate organ morphogenesis via activation of an ADAMTS protease in C. elegans. Dev. Biol. 308: 562–571. [DOI] [PubMed] [Google Scholar]
- Tang X., Wadsworth W. G., 2014. SAX-3 (Robo) and UNC-40 (DCC) regulate a directional bias for axon guidance in response to multiple extracellular cues. PLoS One 9: e110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannoury H., Rodriguez V., Kovacevic I., Ibourk M., Lee M., et al. , 2010. CACN-1/Cactin interacts genetically with MIG-2 GTPase signaling to control distal tip cell migration in C. elegans. Dev. Biol. 341: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Boekhorst V., Friedl P., 2016. Plasticity of cancer cell invasion-mechanisms and implications for therapy. Adv. Cancer Res. 132: 209–264. [DOI] [PubMed] [Google Scholar]
- Te Boekhorst V., Preziosi L., Friedl P., 2016. Plasticity of cell migration in vivo and in silico. Annu. Rev. Cell Dev. Biol. 32: 491–526. [DOI] [PubMed] [Google Scholar]
- Tennessen J. M., Gardner H. F., Volk M. L., Rougvie A. E., 2006. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev. Biol. 289: 30–43. [DOI] [PubMed] [Google Scholar]
- Thomas J. H., Stern M. J., Horvitz H. R., 1990. Cell interactions coordinate the development of the C. elegans egg-laying system. Cell 62: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Topf U., Chiquet-Ehrismann R., 2011. Genetic interaction between Caenorhabditis elegans teneurin ten-1 and prolyl 4-hydroxylase phy-1 and their function in collagen IV-mediated basement membrane integrity during late elongation of the embryo. Mol. Biol. Cell 22: 3331–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A., Nile A. H., Bankaitis V. A., 2014. Sec14-like phosphatidylinositol-transfer proteins and diversification of phosphoinositide signalling outcomes. Biochem. Soc. Trans. 42: 1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowska A., Topf U., Sauder U., Drabikowski K., Chiquet-Ehrismann R., 2008. Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol. Biol. Cell 19: 3898–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S., Weinberg R. A., 2011. Tumor metastasis: molecular insights and evolving paradigms. Cell 147: 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese E., Schocken J., Jacob S., Wimer A. M., Royce R., et al. , 2011. The tailless ortholog nhr-67 functions in the development of the C. elegans ventral uterus. Dev. Biol. 356: 516–528. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R., 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10: 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel B. E., Hedgecock E. M., 2001. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development 128: 883–894. [DOI] [PubMed] [Google Scholar]
- Vuong-Brender T. T. K., Yang X., Labouesse M., 2016. C. elegans embryonic morphogenesis. Curr. Top. Dev. Biol. 116: 597–616. [DOI] [PubMed] [Google Scholar]
- Walck-Shannon E., Reiner D. J., Hardin J., 2015. Polarized Rac-dependent protrusions drive epithelial intercalation in the embryonic epidermis of C. elegans. Development 142: 3549–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shen W., Lei S., Matus D., Sherwood D., et al. , 2014. MIG-10 (Lamellipodin) stabilizes invading cell adhesion to basement membrane and is a negative transcriptional target of EGL-43 in C. elegans. Biochem. Biophys. Res. Commun. 452: 328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chi Q., Sherwood D. R., 2014a MIG-10 (lamellipodin) has netrin-independent functions and is a FOS-1A transcriptional target during anchor cell invasion in C. elegans. Development 141: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Linden L. M., Naegeli K. M., Ziel J. W., Chi Q., et al. , 2014b UNC-6 (netrin) stabilizes oscillatory clustering of the UNC-40 (DCC) receptor to orient polarity. J. Cell Biol. 206: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington R. W., Knecht D. A., 2008. Actin binding domains direct actin-binding proteins to different cytoskeletal locations. BMC Cell Biol. 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Potter C. J., Luo L., Shen K., 2012. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nat. Methods 9: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo J., Kenyon C., 1999. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol. Cell 4: 851–858. [DOI] [PubMed] [Google Scholar]
- Wickstrom S. A., Fassler R., 2011. Regulation of membrane traffic by integrin signaling. Trends Cell Biol. 21: 266–273. [DOI] [PubMed] [Google Scholar]
- Williams J. M., Daniel C. W., 1983. Mammary ductal elongation: differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev. Biol. 97: 274–290. [DOI] [PubMed] [Google Scholar]
- Wong M. C., Schwarzbauer J. E., 2012. Gonad morphogenesis and distal tip cell migration in the Caenorhabditis elegans hermaphrodite. Wiley Interdiscip. Rev. Dev. Biol. 1: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. C., Kennedy W. P., Schwarzbauer J. E., 2014. Transcriptionally regulated cell adhesion network dictates distal tip cell directionality. Dev. Dyn. 243: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Rongali S. C., Miles J. P., Lee K. D., Lee M., 2006. pat-4/ILK and unc-112/Mig-2 are required for gonad function in Caenorhabditis elegans. Exp. Cell Res. 312: 1475–1483. [DOI] [PubMed] [Google Scholar]
- Xu X., Dong C., Vogel B. E., 2007. Hemicentins assemble on diverse epithelia in the mouse. J. Histochem. Cytochem. 55: 119–126. [DOI] [PubMed] [Google Scholar]
- Yan B., Guo Q., Fu F. J., Wang Z., Yin Z., et al. , 2015. The role of miR-29b in cancer: regulation, function, and signaling. Onco Targets Ther. 8: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Weinberg R. A., 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14: 818–829. [DOI] [PubMed] [Google Scholar]
- Yang Q., Roiz D., Mereu L., Daube M., Hajnal A., 2017. The invading anchor cell induces lateral membrane constriction during vulval lumen morphogenesis in C. elegans. Dev Cell 42: 271–285.e3. [DOI] [PubMed] [Google Scholar]
- Yano S., Zhang Y., Miwa S., Tome Y., Hiroshima Y., et al. , 2014. Spatial-temporal FUCCI imaging of each cell in a tumor demonstrates locational dependence of cell cycle dynamics and chemoresponsiveness. Cell Cycle 13: 2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Bais C., Greenwald I., 2004. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science 303: 663–666. [DOI] [PubMed] [Google Scholar]
- Yurchenco P. D., 2011. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 3: a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ward J. D., Cheng Z., Dernburg A. F., 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142: 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel J. W., Hagedorn E. J., Audhya A., Sherwood D. R., 2009. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat. Cell Biol. 11: 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipkin I. D., Kindt R. M., Kenyon C. J., 1997. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90: 883–894. [DOI] [PubMed] [Google Scholar]
- Zucker S., Cao J., Chen W. T., 2000. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene 19: 6642–6650. [DOI] [PubMed] [Google Scholar]