Abstract
Transplantation of neural stem cells (NSCs) is emerging as a new therapeutic approach for stroke. Real-time imaging of transplanted NSCs is essential for successful cell delivery, safety monitoring, tracking cell fate and function, and understanding the interactions of transplanted cells with the host environment. Magnetic resonance imaging (MRI) of magnetic nanoparticle-labeled cells has been the most widely used means to track stem cells in vivo. Nevertheless, it does not allow for the reliable discrimination between live and dead cells. Reporter gene-based MRI was considered as an alternative strategy to overcome this shortcoming. In this work, a class of lentiviral vector-encoding ferritin heavy chain (FTH) and enhanced green fluorescent protein (EGFP) was constructed to deliver reporter genes into NSCs. After these transgenic NSCs were transplanted into the contralateral hemisphere of rats with acute ischemic stroke, MRI and fluorescence imaging (FLI) were performed in vivo for tracking the fate of transplanted cells over a long period of 6 wk. The results demonstrated that the FTH and EGFP can be effectively and safely delivered to NSCs via the designed lentiviral vector. The distribution and migration of grafted stem cells could be monitored by bimodal MRI and FLI. FTH can be used as a robust MRI reporter for reliable reporting of the short-term viability of cell grafts, whereas its capacity for tracking the long-term viability of stem cells remains dependent on several confounding factors such as cell death and the concomitant reactive inflammation.
Keywords: neural stem cells, magnetic resonance imaging, optical imaging, ischemic stroke, ferritin
Introduction
Acute ischemic stroke caused by cerebral artery occlusion can lead to infarction of brain tissue, with acute loss of neurons, astroglia, and oligodendroglia. It remains one of the most common causes of mortality and acquired adult long-term disability worldwide. Despite clinical benefits from systemic thrombolysis or endovascular treatment,1 stroke patients, irrespective of treatment, have a relatively high mortality after the acute phase.2 Stem cell–based therapy is emerging as a promising, alternative therapeutic strategy to improve the functional outcome following ischemic stroke.3 A variety of stem cells have been proposed for the treatment of stroke, including embryonic stem cells (ESCs), neural stem cells (NSCs), bone marrow mononuclear cells (BM-MNCs), mesenchymal stem cells (MSCs), induced pluripotent stem cells (iPSCs), and some immortalized cell lines.4 Among the types of the somatic stem cells, NSCs are the most promising candidate,5,6 as they are considered not only to promote replacement of damaged cells but also to exert immunomodulatory and neuroprotective effects preventing tissue damage and/or rescuing degenerating host cells.7 However, the long-term fates of NSC progeny including the survival, death, proliferation, migration, or differentiation into a specific lineage remain unknown after their transplantation into the host brain. Therefore, in vivo real-time imaging of transplanted stem cells is essential for understanding their interactions with host environments, tracking cell fate and function, and successful delivery and safety monitoring in the clinical setting.8 Of all the current biomedical imaging technologies, magnetic resonance imaging (MRI) of magnetic nanoparticle-labeled cells is the most promising method of choice and has been widely used in preclinical studies and even in several clinical trials.9,10 It is believed that cellular MRI based on direct cell labeling is well suited for real-time, MR-guided delivery of stem cells as well as for monitoring of immediate cell engraftment.11 Nevertheless, it does not permit visualization of subsequent cell proliferation, cell activation, or cell death.12 For tracking the long-term fate of transplanted stem cells, an indirect approach of MR reporter genes that can report on the survival of transplanted cells may become ultimately the method of choice, or a combination of magnetic nanoparticles and reporter genes may be utilized.10
Previously, several MRI reporter genes have been developed and used in neurological, cardiac, and cancer research, such as enzyme-based β-galactosidase,13 tyrosinase,14 divalent metal transporter,14 iron-based transferrin receptor,15 magA,16 ferritin,17 and chemical exchange saturation transfer-based lysine-rich protein.18 Among these candidates, ferritin is an intracellular iron storage protein consisting of 24 heavy (H) and light (L) subunits, which stores and releases the iron to maintain iron homeostasis.11 Overexpression of ferritin traps excess intracellular iron through inducing the expression of the transferrin receptor and increasing intracellular iron uptake19 and causes significant shortening of the T2 relaxation time that is detectable by MRI as voxels with reduced signal intensity. Because ferritin is ubiquitous in most organisms, it may possibly be used clinically.20 Moreover, it was shown that ferritin heavy chain (FTH) alone can function as an MRI reporter.21,22 Recently, a ferritin reporter gene has been successfully used for in vivo tracking of stem/progenitor cells grafted into an infarcted heart.19,23
The use of ferritin as an MRI reporter for in vivo tracking of transplanted NSCs is rare.24 The performance of this appealing reporter gene system in tracking NSCs transplanted for stroke therapy remains unknown. Further study into the possibility of MRI reporter genes is needed before this technology can be used for stem cell transplantation of the stroke brain.25 In this study, NSCs were transduced with a lentiviral vector carrying an MRI reporter gene, FTH and fluorescence imaging (FLI) reporter gene, enhanced green fluorescent protein (EGFP). After these genetically modified NSCs were transplanted into the healthy striatum in rats with acute ischemic stroke, longitudinal dual-modal MRI, and FLI were performed to track the fate of grafted cells over a long period of time. The purpose of our study was to determine the performance of ferritin as an MRI reporter in monitoring the long-term fate of transplanted stem cells in an acute ischemic stroke model.
Materials and Methods
Animals
Thirty adult Sprague-Dawley rats, weighing 250 to 280 g, were obtained from the Animal Experiment Center of Sun Yat-Sen University and were used to establish acute ischemic stroke model. Twelve newly born rats (postnatal 1 d) were used as the donors of NSCs. All animals were housed in a standard animal facility and allowed standard food and water ad libitum. Animals were anesthetized with an intraperitoneal (i.p.) injection of sodium pentobarbital (Sigma-Aldrich, St Louis, MO, USA) at a dose of 40 mg/kg body weight. All procedures adhered to the guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care Committee of Sun Yat-Sen University.
Lentiviral Vector Construction
A replication-defective and self-inactivating lentiviral vector encoding a fusion protein composed of rat FTH linked to the EGFP was constructed to deliver reporter genes into NSCs. To generate recombinant lentiviruses expressing the fusion gene, the rat FTH (GenBank accession number NM-012848) complementary DNA (cDNA) was generated by polymerase chain reaction (PCR) amplification with the primers forward, AgeI 5′-GAGGATCCCCGGGTACCGGTCGCCACCATGACCACCGCGTCTCCCTC-3′ and reverse, AgeI 5′-TCACCATGGTGGCGACCGGGCTCTCATCACCGTGTCC-3′. Then, this cDNA was subcloned into the multiple cloning site (MCS) of pUbi-MCS-EGFP-IRES-Puromycin plasmid (GV218; Gene-Chem, Shanghai, China) under the control of the ubiquitin (Ubi) promoter using In-Fusion cloning to yield the recombinant vector pUbi-FTH-EGFP-IRES-Puromycin lentivirus expressing FTH-EGFP (pLV-FTH-EGFP). The derived construct, pLV-FTH-EGFP, was checked for proper insertion and the absence of unwanted mutations in FTH and flanking sequences by PCR analysis and DNA sequencing. A lentiviral vector without FTH was used for control (mock) transfections. The recombinant LV-FTH-EGFP was produced by cotransfecting the lentiviral transfer vector pLV-FTH-EGFP together with the packaging vector pHelper 1.0 and the envelope vector pHelper 2.0 into human embryonic kidney 293 T packaging cells using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The transgene expression of EGFP was monitored by using fluorescence microscopy, and the expression of ferritin was monitored by Western blot analysis. The supernatant of transfected 293 T cells was collected 48 to 72 h after transfection, clarified by centrifugation, filtered, and stored at −80 °C in small aliquots until use. The functional titer of lentiviral particles was determined in 293 T cells by flow cytometry. Titers were expressed as the number of transduction units per milliliter.
Cell Preparation and Transduction
NSCs were isolated from adjacent tissues around the lateral ventricles in newly born rats. Cells were cultured in StemPro NSC serum-free medium (Life Technologies, Carlsbad, CA, USA) and were characterized by immunostaining for nestin, as previously described.26 NSCs underwent routine passage, and 2 to 3 passages were used in the following experiments. For nestin immunostaining, the neurospheres were embedded in poly-l-lysine (Sigma-Aldrich Co., St Louis, MO, USA), then fixed using 4% paraformaldehyde Follow with the abbreviation in parentheses (PFA) in 0.01 M phosphate-buffered saline (PBS; pH 7.4) for 30 min. After incubation with the primary antibodies against nestin (1:200, Millipore Co., Ltd, Temecula, CA, USA) overnight at 4 °C, the neurospheres were incubated with Alexa Fluor 647-conjugated secondary antibodies (1:200, Life Technologies) for 1 h at 37 °C. Then, cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI; 1:1,000, MP Biomedicals., LLC, Santa Ana, CA, USA) for 8 min at room temperature for counterstaining of nuclei.
For cell transduction, the cultured NSCs were dissociated into single cells. To determine the appropriate condition for infection, 1 × 106 cells were seeded per well in a 24-well plate and infected with LV-FTH-EGFP in 3 different multiplicities of infection (MOI; 1, 10, or 100) for different durations (12, 24, or 36 h) under standard culture conditions (37 °C, 5% CO2). After viral infection, cells were harvested and washed 3 times with PBS. The transduction efficiency was evaluated according to the number of EGFP-expressing cells that were scored under an inverted fluorescence microscope (Nikon, Tokyo, Japan). Further quantitative analysis was performed by using flow cytometry. For flow cytometry, 1 × 106 cells were sorted on an FACScan flow cytometer (FACS Calibur, Becton Dickinson, Mountain View, CA, USA). The percentage of EGFP-positive cells in total cells was used as the transfection efficiency. Nontransduced cells were used as a control for background calibration. The experiments were carried out in triplicate. Data were analyzed using the BD FACS unite TM software (Becton Dickinson).
Cellular Transduction Effect and Stability
After 1 × 106 NSCs were infected with LV-FTH-EGFP under the predetermined appropriate conditions, the transduction effect on cellular iron accumulation and transduction stability was determined. Prussian blue staining, transmission electron microscope (TEM), atomic absorption spectrometry (AAS), and in vitro MRI were performed to assess iron accumulation in stem cells. The messenger RNA (mRNA) level and protein expression level of the FTH gene were further analyzed by a molecular biology assay; 1 × 106 mock-transduced cells served as controls. All experiments were carried out in triplicate.
For an iron accumulation assay, 1 × 106 cells were harvested 1 d after viral infection under predetermined appropriate conditions. Ten microliters of ferric citrate (FAC, 500 μmol/L, Sigma-Aldrich) was added into the medium when cells were infected with LV-FTH-EGFP. For Prussian blue staining, the cells were fixed and then incubated with Prussian blue solution containing 10% hydrochloride and 10% potassium ferrocyanide (II) trihydrate for 30 min at 37 °C. For TEM, the cells were centrifuged into a solid cell pellet and fixed in 2.5% glutaraldehyde solution at 4 °C overnight. After being postfixated in 1% OsO4 for 1 h, the cells were dehydrated and embedded in artificial resin. Ultrathin 50-nm sections were made and stained with uranyl acetate and lead citrate. Sections were observed by using a TEM (JEM-2000EX; JEOL, Tokyo, Japan) at 60 to 80 kV. For AAS, the cells were suspended in 1 mL HCl solution for thorough release and dissolution of Fe. The iron concentration was measured at the specific Fe absorption wavelength (248.3 nm) based on a preestablished calibration curve, and the iron content per cell was determined.
To test the transduction stability of the FTH gene, 1 × 106 cells were harvested for AAS and in vitro MRI to test the iron accumulation capacity at 1, 2, 3, 5, and 7 d after viral infection under predetermined appropriate conditions. FTH mRNA level was determined by real-time PCR (RT-PCR) assay, and the FTH protein expression level was determined by Western blot analysis. For the RT-PCR assay, total RNA was extracted from the cells using TRIzol (Life Technologies) according to the manufacturer’s instructions. After the concentration of total RNA was measured, the cDNA was synthesized using PrimeScript RT Master Mix Kit (Takara Bio Inc., Shiga, Japan). Primers were designed using Primer Premier 5.0 software (Molecular Biology Insights Inc., Cascade, CO, USA), and the sequences of primers were as follows: FTH forward primer: 5′-GAGGATCCCCGGGTACCGGTCGCCACCATGACCACCGCGTCTCCCTC-3′ and reverse primer: 5′-TCACCATGGTGGCGACCGGGCTCTCATCACCGTGT CC-3′; GAPDH forward primer:5′-CAAGGATACTGAGAGCAAGA-3′ and reverse primer: 5′-AGGCCCCTCCTGTTGTTAT-3′. RT-PCR was performed in a 20-μL reaction mixture containing primers, SYBR Green qPCR SuperMix reagent, and 2-μL cDNA sample. The mRNA expression of FTH was quantified using the 2−ΔΔCT analytical method in triplicate using a PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The mRNA level of the Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was measured in each sample as an internal normalization standard.
For Western blot analysis, total proteins were extracted and quantified using radio-immunoprecipitation assay (RIPA) Lysis Buffer (Life Technologies) and bicinchoninic acid protein assay kit, respectively (Life Technologies). Protein samples (25 μg) were separated on 12% sodium dodecyl sulfate polacrylamine gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes. After incubation with blocking tris-buffered saline Tween-20 containing 5% fat-free dry milk for 1 h at room temperature, the membrane was incubated with rabbit anti-FTH antibody (1:1,000, Cell Signaling Technology, Boston, MA, USA) overnight at 4 °C or with mouse anti-GAPDH antibody (1:2,000) as an internal standard for normalizing protein expressions. Horseradish peroxidase (HRP)–conjugated goat anti-rabbit or anti-mouse IgG (1:1,000) were used for 1 h at 37 °C to amplify the signal. Protein signals were detected by using Immobilon Western chemiluminescent detection reagents and chemiluminescence imaging system (Millipore, Temecula, CA, USA).
In Vitro Cytotoxicity Assay
To determine the transduction cytotoxicity, cell viability, apoptosis rate, and differentiation capability were assessed. All experiments were repeated 3 times. Cell viability was determined using the cell counting kit-8 reagent (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer’s protocol. In brief, cells were cultured in flat-bottom 96-well plates at 1 × 104 cells per well with 200-μL culture medium. Cells in 5 wells were transduced with LV-FTH-EGFP or mock under the predetermined appropriate conditions. Cells in other 5 wells were not transduced and served as controls. Until the cells were further incubated for 1 d after viral infection, 10-μL CCK-8 solution was added. After 4 h of incubation, the absorbance at 450 nm was recorded on a microplate reader (SpectraMaxM5; Molecular Devices, Sunnyvale CA, USA).
Cell apoptosis was assessed by Annexin V/propidium iodide (PI) double-staining method and analyzed using a flow cytometry 1 d after viral infection. Briefly, 1 × 106 cells transduced with LV-FTH-EGFP or mock under the predetermined appropriate conditions, and 1 × 106 nontransduced cells were harvested and washed twice with ice-cold PBS. Then, they were resuspended in 200 μL of Annexin-binding buffer solution. Ten microliters of Annexin V-fluorescein isothiocyanate (FITC) and 5 μL PI were added to the cell suspension and incubated for 15 min in the dark. Cells were analyzed using a flow cytometer (FACScalibur, Becton Dickinson, Mountain View, CA, USA).
In order to determine the effect of transduction on the differentiation potentials, 5 × 105 NSCs transduced with LV-FTH-EGFP or mock, and 5 × 105 nontransduced NSCs were cultured under standard conditions (37 °C, 5% CO2) with the culture medium replaced by Dulbecco’s modified eagle’s medium (DMEM; Life Technologies) containing 1% fetal bovine serum follow with the abbreviation in parentheses (FBS), 1% N-2 (Life Technologies), and 1% GlutaMAX (Life Technologies) to induce astrocyte differentiation, by Neurobasal (Life Technologies) containing 2% B27 Serum-Free (Life Technologies) and 1% GlutaMAX to induce neuronal differentiation; or by Neurobasal containing 2% B27 serum-free, 1% GlutaMAX, and 0.1% T3* (Life Technologies) to induce oligodendrocyte differentiation. After 7 d of induction, the expression of cell type–specific markers including glial fibrillary acidic protein (GFAP, specific for astrocytes), neuron-specific nuclear protein (NeuN, specific for neurons), and O4 (specific for oligodendrocytes) were identified by immunofluorescence analysis. Prior to immunostaining, the cells were rinsed in PBS (PH 7.4) twice and fixed with 4% PFA in PBS for 30 min at room temperature. Then, the cells were rinsed in PBS 3 times (10 min for each). For immunofluorescence staining, the cells were incubated with 10% goat serum in PBS in the presence (for intracellular antigens) or absence (for surface markers) of 0.3% Triton X-100 for 30 min at room temperature. After cells were incubated with the primary antibodies against GFAP (1:200, Abcam Ltd, Cambridge, UK), NeuN (1:200, Abcam Ltd. Cambridge, UK), or O4 (1:200, Millipore Co., Ltd, Temecula, CA, USA) for 2 h, then the corresponding Alexa Fluor 647-conjugated secondary antibodies were used for 1 h at 37 °C. Then, cells were incubated with DAPI for 5 min at room temperature for counterstaining of nuclei. Negative controls were established by omitting the primary antibody incubation. The cells were observed by using a laser scanning microscope (LSM710, Carl Zeiss, Jena, Germany).
In Vitro MRI
In vitro MRI was performed on a clinical 1.5-T system (Intera; Philips Medical Systems, Best, The Netherlands) with an 11-cm circular coil and a 3.0-T system (Achieva; Philips Medical Systems) with an 8-channal sense knee coil. The cells were resuspended in 300-μL 2% agarose solution in a 96-well plate. Fast spin echo (FSE) T1- (repetition time [TR]/echo time[TE] = 500/15 ms; number of signal averaged (NSA) = 4, section thickness = 1.5 mm), T2-weighted images (TR/TE = 2000/100 ms; NSA = 3, section thickness = 1.5 mm), and fast field echo (FFE) T2*-weighted images (TR/TE = 300/11.5 ms; NSA = 4, section thickness = 1 mm) were obtained to observe the intracellular iron accumulation mediated by the expression of the FTH gene. T2 relaxation data were acquired by using single-section multispin-echo sequences (TR/TE = 2000/20-80 ms, 4-stepped echoes, NSA = 1; acquisition matrix = 160 × 266) to quantify the effect of iron accumulation. The detailed acquisition parameters and derivation of T2 relaxation times were described previously.26 The values of T2 relaxation times were derived from the T2-mapping images after a circular region of interest (ROI) was selected in each sample. The circular ROI in each sample was drawn as large as possible to cover the whole cell suspension, by one author (X.Z.) who was blinded to the experimental groups. The diameter of the ROI was about 5 mm.
Animal Surgery and Cell Transplantation
The acute ischemic stroke model was established in 30 adult rats, and was induced by middle cerebral artery occlusion (MCAo) using the intraluminal suture, as previously described.26 In brief, a nylon silica gel–coated suture (Model 3500; Jialing Biotech, Guangzhou, China) was inserted via the proximal external carotid artery into the left internal carotid artery. The insertion length of the sutures was 2 cm. After 120 min of occlusion, the suture was withdrawn to restore blood supply to the MCA territory. One day after the MCAo, MRI was performed to confirm the successful establishment of the stroke model. To control the infarct volume of stroke comparably, only the rats with similar areas of ischemic stroke were used in this study. Two days after surgery, 18 animals were randomly assigned to receive stereotactic injection of 5 ×105 NSCs pretransduced with LV-FTH-EGFP (n = 6, FTH-EGFP-NSCs group), equivalent nontransduced NSCs (n = 6, control group), or PBS (n = 6, PBS group). After induction of anesthesia, the cells were injected into the striatum contralateral to the ischemic hemisphere (stereotaxic coordinates: 3.0 mm lateral to bregma, 0.5 mm rostral to bregma, and 6.0 mm deep from the pial surface) using a 28 s gauge needle attached to a 25-μL Hamilton syringe mounted on a microinjector. Before injection, the cells were suspended in 3-μL culture medium, and cell viability was determined to be greater than 90%. The cell suspension was injected at a constant rate of 0.2 μL/min. After injection, the needle was kept in place for an additional 15 min and then slowly withdrawn. At 1, 2, 3, 4, 5, and 6 wk after transplantation, in vivo MRI and FLI were performed to detect the distribution and migration of implanted cells. To determine the FTH expression capacity of transplanted cells, 12 additional animals were randomly assigned to receive stereotactic injection of 5 ×105 NSCs pretransduced with LV-FTH-EGFP (n = 6, FTH-EGFP-NSCs group) and equivalent nontransduced NSCs (n = 6, control group). Three animals in FTH-EGFP-NSCs group and control group were sacrificed each at 1 wk and 6 wk after transplantation for analysis of FTH expression level.
In Vivo MRI
In vivo MRI was performed on a clinical 1.5-T system (Intera; Philips Medical Systems) and a 3.0-T system (Achieva; Philips Medical Systems) with a 50 mm × 50 mm 4-channel phased array rat coil (Shanghai Chenguang Medical Technologies, Shanghai, China). Axial and coronal brain images were obtained. The imaging sequences included FSE T2-weighted imaging (TR/TE = 800/60 ms; NSA = 2), proton density-weighted (PDW) imaging (TR/TE = 3000/20 ms; NSA = 3), and FFE T2*-weighted imaging (TR/TE = 500/18 ms; Flip angel = 20°; NSA = 3). Other acquisition parameters for these sequences were field of view = 60 mm × 60 mm, matrix = 256 × 256, section thickness = 1.0 mm and no intersection gap, number of slices = 9. On T2*-weighted imaging, the signal intensity of cell grafts was measured by using the ROI technique with a minimum size of 50 pixels, and the decrease of signal intensity was normalized to the adjacent normal brain parenchyma. The ROI was drawn to cover all the areas of low signal intensity by an author (X.Z.) in a blinded manner.
In Vivo FLI
FLI was performed on a small animal in vivo FLI system (In-Vivo FxPro; Carestream, MI, USA) immediately after MRI. White light imaging, FLI with 487-nm excitation wavelength and 509-nm emission wavelength and digital X-ray imaging were obtained to detect the grafted cells. The fluorescence intensity of the in vivo imaging tests was quantified using the Carestream MI software by an author (M.C.), who was blinded to the experimental groups.
Therapeutic Effects
To observe the therapeutic effect of grafted cells, the infarct volume was measured, and behavioral tests were performed by an author (L.L.) who was blinded to the experimental groups. The infarct volume was measured on T2-wegihted images with ImageJ software (National Institutes of Health; Bethesda, MD, USA). For each slice, the hyperintense cerebral infarct region and contralateral brain area were manually outlined. Slice hyperintense areas were then summated to generate infarct volume as a percentage of contralateral intact brain volume.27 Behavioral tests were performed using modified neurological severity scores (mNSS) 28 to determine whether the transplanted NSCs could improve sensorimotor deficits, where neurological function was assessed on a scale of 0 to 18 (normal score = 0; maximal deficit score = 18).
Histology and Immunofluorescence Analyses
At 6 wk after transplantation, 3 animals in FTH-EGFP-NSCs group and control group were sacrificed after imaging tests by means of anesthetic overdose with sodium pentobarbital (60 mg/kg body weight) followed by transcardial perfusion with saline and the further 4% PFA in PBS. The brain was removed, postfixed for 1 to 2 h and then cryoprotected in 20% sucrose solution. Frozen brain tissue was cryosectioned in contiguous coronal 12-μm thickness slices and was prepared for Prussian blue staining and immunostaining for nestin, GFAP, NeuN, CD11b (specific for macrophage/microglia) and ferritin. For Prussian blue staining, the slices were immersed in working fluid (water with 10% potassium ferrocyanide and 2% concentrated hydrochloric acid, then mixed) for 30 min. For immunostaining studies, sections were blocked by goat or donkey serum for 30 min at room temperature, followed by incubation with primary antibodies against nestin (1:200), GFAP (1:200), NeuN (1:200), CD11b (1:200, Abcam Ltd. Cambridge, UK), or ferritin (1:200, Cell Signaling Technologies., Danvers, MA, USA) at 4 °C overnight. After washing 3 times with PBS, sections were incubated with Alexa Fluor 647-conjugated secondary antibodies (1:200) at room temperature in the dark for 2 h. Then, the sections were washed 3 times with PBS, followed by incubation with DAPI for 10 min at room temperature. After washing 3 times with PBS, immunoreactivity signals were observed by confocal microscopy (LSM 780; Carl Zeiss, Jena, Germany). RGB (red, green and blue) channels were collected sequentially with a matrix resolution of 1024 × 1024 and 8 averages. Negative control sections from each animal were prepared for immunofluorescence staining in an identical manner except the primary antibodies were omitted.
FTH Expression of Grafted Cells
Three animals in the FTH-EGFP-NSC group and 3 from the control group were sacrificed at 1 wk and 6 wk after transplantation. The brain tissues around the implantation site were obtained using laser microdissection for analysis of the FTH expression level. The FTH mRNA level was determined by RT-PCR assay and FTH protein expression level by Western blot analysis, as described above.
Statistical Analysis
Data were expressed as mean ± standard deviation (SD). Cell viability and apoptosis rates were expressed as average rates. Two independent samples t tests were used to compare T2 value, iron concentration, FTH mRNA level, and protein expression level between cells infected with LV-FTH-EGFP and mock-infected cells. One-way analysis of variance (ANOVA) was used to analyze the differences of viability and apoptosis rates rates among LV-FTH-EGFP-transduced cells, mock-infected cells, and nontransduced cells, followed by least significant difference (LSD) t test for pairwise differences with a confidence interval of 0.95. Two independent samples t tests were used to compare brain FTH mRNA level and protein expression level between the FTH-EGFP-NSC group and control group. The infarct volume and mNSS scores obtained at specific acquisition points were compared by using a repeated-measures ANOVA, followed by the Student–Neuman–Keuls post hoc test for multiple pairwise comparisons among different time points. Statistical analysis was performed by using SPSS 13.0 software for windows (Chicago, IL, USA). P < 0.05 was considered as a statistically significant difference.
Results
Efficacy of NSC Transduction
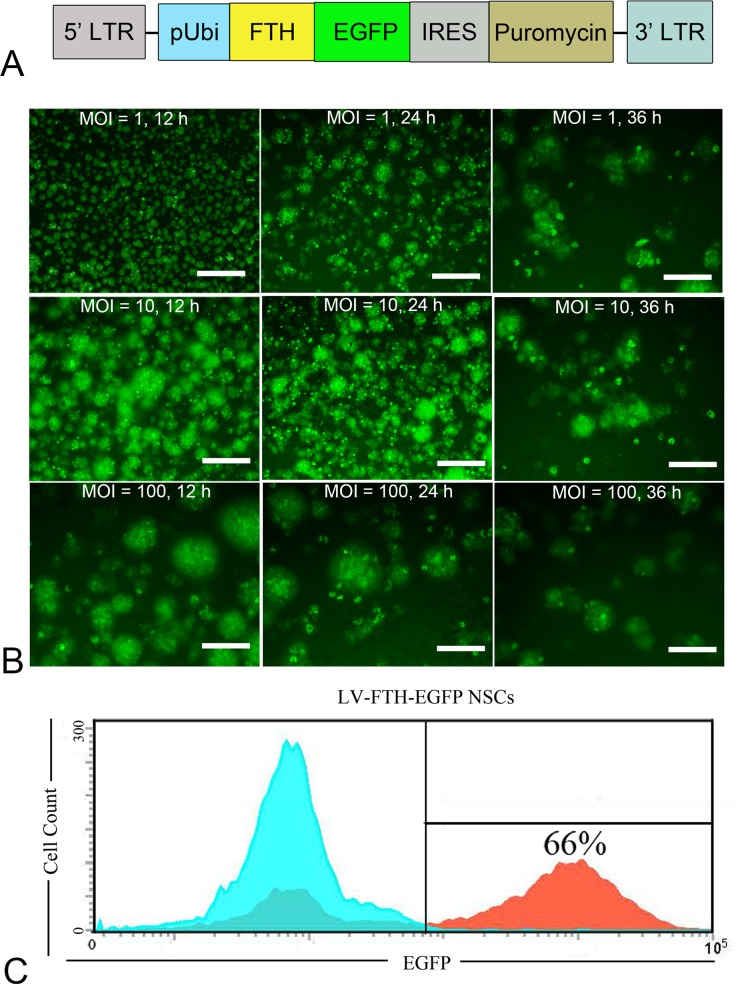
The constructed lentiviral vector carrying FTH and EGFP under the control of a Ubi promotor (LV-FTH-EGFP) is illustrated in Fig. 1A. After cells were transduced with LV-FTH-EGFP with different MOIs for different durations, fluorescence microscopy imaging analysis of EGFP-positive cells showed that LV-FTH-EGFP was able to transduce NSCs. The transduction was most efficient when cells were incubated with viral particles for 24 h at MOI = 10 (Fig. 1B), with an average transduction efficiency of 63.3% ± 3.6% and the highest efficiency was 66% as measured by flow cytometry (Fig. 1C). A 24 h duration and MOI = 10 were deemed the appropriate condition in our study and therefore used to deliver FTH to NSCs in the following experiments.
Fig. 1.
Cell transduction with genetic reporters. (A) A schematic diagram illustrates the constructed lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein (LV-FTH-EGFP), where the expression of FTH and EGFP was under the control of the human ubiquitin (Ubi) promoter, and the antibiotic-resistance gene puromycin was expressed via the IRES. (B) Fluorescence micrograph shows that the transduction efficiency reached the highest level when NSCs were transduced with LV-FTH-EGFP for 24 h at a MOI = 10. Scale bar = 10 μm. (C) Flow cytometry shows that the highest transduction efficiency of LV-FTH-EGFP for NSCs was 66%. LTR, long terminal repeat; EGFP, enhanced green fluorescent protein; FTH, ferritin heavy chain; LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; MOI, multiplicities of infection; NSC, neural stem cells; IRES, internal ribosome entry site.
Effect and Stability of NSC Transduction
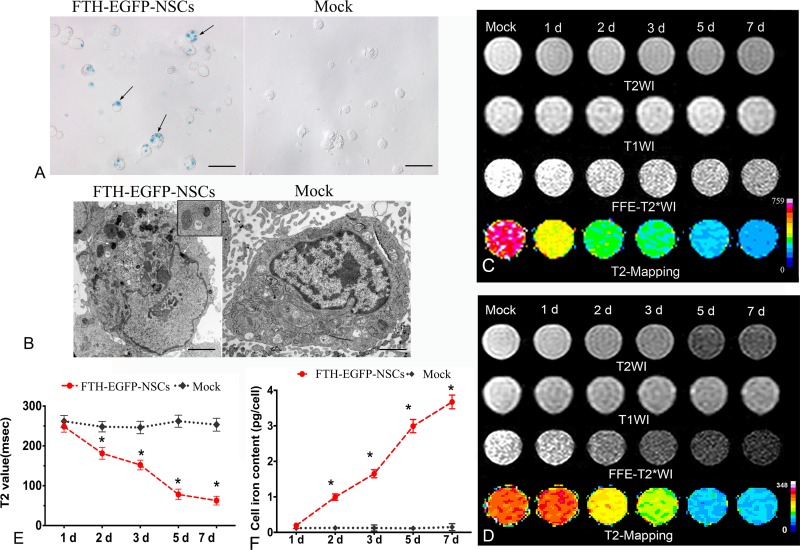
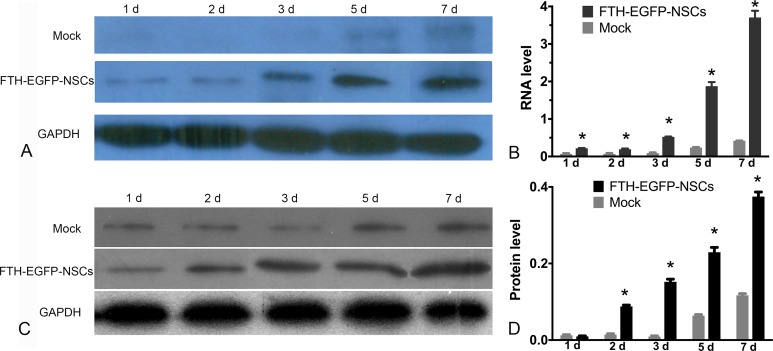
After NSCs were transduced with LV-FTH-EGFP under the appropriate condition, Prussian blue staining showed abundant blue-stained particles inside the cells (Fig. 2A); TEM revealed that the majority of iron particles were located inside cytoplasm (Fig. 2B). In the presence of FAC, the cells transduced with LV-FTH-EGFP showed a gradually reduced hypointense signal on T2- and T2*-weighted imaging, and gradual decrease in T2 values over time both on 1.5-T and 3.0-T MR scanners, as compared with mock-transduced cells. The T2 value reached the lowest level at 7 d after transduction (Fig. 2C and 2D, P < 0.0001). AAS showed that the cellular iron content in cells transduced with LV-FTH-EGFP increased in a time-dependent manner, from 0.168 ± 0.023 pg/cell at 1 d to 3.5378 ± 0.0985 pg/cell at 7 d after transduction, which were significantly higher than that in mock-transduced cells (0.1262 ± 0.0385 pg/cell and 0.149 ± 0.1376 pg/cell, respectively; Fig. 2E, P <0.0001). RT-PCR and Western blot analyses confirmed that FTH expression remained stable throughout a 7-d observation period (Fig. 3).
Fig. 2.
Cellular transduction effect and stability. (A) Prussian blue staining micrograph shows numerous blue-stained particles inside the cytosol of NSCs transduced with LV-FTH-EGFP (FTH-EGFP-NSCs), while no such particles were in mock-transduced NSCs (Mock). Scale bar = 10 μm. (B) Transmission electron microscopy micrographs demonstrate many iron particles in the cytosol of LV-FTH-EGFP-transduced NSCs (FTH-EGFP-NSCs, white arrows) but not in mock-transduced NSCs (Mock). The inserted magnified view shows a single iron-positive organelle. Scale bar = 2 μm. (C) In vitro MRI at 1.5 T and (D) 3.0 T both show a gradually decreased hypointense signal in cells transduced with LV-FTH-EGFP on T2- and T2*-weighted imaging over time. (E) T2 value decreased and (F) cellular iron content increased in a time-dependent manner in cells transduced with LV-FTH-EGFP but not in mock-transduced cells. LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; MRI, magnetic resonance imaging; NSCs, neural stem cells. *P < 0.05 compared with mock-transduced cells.
Fig. 3.
In vitro molecular biology assay of stability of FTH gene expression. (A) RT-PCR and (C) Western blot analysis show that FTH expression in LV-FTH-EGFP-transduced NSCs was stably increased throughout a 7-d observation period. Graphs show the densitometric analysis of the FTH mRNA (B) and FTH protein (D) expression levels in LV-FTH-EGFP-transduced NSCs (FTH-EGFP-NSCs) and mock-transduced NSCs (Mock). FTH, ferritin heavy chain; RT-PCR, Real-time PCR; LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NSCs, neural stem cells. *P < 0.05 compared with mock-transduced cells.
In Vitro Cytotoxicity Assay
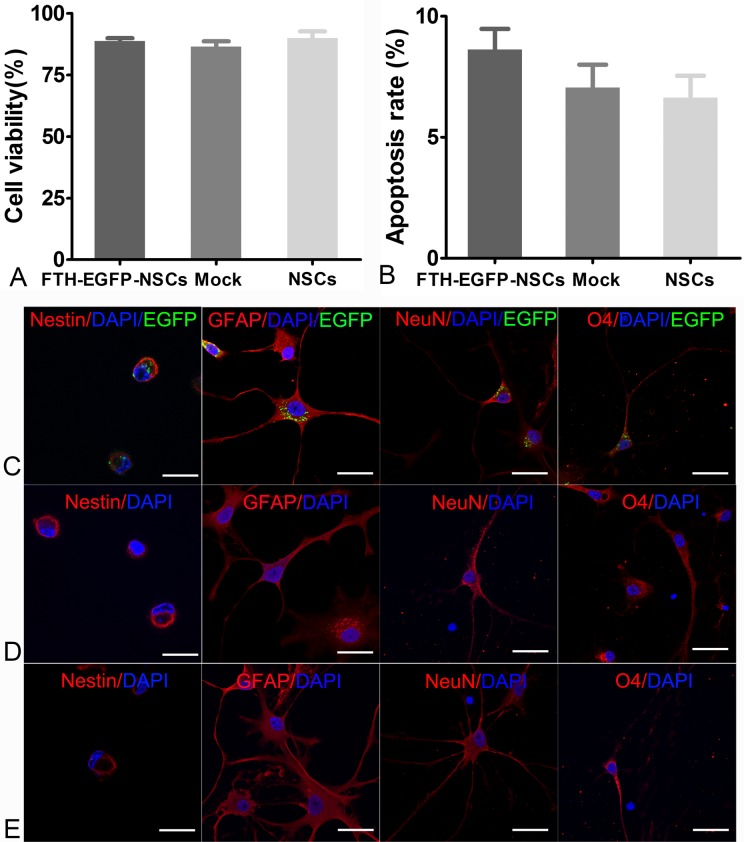
CCK8 assay showed that there was no significant difference in cell viability among LV-FTH-EGFP-transduced cells, mock-transduced cells, and nontransduced cells (P > 0.05; Fig. 4A). Flow cytometry showed that the apoptosis rates of LV-FTH-EGFP-transduced cells, mock-transduced cells, and nontransduced cells were 8.63% ± 0.85%, 7.05% ± 0.94%, and 6.63% ± 0.91%, respectively. No significant differences were found among the 3 groups (P > 0.05 Fig. 4B). LV-FTH-EGFP-transduced cells, mock-transduced cells, and nontransduced cells all showed positive staining for GFAP, NeuN, and O4 after induction of differentiation, which indicated an intact multilineage differentiation potential (Fig. 4C).
Fig. 4.
In vitro cytotoxicity assay of transduction. Graphs show no differences in cell viability (A) or apoptosis rate (B) among LV-FTH-EGFP-transduced cells (FTH-EGFP-NSCs), mock-transduced cells (Mock), or nontransduced cells (NSCs). Immunofluorescence micrographs show that LV-FTH-EGFP-transduced cells (C), mock-transduced cells (D), and nontransduced cells (E) all were positive for nestin and could differentiate into GFAP-positive, NeuN-positive, and O4-positive neural lineage cells after 7 d of induction differentiation. Scale bar = 10 μm. GFAP, glial fibrillary acidic protein; LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; NeuN, neuron-specific nuclear protein. NSCs, neural stem cells; DAPI, 4',6-diamidino-2-phenylindole.
In Vivo MRI and FLI Tracking
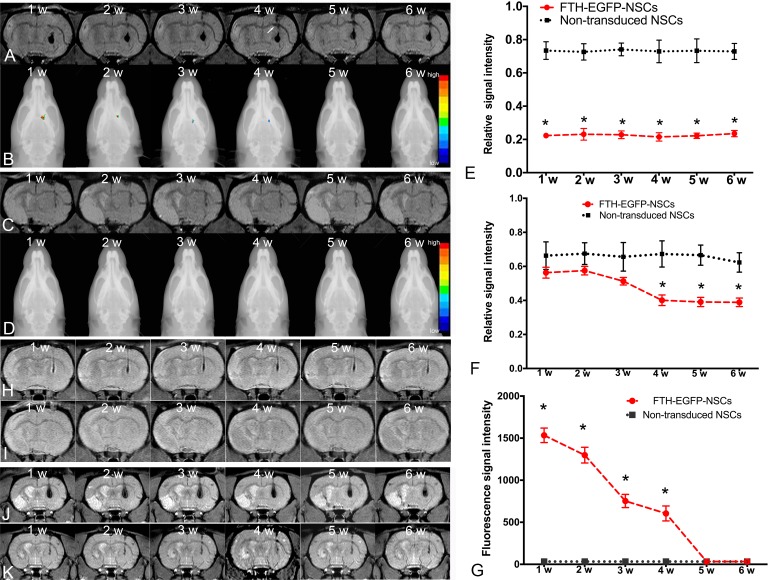
On MRI, 1 wk after transplantation, cells transduced with LV-FTH-EGFP were detected as strongly hypointense areas in the striatum contralateral to the infarcted hemisphere on T2*-weighted images both at 3.0 T (Fig. 5A) and 1.5 T (Fig. 5J) MR scanners. These hypointense signals remained consistent until 6 wk after injection on MRI. At 4 wk after transplantation, the corpus callosum contralateral to the infarct lesion displayed linear hypointense signal on T2*-weighted imaging (Fig. 5A). No signal representing migrating cells was found in the vicinity of the infarcted area. Graphs show the relative signal intensities of the striatum (Fig. 5E) and the corpus callosum (Fig. 5F) measured at 3.0 T MRI in animals grafted with LV-FTH-EGFP-transduced cells and nontransduced cells (Fig. 5H). On PDW images at 3.0 T, the cells transduced with LV-FTH-EGFP also appeared as a persistent, but less obvious hypointense area in the striatum compared to T2*-weighted imaging. On FLI, the grafted cells pretransduced with LV-FTH-EGFP were detected as a focal fluorescence signal 1 wk after transplantation, then these signals gradually declined and eventually disappeared by 5 wk after transplantation (Fig. 5B and G). No positive signals were found in animals grafted with nontransduced cells on T2*-weighted images (Fig. 5C), PDW images (Fig. 5I) at 3.0-T and T2*-weighted images (Fig. 5K) at 1.5 T as well as on FLI images (Fig. 5D).
Fig. 5.
In vivo MR imaging and fluorescence imaging tracking of grafted cells. Coronal T2*-weighted (A) and PDW images (H) obtained at 3.0 T MR scanner show a persistent hypointense area in the implantation site until 6 wk after transplantation in the implantation site of animals grafted with LV-FTH-EGFP-transduced NSCs. Hypointense area on PDW images was less pronounced than that on T2*-weighted images. (B) Fluorescence images overlaid on radiograph show gradually declined focal fluorescence signal until 4 wk after transplantation in the implantation site of animals grafted with LV-FTH-EGFP-transduced NSCs. At 4 wk after transplantation, a spotty hypointense area was found in the corpus callosum contralateral to the infarct region on T2*-weighted images in animals grafted with LV-FTH-EGFP-transduced NSCs (arrow). No such developmental hypointense signal or fluorescence signal were observed in animals grafted with nontransduced NSCs on T2*-weighted (C), PDW images (I), and fluorescence images (D). Graphs show relative signal intensity of the striatum (E) and corpus callosum (F) on T2*-weighted images at 3.0 T and relative fluorescence signal intensity in the striatum (G) in animals grafted with LV-FTH-EGFP-transduced NSCs (FTH-EGFP-NSCs) and with nontransduced NSCs. * P < 0.05 compared with animals transplanted with nontransduced NSCs. (J) Coronal T2*-weighted images obtained at 1.5 T MR scanner also show a persistent hypointense area in the implantation site until 6 wk after transplantation in the implantation site of animals grafted with LV-FTH-EGFP-transduced NSC, while no obvious hypointense signal was seen in animals grafted with nontransduced NSCs (K). LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; MR, magnetic resonance; PDW, proton density-weighted.
Therapeutic Effects of Transduced NSCs
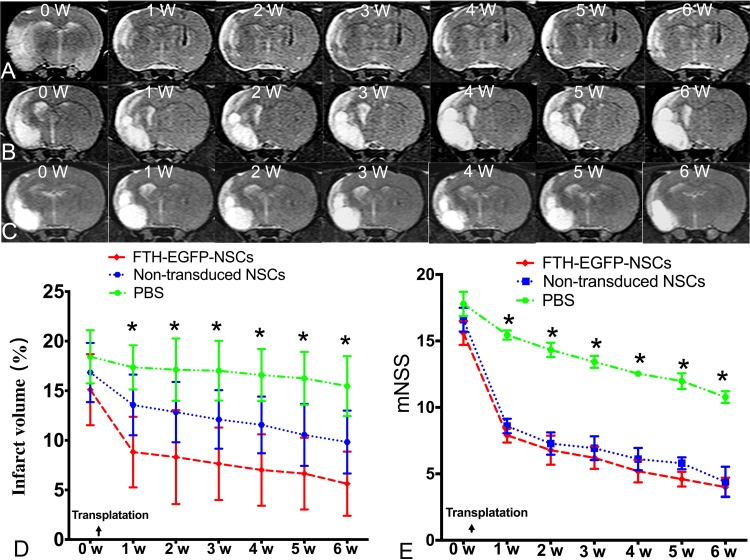
On T2-weighted imaging, the infarcted region in animals grafted with transduced cells, nontransduced cells and PBS all appeared as hyperintense signals (Fig. 6A to C). The infarct volume showed a slow decline from 1 wk to 6 wk after transplantation in both the FTH-EGFP-NSC group and control group (Fig. 6D). Behavioral tests showed a gradual decrease in mNSS scores beginning at 1 wk after transplantation in both the FTH-EGFP-NSC group and control group compared with the PBS group. Both the infarct volume and mNSS scores were significantly different in animals grafted with LV-FTH-EGFP-transduced and nontransduced cells, compared to that grafted with PBS 1 wk after transplantation (P < 0.05). However, there were no significant differences between animals grafted with LV-FTH-EGFP-transduced cells and with nontransduced cells at any time point (P > 0.05; Fig. 6E). These data indicated a gradual improvement of the sensorimotor deficits in animals grafted with transduced and nontransduced cells.
Fig. 6.
Therapeutic effects of transduced NSCs. Coronal T2-weighted images show that the infarcted area in animals grafted with LV-FTH-EGFP-transduced NSCs (A), nontransduced cells (B) and PBS (C) appeared as hyperintense signal. Graphs show that the measured infarct volume (D) and modified neurological severity score (mNSS, E) in animals grafted with LV-FTH-EGFP-transduced NSCs (FTH-EGFP-NSCs) and nontransduced NSCs. LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; NSCs, neural stem cells; PBS, phosphate-buffered saline. *P < 0.05 compared with animals grafted with FTH-EGFP-NSCs and nontransduced NSCs.
Histological Iron Localization and Immunofluorescence-Detected Colocalizations
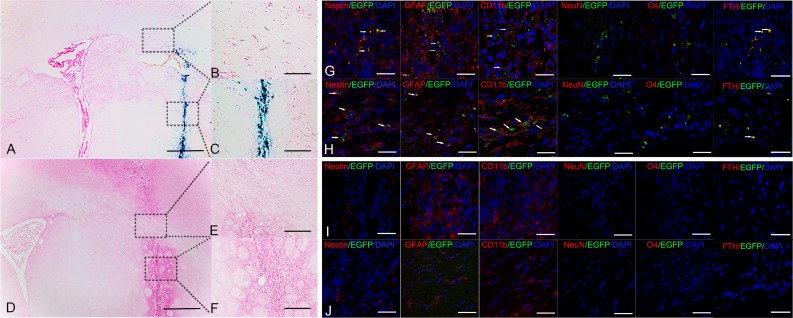
At 6 wk after transplantation, Prussian blue staining showed many positive cells and blue-stained extracellular particles at the injection site in the FTH-EGFP-NSC group (Fig. 7A and C). A small number of positive cells was found in corpus callosum contralateral to the infarct region, which correlated well with the findings on the T2*-weighted images (Fig. 7B). In the control group, no positive cells were observed by Prussian blue staining (Fig. 7D to F). Immunofluorescence staining showed that there were few viable, nestin+ cells, considerable FTH+/EGFP+ cells and GFAP+/EGFP+ astrocytes, and many CD11b+/EGFP+ macrophages/microglia in the injection site (Fig. 7G) and corpus callosum (Fig. 7H) contralateral to the infarct region in the FTH-EGFP-NSC group. This indicated that a small portion of grafted cells were viable, and grafted cells mainly differentiated into GFAP+/EGFP+ astrocytes. A massive number of cells died and were then phagocytized by macrophages/microglia. A low number of grafted cells migrated toward the corpus callosum, where they mainly differentiated into EGFP+/GFAP+ astrocytes. No EGFP+/NeuN+ or EGFP+/O4+ oligodendrocytes were found at the injection site or the corpus callosum. Similar findings were observed in the control group (Fig. 7I and J).
Fig. 7.
Histological studies of the grafted cells. (A) Prussian blue staining micrographs show many positive cells and extracellular blue-stained particles in the implantation site (C) and a few positive cells in corpus callosum (B) contralateral to the infarct region in animals grafted with LV-FTH-EGFP-transduced NSCs at 6 wk after transplantation. (D) Prussian blue staining micrographs show no positive cells in the implantation site (F) and corpus callosum (E) contralateral to the infarct region in animals grafted with nontransduced NSCs at 6 wk after transplantation. (A, D, Bar = 50 μm (A and B); Bar = 25 μm (B, C, E, and F). Confocal microscopy micrographs show a few Nestin+/EGFP+ cells (arrows), considerable FTH+/EGFP+ cells (arrows), many GFAP+/EGFP+ cells (arrows), and CD11b+/EGFP+ cells (arrows) in the injection site (G) and the corpus callosum (H) in animals grafted with LV-FTH-EGFP-transduced NSCs at 6 wk after transplantation. Confocal microscopy micrographs show no Nestin+/EGFP+ cells but Nestin+, GFAP+, and CD11b+ cells in the injection site (I), and GFAP+, CD11b+ cells in the corpus callosum (J) in animals grafted with nontransduced cells. Scale bar = 10 μm (G-J). LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; NSCs, neural stem cells; GFAP, glial fibrillary acidic protein; NeuN, neuronspecific nuclear protein; DAPI, 4',6-diamidino-2-phenylindole.
FTH Expression of Grafted Cells
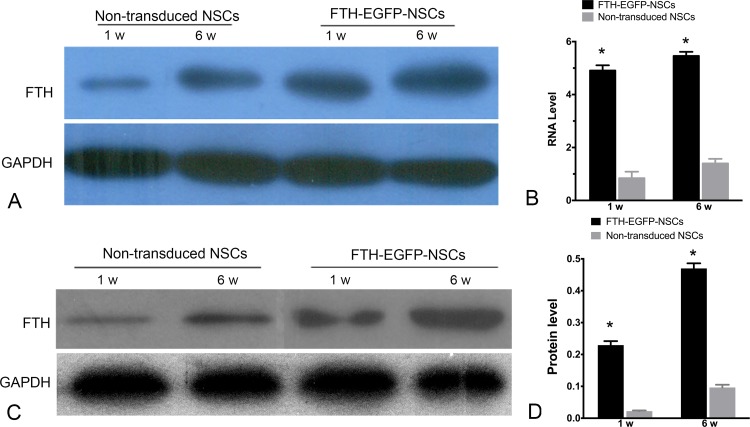
RT-PCR and Western blot analyses showed that FTH RNA and protein expression levels of cell grafts in FTH-EGFP-NSC group increased from 1 wk to 6 wk after transplantation, which were significantly higher than that in the control group (P < 0.0001; Fig. 8).
Fig. 8.
In vivo FTH expression of grafted cells. (A) RT-PCR and (B) Western blot analysis show high FTH expression in animals grafted with LV-FTH-EGFP-transduced NSCs (FTH-EGFP-NSCs) compared with animals grafted with nontransduced NSCs at 1 and 6 wk after transplantation. Graphs show the densitometric analysis of FTH RNA levels (B) and protein levels (D) in animals grafted with LV-FTH-EGFP-transduced NSCs (FTH-EGFP-NSCs) and with nontransduced cells. FTH, ferritin heavy chain; LV-FTH-EGFP, lentiviral vector encoding ferritin heavy chain and enhanced green fluorescent protein; RT-PCR, real-time PCR; NSCs, neural stem cells. *P < 0.05 compared with nontransduced cells.
Discussion
Our results showed that NSCs could be successfully transduced with a LV-FTH-EGFP viral vector to express the FTH gene stably. This genetic modification could lead to intracellular iron acumination that makes cells visible on MRI, but without noticeable detrimental effects on the cell viability, apoptosis, or multilineage differentiation capacity. After transplantation, the distribution and migration of the grafted, transduced cells in stroke could be tracked in vivo by MRI and FLI in a well-matched manner until 4-wk follow-up. Thereafter, a discrepancy of detectable biological distribution of grafted cells was observed.
Stroke is an extremely enticing target for stem, progenitor, or engineered cell therapies because of an urgent need to find therapies that protect at-risk brain cells as well as to promote functional recovery after the initial ischemic event.29 Imaging of cell tracking and the host response are relevant to the development of cell-based, biological, and pharmacological restorative therapies for the treatment of stroke. Use of imaging is also strongly encouraged in clinical trials to provide as much information as possible to assess vascular/structural lesions, infarct size, cell viability, location, the success and safety of implantation, and inflammation.30 Cellular MRI may be used to monitor the delivery and tracking of cells and improve understanding of the host response to restorative therapies.29 However, there are several limitations to cellular labeling with magnetic nanoparticles for long-term MRI. Most importantly, cellular labeling with magnetic nanoparticles do not allow for the reliable discrimination by MRI between live and dead cells,10,31 though a long-term cell tracking (even up to 6 months) has been reported by using this method.32 The MRI signal from magnetic particles may be compounded by the extracellular particles or particles engulfed by host macrophages if the transplanted cells died.10,31 Moreover, the quantification of labeled cells by MRI is an indirect technique. Signal change is the result of the overall concentration of magnetic nanoparticles and cannot reflect the actual total number of cells. The last dilution of the iron oxide label is a possible limitation for long-term tracking of cell survival, as the MR signal is lost over time due to cellular proliferation, especially with rapidly dividing cells. There are also concerns that stem cells may divide asymmetrically, leading to an unequal distribution of label with 1 daughter cell receiving most of the nanoparticles. A way to circumvent these problems may involve the modification of target cells with a transgenic reporter, such as ferritin, transferrin receptor, and MagA, with which cells can be engineered to produce iron-containing proteins, resulting in MRI contrast in the transplanted cells.12 However, overexpression of the transferrin receptor could lead to iron-catalyzed free radical formation via the Fenton reaction and could be potentially toxic to cells.33 MagA is a gene in magnetotactic bacteria known to be involved with iron transport. Intracellular particles of biogenic magnetite (Fe3O4) synthesized by the MagA gene can also be used as MRI reporter genes.16,34 However, a recent study indicated that MagA is not involved in magnetosome formation.35
Ferritin, the ubiquitous iron storage protein, is probably the most popular reporter gene for MRI. Overexpression of the heavy chain alone, or in combination with the light chain of ferritin or the transferrin receptor, results in the elevation of intracellular ferritin iron stores, and a corresponding change in R2 relaxation.36 It was first shown that overexpression of ferritin in glioma tumors 22 and in adenocarcinoma cells 17 generated sufficient inhomogeneities in local magnetic field to be visualized on T2- and T2*-weighted images. The Fe3+ stored in ferritin does not participate in the Fenton reaction; thus, it is not expected to be toxic.17 Despite the many advantages of the ferritin reporter, its use in tracking the long-term fate of stem cell grafts is still inadequate. Initially, ferritin proved successful as an MRI reporter in mouse neural stem cells implanted in the intact brain.24 In this study, the MRI reporter FTH and FLI reporter EGFP were transduced into NSCs via a lentiviral vector construct encoding FTH gene and EGFP gene. The transduction efficiency achieved was as high as 66%. The iron content in the lentiviral vector-transduced cells reached at the order of pg/cell, which is significantly higher than the order of fg/cell reached in cells transduced by a plasmid vector.24 Lentiviral vectors are capable of transducing dividing and nondividing cells with high efficiency, such as neurons and NSCs. They integrate into the genomic DNA of target cells and allow long-term expression of a reporter or therapeutic gene. Most importantly, the cDNA that is inserted into the host genome is duplicated at the time of mitosis together with the cellular chromosomes and is transmitted to the progeny of transduced cells, thus enabling long-term persistence of gene expression with cell division.37–39 In our study, overexpression of FTH had no detrimental effects on cellular proliferation, viability, or multilineage differentiation. After transplanted into brain, there was no tumor formation over a 6-wk observation period. This indicates that FTH can function as an efficient and safe reporter gene to track cell survival and proliferation of stem/progenitor cells in vivo.
In our study, the distribution and migration of grafted, LV-FTH-EGFP-transduced NCSs in stroke was detectable by MRI without the need for exogenously administered iron over a relatively long period. The cell grafts were detected as a persistent hypointense signal on T2 and T2*-weighted imaging up to 6 wk after transplantation. Histology showed that there were a few viable stem cells and many grafted cells that differentiated into astrocytes at 6 wk after transplantation. These findings suggest that the persistent iron-dependent signal on T2*-weighted imaging can overestimate the the true size of viable cell grafts. Given the fact that the number of transplanted stem cells that acutely home and survive in host organs is generally quite low,40 the gradual reduction in graft size is most likely resultant from the progressively decreased survival of grafted cells. It is noted that histologically many extracellular iron particles and microglia/macrophages were present in the implantation site at 6 wk after transplantation. These indicate that the long-term cell viability reported by the FTH gene might be overestimated. Its accuracy can be affected by several factors, in particular, cell death and concomitant microglia/macrophage accumulation potentially activated by immunoreaction. In contrast, cell tracking in the infarcted mouse heart with the ferritin reporter gene showed that the signal attenuation by ferritin-containing grafts persists for at least 1 mo and correlates well with the histologically determined graft size.21 In that study, the transduction efficacy was 20%, and myocardial inflammation was mild as demonstrated by the presence at the scattered macrophages in the myocardium of infarct border zone. If the transplantation area is undergoing an inflammatory response, iron-based reporter proteins in dead or live cells can be phagocytosed by macrophages, resulting in false-positive signals, similar to that with direct magnetic nanoparticle labels.20 Collectively, ferritin reporter gene-based MRI has the advantages of the long-term tracking of the special distribution and migration of transplanted stem cells. Its capacity to report the actual survival of transplanted cells might be affected by the immunoreaction in the host organ.
Transplantation of NSCs via different delivery routes is considered as a promising approach for reducing the size of the infarction and neurological function recovery in ischemic stroke.3,4 However, the optimal cell delivery route remains unresolved so far. At present, intravenous, intra-arterial, and intracerebral administration have been the most frequently used delivery routes.3 Intravenous transplantation is the most commonly used technique in both preclinical and clinical studies41 because it is relatively easy to perform, yields better distribution into the injured brain areas, and avoids the process of invasive surgery.3,42 Nevertheless, most of the infused cells through the intravenous (i.v.) delivery route are rapidly trapped in the lungs, resulting in a low number of grafted cells in the brain. Intra-arterial administration may increase migration of the cells to the ischemic hemisphere.41 However, both i.v. and intra-arterial cell transplantations suffer from a number of risks and complications, for example, an instant immune response against the nonautologous cells and a microembolism after systemic stem cell injection.3,42 On the other hand, intracerebral delivery is the most efficient route for cell transplantation and allows the spatially precise deposition of cells within or next to a lesion, although it is invasive.3 In addition, it is superior to other delivery approaches for MRI because of the largest absolute cell numbers reaching the infarcted brain. In this study, intracerebral delivery of NSCs was performed to obtain enough cells to confer therapeutic effects for on ischemic stroke and for MRI.
The main rationale to employ cell-based therapies in ischemic stroke is to replace infarcted CNS tissue in an organotypic appropriate manner.43 In our study, both transduced cells and nontransduced cells exerted a therapeutic effect on ischemic stroke as the recipient animals showed gradual improvement in functional recovery and reduction in infarction volume starting from 1 wk after transplantation, compared with the PBS group. However, histology results showed that massive cells death was present in implantation site, and a few stem cells migrated toward the lesion. Moreover, most of the surviving cells differentiated into astrocytes at the graft site and migration site. These findings indicated that directed differentiation of stem cells might not play a critical role in stem cell–based therapy for stroke.44 The observed cellular mechanisms of action might be presumably mediated by nondirect substitution or a paracrine mechanism to promote functional benefits.3,40,45
There were several limitations in our study. First, exogenous NSCs appear to have a limited long-term survival rate after transplantation. It has been reported that 70% to 90% of implanted cells in the striatum die after transplantation, and the survival rate was only 1% to 32%.45 Coupling therapeutic genes to an FTH reporter gene would be needed in the future to promote the survival of transplanted NSCs. Secondly, NSCs were transplanted into immunocompetent rats, and no immunosuppressive medication was administered in our study. However, our results clearly described the ability of the FTH reporter gene to track the fate of allogeneic stem cells. The occurrence of an immune response/inflammation at the graft site would affect the capacity of FTH-based MR reporter to monitor the long-term viability of stem cell grafts. A more sensitive MRI reporter gene, and particularly one that can yield a gain of signal rather than signal attenuation, would be more favorable for under such conditions.
Conclusion
In summary, our study results suggest that the FTH gene can be safely and efficiently delivered to NSCs via a lentiviral vector. The overexpression of reporter genes enables transduced cells to be clearly visible on MRI and FLI. FTH can act as a robust MRI reporter to track the distribution and migration of grafted cells for acute ischemic stroke. In vivo tracking of the long-term viability of stem cells with the use of such an iron-based reporter gene could be compounded by several factors, such as the death of stem cells themselves and the reactive immune response and inflammation of host tissues.
Footnotes
Authors’ Note: Fang Zhang and Xiaohui Duan contributed equally to this work.
Ethical Approval: The experimental protocols were approved by the Institutional Animal Care and Use Committee of the Sun Yat-Sen University.
Statement of Human and Animal Rights: This study was conducted in accordance with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health, Bethesda, Maryland, USA.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: This study was supported by the National Natural Science Foundation (Grant Nos: 81371607, 81571739) and National Basic Research Program of China (Grant No: 2015CB755500) and Natural Science Foundation of Guangdong Province, China (Grant No: 2014A030312018) and Elite Young Scholars Program of Sun Yat-sen Memorial Hospital (Grant No: J201403).
ORCID iD: Jun Shen http://orcid.org/0000-0001-7746-5285
References
- 1. Chen CJ, Ding D, Starke RM, Mehndiratta P, Crowley RW, Liu KC, Southerland AM, Worrall BB. Endovascular vs medical management of acute ischemic stroke. Neurology. 2015;85(22):1980–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Appelros P, Terént A. Thrombolysis in acute stroke. Lancet. 2015;385(9976):1394. [DOI] [PubMed] [Google Scholar]
- 3. Leong WK, Lewis MD, Koblar SA. Concise review: preclinical studies on human cell-based therapy in rodent ischemic stroke models: where are we now after a decade? Stem Cells. 2013;31(6):1040–1043. [DOI] [PubMed] [Google Scholar]
- 4. Gutierrez-Fernandez M, Rodriguez-Frutos B, Ramos-Cejudo J, Otero-Ortega L, Fuentes B, Diez-Tejedor E. Stem cells for brain repair and recovery after stroke. Expert Opin Biol Ther. 2013;13(11):1479–1483. [DOI] [PubMed] [Google Scholar]
- 5. Flax JD, Aurora S, Yang C, Simonin C, Wills AM, Billinghurst LL, Jendoubi M, Sidman RL, Wolfe JH, Kim SU, et al. Engraftable human neural stem cells respond to developmental cues, replace neurons, and express foreign genes. Nat Biotechnol. 1998;16(11):1033–1109. [DOI] [PubMed] [Google Scholar]
- 6. Martino G, Pluchino S. The therapeutic potential of neural stem cells. Nat Rev Neurosci. 2006;7(5):395–406. [DOI] [PubMed] [Google Scholar]
- 7. Kokaia Z, Martino G, Schwartz M, Lindvall O. Cross-talk between neural stem cells and immune cells: the key to better brain repair? Nat Neurosci. 2012;15(8):1078–1087. [DOI] [PubMed] [Google Scholar]
- 8. Daadi MM, Li Z, Arac A, Grueter BA, Sofilos M, Malenka RC, Wu JC, Steinberg GK. Molecular and magnetic resonance imaging of human embryonic stem cell-derived neural stem cell grafts in ischemic rat brain. Mol Ther. 2009;17(7):1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edmundson M, Thanh NT, Song B. Nanoparticles based stem cell tracking in regenerative medicine. Theranostics. 2013;3(8):573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cromer Berman SM, Walczak P, Bulte JW. Tracking stem cells using magnetic nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2011;3(4):343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srivastava AK, Bulte JW. Seeing stem cells at work in vivo. Stem Cell Rev. 2014;10(1):127–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kircher MF, Gambhir SS, Grimm J. Noninvasive cell-tracking methods. Nat Rev Clin Oncol. 2011;8(11):677–688. [DOI] [PubMed] [Google Scholar]
- 13. Gulaka PK, Yu JX, Liu L, Mason RP, Kodibagkar VD. Novel S-Gal((R)) analogs as (1)H MRI reporters for in vivo detection of beta-galactosidase. Magn Reson Imaging. 2013;31(6):1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qin C, Cheng K, Chen K, Hu X, Liu Y, Lan X, Zhang Y, Liu H, Xu Y, Bu L, et al. Tyrosinase as a multifunctional reporter gene for Photoacoustic/MRI/PET triple modality molecular imaging. Sci Rep. 2013;3:1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weissleder R, Moore A, Mahmood U, Bhorade R, Benveniste H, Chiocca EA, Basilion JP. In vivo magnetic resonance imaging of transgene expression. Nat Med. 2000;6(3):351–355. [DOI] [PubMed] [Google Scholar]
- 16. Goldhawk DE, Lemaire C, McCreary CR, McGirr R, Dhanvantari S, Thompson RT, Figueredo R, Koropatnick J, Foster P, Prato FS. Magnetic resonance imaging of cells overexpressing MagA, an endogenous contrast agent for live cell imaging. Mol Imaging. 2009;8(3):129–139. [PubMed] [Google Scholar]
- 17. Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005;11(4):450–454. [DOI] [PubMed] [Google Scholar]
- 18. Gilad AA, McMahon MT, Walczak P, Winnard PT, Jr, Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25(2):217–219. [DOI] [PubMed] [Google Scholar]
- 19. Naumova AV, Reinecke H, Yarnykh V, Deem J, Yuan C, Murry CE. Ferritin overexpression for noninvasive magnetic resonance imaging-based tracking of stem cells transplanted into the heart. Mol Imaging. 2010;9(4):201–210. [PMC free article] [PubMed] [Google Scholar]
- 20. Naumova AV, Modo M, Moore A, Murry CE, Frank JA. Clinical imaging in regenerative medicine. Nat Biotechnol. 2014;32(8):804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campan M, Lionetti V, Aquaro GD, Forini F, Matteucci M, Vannucci L, Chiuppesi F, Di Cristofano C, Faggioni M, Maioli M, et al. Ferritin as a reporter gene for in vivo tracking of stem cells by 1.5-T cardiac MRI in a rat model of myocardial infarction. Am J Physiol Heart Circ Physiol. 2011;300(6): H2238–H2250. [DOI] [PubMed] [Google Scholar]
- 22. Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7(2):109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naumova AV, Yarnykh VL, Balu N, Reinecke H, Murry CE, Yuan C. Quantification of MRI signal of transgenic grafts overexpressing ferritin in murine myocardial infarcts. NMR Biomed. 2012;25(10):1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deans AE, Wadghiri YZ, Bernas LM, Yu X, Rutt BK, Turnbull DH. Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn Reson Med. 2006;56(1):51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gera A, Steinberg GK, Guzman R. In vivo neural stem cell imaging: current modalities and future directions. Regen Med. 2010;5(1):73–86. [DOI] [PubMed] [Google Scholar]
- 26. Wen X, Wang Y, Zhang F, Zhang X, Lu L, Shuai X, Shen J. In vivo monitoring of neural stem cells after transplantation in acute cerebral infarction with dual-modal MR imaging and optical imaging. Biomaterials. 2014;35(16):4627–4635. [DOI] [PubMed] [Google Scholar]
- 27. Zhang F, Guo RM, Yang M, Wen XH, Shen J. A stable focal cerebral ischemia injury model in adult mice: assessment using 7 T MR imaging. AJNR Am J Neuroradiol. 2012;33(5):935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J, Li Y, Wang L, Zhang Z, Lu D, Lu M, Chopp M. Therapeutic benefit of intravenous administration of bone marrow stromal cells after cerebral ischemia in rats. Stroke. 2001;32(4):1005–1011. [DOI] [PubMed] [Google Scholar]
- 29. Participants. Stem Cell Therapies as an Emerging Paradigm in Stroke (STEPS): bridging basic and clinical science for cellular and neurogenic factor therapy in treating stroke. Stroke. 2009;40(2):510–515. [DOI] [PubMed] [Google Scholar]
- 30. Savitz SI, Cramer SC, Wechsler L, STEPS 3 Consortium. Stem cells as an emerging paradigm in stroke 3: enhancing the development of clinical trials. Stroke. 2014;45(2):634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kraitchman DL, Bulte JW. Imaging of stem cells using MRI. Basic Res Cardiol. 2008;103(2):105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Z, Li C, Yang S, Xu J, Shen Y, Xie X, Dai Y, Lu H, Gong H, Sun A, et al. Magnetic resonance hypointensive signal primarily originates from extracellular iron particles in the long-term tracking of mesenchymal stem cells transplanted in the infarcted myocardium. Int J Nanomedicine. 2015;10:1679–1690.25767388 [Google Scholar]
- 33. Kotamraju S, Chitambar CR, Kalivendi SV, Joseph J, Kalyanaraman B. Transferrin receptor-dependent iron uptake is responsible for doxorubicin-mediated apoptosis in endothelial cells: role of oxidant-induced iron signaling in apoptosis. J Biol Chem. 2002;277(19):17179–17187. [DOI] [PubMed] [Google Scholar]
- 34. Zurkiya O, Chan AW, Hu X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magn Reson Med. 2008;59(6):1225–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uebe R, Henn V, Schuler D. The MagA protein of Magnetospirilla is not involved in bacterial magnetite biomineralization. J Bacteriol. 2012;194(5):1018–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vande Velde G, Himmelreich U, Neeman M. Reporter gene approaches for mapping cell fate decisions by MRI: promises and pitfalls. Contrast Media Mol Imaging. 2013;8(6):424–431. [DOI] [PubMed] [Google Scholar]
- 37. Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. [DOI] [PubMed] [Google Scholar]
- 38. Consiglio A, Gritti A, Dolcetta D, Follenzi A, Bordignon C, Gage FH, Vescovi AL, Naldini L. Robust in vivo gene transfer into adult mammalian neural stem cells by lentiviral vectors. Proc Natl Acad Sci U S A. 2004;101(41):14835–14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reumers V, Deroose CM, Krylyshkina O, Nuyts J, Geraerts M, Mortelmans L, Gijsbers R, Van den Haute C, Debyser Z, Baekelandt V. Noninvasive and quantitative monitoring of adult neuronal stem cell migration in mouse brain using bioluminescence imaging. Stem Cells. 2008;26(9):2382–2390. [DOI] [PubMed] [Google Scholar]
- 40. Dimmeler S, Ding S, Rando TA, Trounson A. Translational strategies and challenges in regenerative medicine. Nat Med. 2014;20(8):814–821. [DOI] [PubMed] [Google Scholar]
- 41. Cui LL, Kerkelä E, Bakreen A, Nitzsche F, Andrzejewska A, Nowakowski A, Janowski M, Walczak P, Boltze J, Lukomska B, et al. The cerebral embolism evoked by intra-arterial delivery of allogeneic bone marrow mesenchymal stem cells in rats is related to cell dose and infusion velocity. Stem Cell Res Ther. 2015;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boltze J, Arnold A, Walczak P, Jolkkonen J, Cui L, Wagner DC. The dark side of the force - constraints and complications of cell therapies for stroke. Front Neurol. 2015;6:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haas S, Weidner N, Winkler J. Adult stem cell therapy in stroke. Curr Opin Neurol. 2005;18(1):59–64. [DOI] [PubMed] [Google Scholar]
- 44. Bliss T, Guzman R, Daadi M, Steinberg GK. Cell transplantation therapy for stroke. Stroke. 2007;38(suppl 2):817–826. [DOI] [PubMed] [Google Scholar]
- 45. Hyun JS, Tran MC, Wong VW, Chung MT, Lo DD, Montoro DT, Wan DC, Longaker MT. Enhancing stem cell survival in vivo for tissue repair. Biotechnol Adv. 2013;31(5):736–743. [DOI] [PubMed] [Google Scholar]