Abstract
Traumatic brain injury (TBI) may cause neurological damage, but an effective therapy and the associated mechanisms of action have not yet been elucidated. A TBI model was established using the modified Feeney method. A2B5+ cells, an oligodendroglial progenitor, were acquired from induced pluripotent stem cells (iPSCs) by mouse embryonic fibroblasts and were transplanted into the injured site. The neurological severity score (NSS) was recorded on 3 d, 7 d, 11 d, 15 d, and 19 d. Seven days after transplantation, oligodendrocytes 2 (Olig2) and myelin basic protein (MBP) were detected by immunohistochemistry (IHC) and Western blot (WB), and long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) were screened by microarray technology. Moreover, we took an intersection of the differentially expressed lncRNAs or mRNAs and scanned 10 kb upstream and downstream of the common lncRNAs. Meanwhile, Gene Ontology (GO) and pathway analysis on mRNAs was performed in the A2B5+ iPSC group. A2B5+ iPSCs survived and migrated around the injury site and differentiated into oligodendrocytes. Meanwhile, the increase in Olig2 and MBP were higher in A2B5+ cell-engrafted rats than that in TBI rats. However, the NSSs in the A2B5+ iPSC group were lower than that in the TBI group. Between the TBI and sham groups, 270 lncRNAs and 1,052 mRNAs were differently expressed (P < 0.05, fold change (FC) > 2), while between the A2B5+ iPSC and TBI groups, 83 lncRNAs and 360 mRNAs were differently expressed (P < 0.05, FC > 2). Meanwhile, 37 lncRNAs and 195 mRNAs were simultaneously changed in the 2 parts. Using bioinformatic analysis, we found the crucial lncRNA and mRNA were ENSRNOT00000052577 and Kif2c in the TBI brain with cell transplantation. This study demonstrated that A2B5+ iPSC grafts effectively improved neurological function, and the mechanism of action was associated with lncRNA and mRNA expression. Therefore, A2B5+ iPSC transplantation could be considered as a new method for the treatment of TBI, and ENSRNOT00000052577 and Kif2c may be new molecular targets or markers for functional improvement.
Keywords: A2B5+ iPS, transplantation, traumatic brain injury, lncRNA, mRNA
Introduction
Traumatic brain injury (TBI) is one of the leading causes of death and permanent disability in young adults of industrialized countries.1,2 The common disabilities of patients include sensory and motor deficits and cognitive problems,3 which are commonly induced by several pathological events such as necrosis, apoptosis, and excessive autophagy.4,5 Disconnection of neural circuits from axonal injury in the central nervous system (CNS) is the chief reason for neurological impairments,6,7 Therefore it is necessary to search for an effective therapy to reduce TBI-related disabilities. To date, biological therapy based on stem cell transplantation has been used in experimental research of CNS disorders like TBI.8–10 However, treatment efficiency and the underlying mechanisms of action remain to be determined.
Induced pluripotent stem cells (iPSCs), which are from somatic cells by the ectopic expression of defined transcription factors, can differentiate into cells of all 3 germ layers both in vitro and in vivo.11,12 As a potential cell source, iPSCs may be useful for the treatment of several diseases, but this needs to be confirmed in experimental animals before clinic usage. Recent evidence indicated that transplantation of neural precursor cells (NPCs), especially oligodendrocyte progenitor cells (OPCs), could prevent axonal degeneration and promote nerve repair/remyelination in the injured CNS.13–15 A2B5, a cell surface ganglioside, is the best characterized marker of developing oligodendroglial progenitors.16–18 Therefore, A2B5+ iPSCs may have the capacity to improve brain injury by differentiating into oligodendrocytes.4 Based on these findings, we transplanted A2B5+ iPSCs into the damaged area of the brain to promote the restoration of neurologic function and explored the associated molecular mechanisms.
Long noncoding RNAs (lncRNAs), consisting of over 200 nucleotides in length,19,20 have been shown to play vital roles in epigenetic mechanisms and gene regulatory networks of chromatin modification,21 and transcriptional and posttranscriptional regulation22 by binding with noncoding RNAs, genes and transcription factors.23 Recently, it was reported that lncRNAs have important and diverse functions in the development and progression of various diseases24 such as the progression of cancers,25 cardiovascular diseases,26 and neurodegenerative diseases.27,28 Specifically, lncRNAs have been shown to exhibit strong expression in developing and adult brains20,27,29 and are involved in neural differentiation and synaptic plasticity.27,30 Many studies have resulted in considerable progress in understanding of the relationship between lncRNAs and brain physiology and diseases.19,20 However, the role of lncRNAs in TBI rats, and whether the mechanism of A2B5 transplantation is involved in the expression of lncRNA is unclear and needs to be determined.
Together, the aims of this study were to determine the effectiveness of A2B5+ iPSC transplantation and explain the molecular mechanisms involved for the treatment of TBI. Therefore, we evaluated the effect of A2B5+ IPSC transplantation in the TBI brain and described the lncRNA and mRNA expression profile using microarray analysis in TBI and A2B5+ iPSC transplantation groups.
Materials and Methods
Animals
Thirty female Sprague-Dawley rats, weighing 200 to 240 g, were provided by the Laboratory Animal Centre of Kun Ming Medical University. All rats were housed in the laboratory animal room, which was maintained at 25°C ± 1°C with 65% ± 5% humidity on a 12-hour light/dark cycle (lights on from 07:30 to 19:30) for at least 1 wk before the experiments and given free access to fresh water and food. All animal procedures were approved by the Animal Care and Welfare Committee of Kunming Medical University. In addition, all experiments followed the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. All rats were randomly divided into 3 groups: a sham group (rats with brain exposed but not subjected to weight impact injury and receiving medium injections instead of cultured cells), a TBI group (rats subjected to TBI and receiving medium treatment), a A2B5+ iPSC group (rats subjected to TBI and receiving A2B5+ iPSC transplantation).
TBI model
In this study, the TBI model was established by the revised Feeney method.10 All rats were anesthetized by intraperitoneal (i.p.) injection of 3.6% chloral hydrate (1 mL/100 g; Sigma-Aldrich, St. Louis, MO, USA) and then their heads were fixed in a stereotaxic frame. The right parietal bone was exposed. A bore was drilled into the skull 2.5 mm away from the sagittal suture and 1.5 mm away from the arcuate suture, not touching the dura mater. The skull was removed until the diameter of the “bone window” was enlarged to 5.0 mm. The cerebral cortex was exposed and subjected to an impact from a 20 cm high position along the guide bar by a 50 g hammer, which resulted in a predominantly focal injury of the right cerebral cortex. Then, the exposed dura was covered with bone wax and the scalp sutured. Rats were given food and water ad libitum and received injections of penicillin to prevent infection.
The Production, Culture, Differentiation, and Transplantation of A2B5+ iPSCs
Procedures for the culture and differentiation of mouse iPSCs in vitro have been described previously.31,32 In brief, mouse embryonic fibroblasts (MEFs) were suspended in Dulbecco's modified Eagle's medium (DMEM) (Hyclone, Logan, UT, USA) with 10% Fetal Bovine Serum (FBS) (Invitrogen, Carlsbad, CA, USA) and adjusted at a density of 1 × 105 cells at 37 °C with 5% CO2 and then were transferred twice with a retrovirus containing 4 transcription factors (Oct3/4, Sox2, c-Myc, and Klf4) in 2 d. After approximately 4 d of culturing, the cell growth on the MEF cell layer (Global Stem, Gaithersburg, MD, USA) was passaged and inoculated in DMEM with 20% FBS containing mitomycin. One day later, the medium was replaced by embryonic stem cell (ESC) specialized medium made by combining knockout DMEM (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), 2 mM GlutaMAX (Invitrogen), 100 mM nonessential amino acid (NEAA; Invitrogen), 20% serum replacement (SR; Invitrogen), 1,000 U/mL leukemia inhibitory factor (LIF) (Millipore, Billerica, MA, USA), and 100 U/mL penicillin/streptomycin (Hyclone) and exchanged every day. The induced pluripotent stem cell (iPSC) colonies were digested by collagenase IV (1 mg/mL in DMEM/F12, Invitrogen) for 15 min at 37 °C and isolated from the MEF cell layer every 4 to 6 d, then cultured in stem cell culture medium made by knockout DMEM (Invitrogen), 20% SR (Invitrogen), 2 mM GlutaMAX (Invitrogen), 100 mM NEAA (Invitrogen), and 100 U/mL penicillin/streptomycin for 4 d. Afterward, the iPSCs were treated with retinoic acid (all-trans retinoic acid [RA], 500 nM; Sigma-Aldrich) and purmorphamine (1 μm, Calbiochem, La Jolla, CA, USA) for 4 d. As above, the medium was exchanged every day. On the eighth day of differentiation, the medium was replaced by A2B5 medium (50% DMEM/F12 [Invitrogen], 50% neurobasal [Invitrogen], 1% N2 [Invitrogen], 2% B27 [Invitrogen], 2 mM GlutaMAX [Invitrogen], 100 mM NEAA [Invitrogen], 100 U/mL penicillin/streptomycin, 10 ng/mL human epidermal growth factor (EGF) [PeproTech EC, London, UK], and 20 ng/mL human fibroblast growth factor 2 (FGF2) [PeproTech EC]) and culture was continued for 8 to 10 d. According to the manufacturer’s instructions, the anti-A2B5 MicroBeads mouse kit (Miltenyi Biotec, Bergisch Gladbach, Germany) was used to purify A2B5-+ cells. Purified A2B5+ cells were inoculated in poly-d-lysine (PDL)-coated slide flasks with A2B5 medium, and half of the medium was exchanged every other day. In order to label A2B5 cells, green fluorescent protein (GFP) was introduced into the HIV vector and transferred into A2B5 cells. The cells were identified by immunofluorescent staining with an A2B5 antibody (1:200; Abcam, Cambridge, UK) and observed under a fluorescent microscope (Leica Microsystems, Wetzlar, Germany). Once cultured A2B5+ iPSCs cells were transduced with the Green fluorescent protein (GFP) gene or stained by anti-GFP antibody (1:1,000; Abcam), they can emit green fluorescence under the microscope. After the labeled cell suspension was prepared, it was stored at 4 °C and transplanted into the host within 4 h. A2B5+ iPSCs were transplanted into the damaged cortex through local injection immediately after TBI. The cell density was adjusted to 1 × 108/mL before cell transplantation. A Five microliter A2B5+ iPSC suspension, (about 5 × 105 cells), was implanted into the paracele by a 25-gauge micro-injector with the help of a stereotaxic instrument. The suspension was injected with a constant velocity within 2 min. The micro-injector was inserted, respectively, into 4 locations: 3 mm rostral, caudal, left, and right from the epicenter of the injured site. For each location, the cell suspension was injected at 1 μL/min at 3 depth points, viz., 2 mm, 1.5 mm, and 1 mm from the surface of the cortex. The micropipette was left for 1 min to minimize efflux of the inoculum after pipette withdrawal. Therefore, the A2B5+ iPSC group was transplanted as described above. The TBI group was injected with only medium. The sham group was only subjected to sham surgery (opening of the scalp and skull without TBI) and medium injection. Three days before operation, all of the rats were injected intraperitoneally daily with cyclosporine A (1 mg/kg/ d; Novartis, Basel, Switzerland). The injection continued daily after the operation until the day of sample harvesting in order to suppress immunologic rejection of the transplant.
Survival and Differentiation of Transplanted A2B5+ iPSCs by Immunohistochemistry
At 1 wk after TBI, the rats were anesthetized by i.p. injection of 3.6% chloral hydrate (1 mL/100 g; Sigma-Aldrich). Then, samples were harvested and fixed by paraformaldehyde (PFA) for immunohistochemistry. Samples were put into 4% paraformaldehyde (PFA) phosphate buffer and postfixed until they were processed for immunohistochemistry. The postfixed samples were dehydrated with 20% sucrose (Sigma-Aldrich) at 4 °C overnight, followed by sectioning into 15-µm sections. Then, a 2-step immufluorescent staining method was used to detect the survival and differentiation of the transplanted A2B5+cells emitting GFP (1:1,000; Abcam) fluorescence using an oligodendrocytes 2 (Olig2) antibody (1:500; Abcam). Meanwhile, the expression of MBP was detected using an MBP antibody (1:500; Abcam); simultaneously, we detected the GFP fluorescence to monitor myelinogenesis. The sections were placed under a fluorescence microscope (Leica Microsystems, Wetzlar, Germany) to observe the implanted cells.
Western Blot
Seven days after A2B5+ iPSC transplantation, the tissue from engrafted cortices were harvested and homogenized on ice in radioimmunoprecipitation assay (RIPA) lysis solution and centrifuged at 12,000 g force for 30 min. The supernatant was obtained and stored at −80 °C for later use. The protein concentration was determined by bicinchonininic acid (BCA) assay (Sigma-Aldrich). A 20 µL aliquot of the samples was loaded into each lane and electrophoresed on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel at a constant voltage of 100 V for 1.5 h. Proteins were transferred from the gel to a plyvinylidene fluoride (PVDF) membrane at 20 V for 18 min. The membrane was blocked at room temperature for 1 h with PBS containing 5% skim milk. It was incubated with the primary antibody for MBP (1:1,000; rabbit Santa Cruz Biotechnology, Santa Cruz, CA, USA) on the shaker overnight at 4 °C. After washing 3 times for 10 min each, the membrane was incubated with a horse radish peroxidase (HRP)-conjugated goat anti-rabbit MBP antibody (1:5,000; Vector Laboratories, Lowellville, OH, USA) for 2 h at room temperature and washed as described above. The membrane was developed using an enhanced chemiluminescence (ECL) kit and photographed in a Bio-Gel Imagining System (Bio-Rad, Hercules, CA, USA) equipped with Genius synaptic gene tool software (quantity one4.62). Densitometry analysis for MBP was carried out with β-actin (1:500, Santa Cruz Biotechnology) as an internal control.
Neurological Severity Score Assessment
An NSS was used to evaluate the degree of impaired cerebral function of TBI rats.10,33 Total scores ranged from 0 to 25, which were shown in Table 1. According to its severity, an impaired performance on any activities received a score. Assessment for NSS was conducted on 3 d, 7 d, 11 d, 15 d, and 19 d. All behavioral and neurological test scores were recorded by professional investigators who were blind to the experiment. Animals were returned to their cages with free food and water after completion of the NSS test.
Table 1.
Grading of Neurological Severity Score.
| Points | ||
|---|---|---|
| At 3 d | Other Times | |
| Hemiplegia: Inability of rat to resist forced changes in position | 1 | 1 |
| Flexion of hind limb when raised by the tail | 1 | 1 |
| Inability to walk straight when placed on the floor | 1 | 1 |
| Inability to walk | 1 | 1 |
| Reflexes | ||
| Loss of right reflex for | ||
| 20 min | 1 | |
| 40 min | 1 | |
| >60 min | 1 | 3 |
| Limb reflexes: Loss of placing reflexes | ||
| Left forelimb | 1 | 1 |
| Right forelimb | 1 | 1 |
| Left hind limb | 1 | 1 |
| Right hind limb | 1 | 1 |
| Startle reflex | 1 | 1 |
| Clinical grade | ||
| Loss of seeking behavior | 1 | 1 |
| Prostration | 1 | 1 |
| Inability to exit from circle (50 cm in diameter) when left in its center for | ||
| 30 min | 1 | |
| 60 min | 1 | |
| >60 min | 1 | 3 |
| Functional test: Failure in beam balancing test (1 cm wide) | ||
| Balances with steady posture; paws on top of beam | 0 | 0 |
| Grasps sides of beam and/or has shaky movement | 1 | 1 |
| One or more paws slip off beam | 1 | 1 |
| Attempts to balance on beam but falls off | 1 | 1 |
| Drapes over beam and/or hangs on beam and falls off | 1 | 1 |
| Falls off beam with no attempt to balance or hang on | 1 | 1 |
| Failure in beam walking task | ||
| 2.5 cm wide | 1 | 1 |
| 5.0 cm wide | 1 | 1 |
| 8.5 cm wide | 1 | 1 |
| Maximum points | 25 | 25 |
RNA Labeling, Array Hybridization, and Microarray Analysis
At 1 wk after TBI, the rats were anesthetized by i.p. injection of 3.6% chloral hydrate (1 mL/100 g; Sigma-Aldrich). Then, samples were harvested from the injured side of the brain from the living body of rats and frozen freshly in liquid nitrogen. The lncRNA microarray analysis was performed by KangChen Bio-tech (Shanghai, PR China). After the brain tissues were kept in liquid nitrogen, the total RNA was isolated using Trizol reagent according to the manufacturer’s guideline. The Agilent Array platform was employed for microarray analysis. Sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols with minor modifications. In brief, RNA was purified from the total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre, Madison, WI, USA). Then, RNA was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts and simultaneously labeled using a fluorescent probe such Cy3. After purification, the fluorescent cRNA probe along the entire length of the transcripts was hybridized onto the rat lncRNA Array v2.0 (8 × 60 K, ArrayStar). After washing the slides, the arrays were scanned by the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1, Santa Clara, CA, USA) was used to analyze the acquired array images. The random variance model (RVM) t-test efficiently increases the degrees of freedom in small samples,34 2 Class Dif method and RVM t tests were used to identify the differentially expressed lncRNAs and genes between TBI and sham groups and A2B5+ iPSC and TBI groups. After the significant analysis and false discovery rate analysis were performed, a P value < 0.05 was regarded as a significant difference35.
Statistical Analysis
SPSS 17.0 software was used for statistical analysis. Data were expressed as mean ± standard deviation Provide the abbreviation in parentheses following the spelled out form. It is (SD) from at least 3 independent experiments. For multiple group comparison, the experimental results were statistically analyzed for significant differences using one-way analysis of variance (ANOVA). Student’s t tests were used to analyze the data with a 2-tailed distribution. P value < 0.05 was considered statistically significant.
Results
Identification of A2B5+ Cells in In Vitro Culture
MEFs were transferred twice with the retrovirus containing that expressed 4 transcription factors (Oct3/4, Sox2, c-Myc, and Klf4) to induce somatic cells into iPSCs. Subsequently, iPSCs were cultured with A2B5 medium and A2B5+ iPSCs were collected for the transplantation. A2B5+ iPSCs have been successfully identified and isolated by A2B5 immunostaining (Fig. 1), confirming that they were A2B5+ iPSCs. In order to track the transplanted A2B5+ iPSCs, GFP was used to label the transplanted cells (Fig. 1). All the cells emitted green (GFP) and blue fluorescence (4',6-diamidino-2-phenylindole [DAPI] labeling) under fluorescence microscopy (Fig. 1).
Fig. 1.
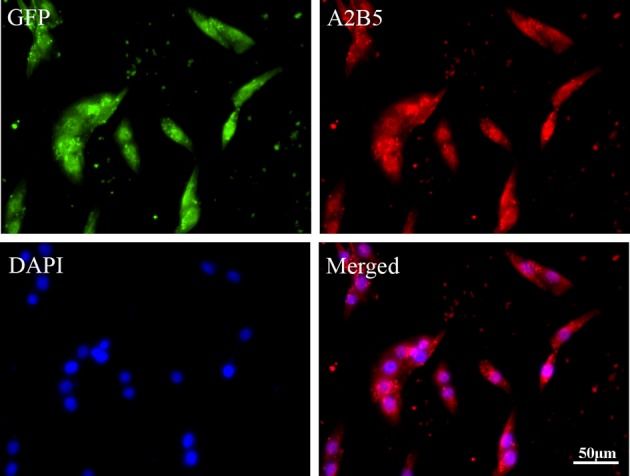
Identification of A2B5+ cells in vitro. GFP was used to label the transplanted cells. A2B5+ immunostaining was used to identify the A2B5+ iPSCs. All the cells emitted blue fluorescence in nuclei after DAPI labeling under fluorescence microscopy. iPS, induced pluripotent stem.
Survival, Migration, Differentiation of Transplanted A2B5+Cells In Vivo
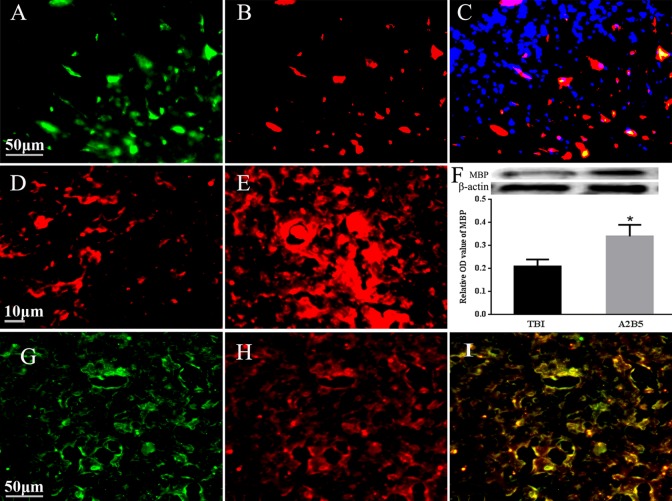
After transplantation, several A2B5-positive cells were detected in the brain surrounding the epicenter, and exhibited Olig2 staining, confirming they could differentiate into OPCs. Moreover, we determined the capacity of grafts for remyelination. MBP staining showed that only a few cells were positive for MBP expression in the TBI brain, while the staining of MBP was largely increased in the A2B5+ cell-transplanted group, which corresponded with the results of WB detection (Fig. 2)
Fig. 2.
Detection of differentiation on engrafted A2B5+ cells in the brain. (A) Engrafted A2B5+ cells; (B) Olig2+ cells; (C) merged picture from A2B5+ cells and Olig2+ cells plus DAPI stain; (D) MBP staining in TBI brain; (E) MBP staining in A2B5+ grafts after TBI; (F) the comparison of MBP expression indicated by WB. *Compared with TBI group, P < 0.05. G, H, and I are to show the myelinogenisis from transplanted A2B5+ cells. (G) GFP recognition from myelin sheath, (H) MBP staining, (I) merged piture to show myelin sheath are resulted from grafted A2B5+ cells. Scale bar = 50 μm, shown in A, G, and 10 μm in D. Olig2, oligodendrocytes 2; MBP, myelin basic protein; TBI, traumatic brain injury.
The Results of the NSS
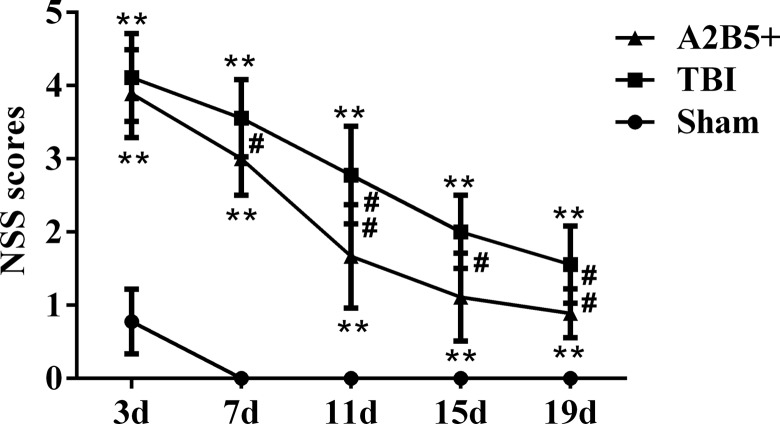
After TBI, the NSSs in TBI and A2B5+ groups were significantly higher than that of the sham group on 3 d, 7 d, 11 d, 15 d, and 19 d postoperation, and all P values were less than 0.001. After A2B5+ iPSC transplantation, the NSSs of the A2B5+ group had no statistical significance compared with that of the TBI group on 3 d, while it decreased significantly after 7 d, and was significantly lower than that of TBI group on 7 d, 11 d, 15 d, and 19 d. Meanwhile, as time went on, the degree of brain injury was gradually restored. In detail, compared with the NSS on 3 d, the NSSs of the TBI group were significantly decreased on 7 d and then continually decreased on 11 d, 15 d, and 19 d. Comparatively, in the cell transplantation group, the NSSs were also decreased on 3 d to 19 d. Moreover, the NSSs at most time points were lower than that in TBI group, which showed that A2B5+ iPSC transplantation could promote recovery of neurological function after TBI (Fig. 3).
Fig. 3.
Results of the NSSs. **Comparison between TBI group and sham group or between A2B5+ group and sham group, P < 0.01; #Comparison between A2B5+ group and TBI group, P < 0.05; and ## Comparison between A2B5+ group and TBI group, P < 0.01. NSS, neurological severity score; TBI, traumatic brain injury.
Overview of the lncRNA Profiles in Brain Tissue of Rats
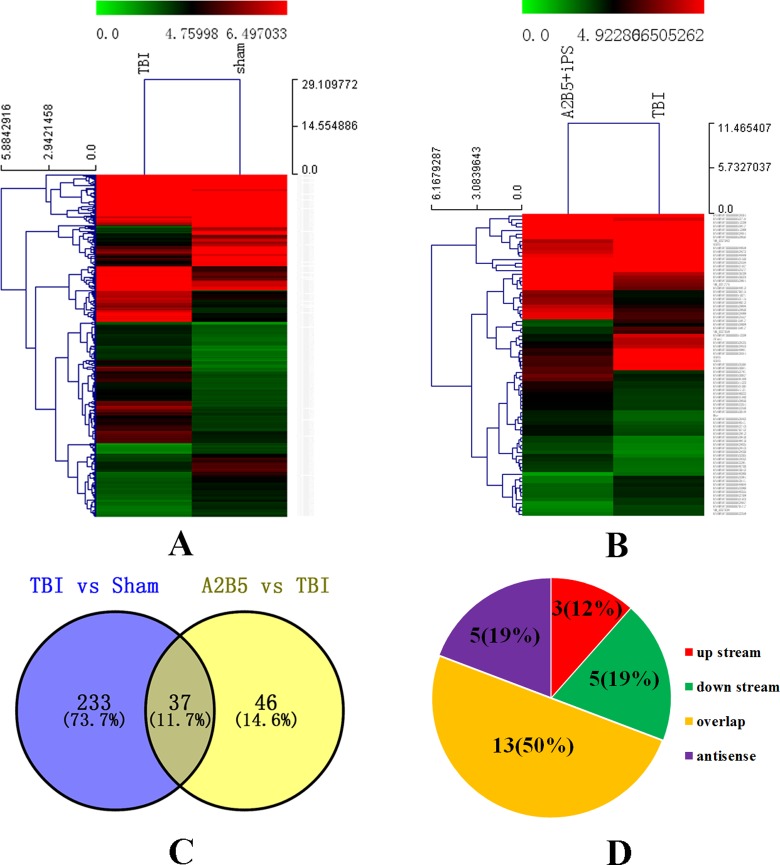
To profile differentially expressed lncRNAs in brain tissue from the sham group, TBI group, and A2B5+ iPSC group, we generated the rat model of TBI and transplanted the rats with A2B5+ iPSCs. On the seventh day after cell transplantation, the animals were sacrificed for microarray analysis to detect lncRNA and mRNA expression. First, variations in lncRNAs and mRNA expression levels between the TBI group and sham group were shown in Figure 4A. Of these, 270 differentially expressed lncRNAs were detected in brain tissue from the TBI group with a fold change of at least 2 (P < 0.05). Among them, 224 lncRNAs were upregulated and 46 lncRNAs were downregulated (P < 0.05). Nine lncRNAs exhibited a high fold change of at least 5-fold, with 7 upregulated and 2 downregulated. More specifically, ENSRNOT00000063536 (fold change: 12.880, P < 0.01) was the most upregulated lncRNA and ENSRNOT00000053950 was the most downregulated lncRNA (fold change: 28.833, P < 0.01). Comparatively, lncRNA expression profiles from the A2B5+ iPSC group and TBI group were shown in Figure 4B. Overall, 83 differentially expressed lncRNAs were detected in brain tissue with a fold change of at least 2 (P < 0.05). Among them, 48 lncRNAs were upregulated and 35 lncRNAs were downregulated (P <0.05). Of these changed genes, ENSRNOT00000052716 (fold change: 4.529, P < 0.01) was the most upregulated lncRNA and ENSRNOT00000053394 was the most downregulated lncRNA (fold change: 3.455, P < 0.01).
Fig. 4.
Differentially expressed lncRNAs in TBI group and in A2B5+ iPSC group. (A) The hierarchical clustering of differentially expressed lncRNAs in TBI group versus sham group. (B) The hierarchical clustering of differentially expressed lncRNAs in A2B5+ iPSC group versus TBI group. (C) The result of intersection of differentially expressed lncRNAs in TBI group and in A2B5+ iPSC group. (D) The location relationship of common differentially expressed lncRNAs and potential target genes. The intensity of the color scheme corresponds to the log2 expression values. Red indicates high relative expression, and blue indicates low relative expression. lncRNAs, long noncoding RNAs; iPSC, induced pluripotent stem cell.
We found that there were 37 differentially expressed lncRNA which were simultaneously changed in the A2B5+ iPSC group versus the TBI group and the TBI group versus the sham group (Fig. 4C; Table 2). Then, the potential target genes of the simultaneously differentially expressed lncRNAs were found through scanning the 10 kb upstream and downstream lncRNAs. The results showed that there were 3 mRNAs that were located in upstream, 13 mRNAs overlapped with lncRNAs, 5 mRNAs were downstream, and 5 mRNAs were located in the antisense strand (Fig. 4D; Table 3).
Table 2.
Cochanged lncRNAs in TBI Group and in A2B5+ iPSC Group.
| Change Style | lncRNA | |
|---|---|---|
| Down → up | ENSRNOT00000052716 | ENSRNOT00000053450 |
| ENSRNOT00000052781 | Bsr | |
| ENSRNOT00000053940 | ENSRNOT00000062604 | |
| ENSRNOT00000054239 | ENSRNOT00000069489 | |
| ENSRNOT00000052682 | ENSRNOT00000053950 | |
| ENSRNOT00000062467 | ENSRNOT00000053023 | |
| ENSRNOT00000053115 | ENSRNOT00000052988 | |
| ENSRNOT00000062557 | ||
| Up → down | ENSRNOT00000062867 | ENSRNOT00000053909 |
| ENSRNOT00000062864 | ENSRNOT00000068981 | |
| Zfas1 | ENSRNOT00000062664 | |
| ENSRNOT00000068968 | ENSRNOT00000045917 | |
| ENSRNOT00000052956 | ENSRNOT00000062872 | |
| ENSRNOT00000069355 | ENSRNOT00000053394 | |
| ENSRNOT00000054299 | ENSRNOT00000062853 | |
| ENSRNOT00000053390 | ENSRNOT00000068889 | |
| GAS5 | ENSRNOT00000052577 | |
| ENSRNOT00000053506 | ENSRNOT00000053061 | |
| ENSRNOT00000062709 | ENSRNOT00000053463 | |
Note. Down → up represents that lncRNAs are downregulated in TBI group and upregulated in A2B5+ iPSC group. Up → down represents that lncRNAs are upregulated in TBI group and downregulated in A2B5+ iPSC group. lncRNAs, long noncoding RNAs; TBI, traumatic brain injury; iPSC, induced pluripotent stem cell.
Table 3.
The mRNAs Near the Cochanged lncRNAs.
| lncRNA Accession | TBI versus sham | A2B5 versus TBI | mRNA Name | Location |
|---|---|---|---|---|
| Zfas1 | Up | Down | Znfx1 | Antisense |
| ENSRNOT00000069489 | Down | Up | Hdac4 | Antisense |
| ENSRNOT00000069355 | Up | Down | Dnm3 | Antisense |
| ENSRNOT00000054239 | Down | Up | LOC690114 | Antisense |
| ENSRNOT00000053115 | Down | Up | Cyfip1 | Antisense |
| ENSRNOT00000053506 | Up | Down | Acadm | Up stream |
| ENSRNOT00000053463 | Up | Down | Slc25a5 | Up stream |
| ENSRNOT00000052577 | Up | Down | Kif2c | Up stream |
| ENSRNOT00000068968 | Up | Down | Ubap2 | Overlap |
| ENSRNOT00000068889 | Up | Down | Nop58 | Overlap |
| ENSRNOT00000062872 | Up | Down | Rps3a | Overlap |
| ENSRNOT00000062664 | Up | Down | Rps3a | Overlap |
| ENSRNOT00000062604 | Down | Up | Slc25a3 | Overlap |
| ENSRNOT00000054299 | Up | Down | Rpl7a | Overlap |
| ENSRNOT00000053940 | Down | Up | Zfand6 | Overlap |
| ENSRNOT00000053463 | Up | Down | RGD1359290 | Overlap |
| ENSRNOT00000053450 | Down | Up | Taf1d | Overlap |
| ENSRNOT00000053394 | Up | Down | Eef2 | Overlap |
| ENSRNOT00000052577 | Up | Down | Rps8 | Overlap |
| ENSRNOT00000052956 | Up | Down | Eif4a2 | Overlap |
| ENSRNOT00000052682 | Down | Up | Fam178a | Overlap |
| ENSRNOT00000054299 | Up | Down | Surf2 | Down stream |
| ENSRNOT00000053506 | Up | Down | Rabggtb | Down stream |
| ENSRNOT00000053506 | Up | Down | Msh4 | Down stream |
| ENSRNOT00000053463 | Up | Down | RGD1562987 | Down stream |
| ENSRNOT00000053394 | Up | Down | Dapk3 | Down stream |
Abbreviation: lncRNAs, long noncoding RNAs; mRNA, messenger RNAs; TBI, traumatic brain injury.
Overview of Coding Gene Profiles in Brain Tissue of Rats
With the aim to get more credible biological functions, the microarray analysis also pointed out the expression profile of coding genes. First, a total of 1,052 coding transcripts (mRNA) were differentially expressed between the TBI and sham groups (>2-fold, P < 0.05; Fig. 5A). Among them, 921 mRNAs were upregulated and 131 mRNAs were downregulated (P < 0.05). In addition, a total of 54 mRNAs were upregulated with more than a 5 fold change. Secondly, variations in mRNA expression levels of the A2B5+ iPSC group compared to the TBI group are shown in Figure 5B. Of these, 360 differentially expressed mRNAs were detected in the A2B5+ iPSC group with a fold change of at least 2 (P < 0.05). Among them, 77 mRNAs were upregulated and 283 mRNAs were downregulated (P < 0.05). The numbers of upregulated mRNAs and downregulated mRNAs whose fold change were more than 10 times were 6 and 10.
Fig. 5.
Differentially expressed mRNAs in the TBI group and in the A2B5+ iPSC group. (A) When compared with the sham group, the heat map of differentially expressed mRNAs in the TBI group is shown in A. (B) When compared with the TBI group, the heat map of differentially expressed mRNAs in the A2B5+ iPSC group is shown in B. (C) The result of intersection of common differentially expressed mRNAs in the TBI group and in the A2B5+ iPSC group. (D) Through scanning the upstream and downstream 10 kb of the simultaneously differentially expressed mRNAs, the location relationship of mRNAs and potential lncRNAs is shown in D. The intensity of the color scheme corresponds to the log2 expression values. Red indicates high relative expression, and blue indicates low relative expression. mRNA, messenger RNAs; TBI, traumatic brain injury; iPSC, induced pluripotent stem cell.
The combined analysis of different mRNAs in the A2B5+ iPSC group versus the TBI group and the TBI group versus the sham group showed that 195 mRNAs were simultaneously changed (Fig. 5C; Table 4). One hundred sixty-two mRNAs were upregulated in the TBI group versus the sham group, but they were downregulated in the A2B5+ iPSC group versus the TBI group. However, one mRNA, Slc19a3, was downregulated TBI group versus sham group but upregulated in A2B5+ iPS group versus TBI group. Two mRNA, Naaa and Lcn2, were upregulated at the same time in the TBI group versus sham group and A2B5+ iPS group versus TBI group. Then, we found out the lncRNAs that regulated those mRNAs from the differentially expressed lncRNAs in the microarray. The results showed that 3 lncRNAs were located in upstream of their mRNAs, 7 lncRNAs were overlapped with mRNAs, 6 lncRNAs were located in downstream, and 5 lncRNAs were located in the antisense strand (Fig. 5D; Table 5).
Table 4.
Cochanged mRNAs in TBI Group and in A2B5+ iPSC Group.
| Change Style | Gene | |||
|---|---|---|---|---|
| Down → up | Slc19a3 | |||
| Up → down | Pbk | Tnfsf10 | Oip5 | RT1-M6-2 |
| LOC689064 | Tctex1d2 | Ttk | Kif20a | |
| Ccl2 | Hmmr | Hmox1 | Nqo2 | |
| LOC100362296 | Kdelr3 | Slc4a1 | Cenpq | |
| Lgals5 | Diaph3 | LOC679711 | Tuba1c | |
| Ly49si1 | Alas2 | Kif4a | Metrn | |
| Lgals3 | Cyp2j10 | Tpx2 | Mmp8 | |
| Anxa1 | Ccr5 | Clec5a | Gas2l3 | |
| Ccr2 | Rgs18 | Nusap1 | Bub1 | |
| Mmp12 | Gpr34 | Hba-a2 | MGC108823 | |
| Nuf2 | Cmc1 | Prc1 | Cenpf | |
| LOC287167 | Ptplad2 | Aurkb | LOC691543 | |
| Gpnmb | Rasgrp3 | Hbb-b1 | RGD1565648 | |
| Ckap2 | Ifi30 | Alox15 | Cav1 | |
| Clec7a | Echdc1 | S100a11 | Pter | |
| Hbb | Fam46c | Ms4a6b | Ccdc152 | |
| LOC691666 | Kif2c | Cdk1 | RGD1309362 | |
| Stk32a | Lpar4 | Sgol2 | Rrm1 | |
| LOC100364306 | Pmp2 | Asb15 | Cd14 | |
| Cks2 | Ns5atp9 | Ccna2 | Ptgdrl | |
| Slc39a8 | Gcnt1 | Ifitm1 | Fabp7 | |
| Fcgr2a | Gbp5 | Pld5 | Arl11 | |
| Plin2 | Cd22 | Acot1 | RGD1306001 | |
| Cd53 | Msr1 | Klra7 | Lpar6 | |
| Postn | Cyp2j4 | Ndc80 | Mt1a | |
| Fgl2 | Glipr1 | Ccnb1 | Frrs1 | |
| Irak4 | LOC679994 | Lsp1 | S100a3 | |
| Plp2 | Ms4a4a | Arhgap11a | Rarres2 | |
| Gadd45a | Pcna | Lgals1 | Lilrb3l | |
| Cdc25b | Mlf1ip | Cenpi | Cd180 | |
| Ccnb2 | Creg1 | Rsad2 | Emp3 | |
| Cd7 | Kpna2 | Fam105a | Gpr65 | |
| Spp1 | Lhfpl2 | Tyrobp | Dusp12 | |
| Top2a | Hapln1 | Olr1 | Slc35d2 | |
| Ccl7 | Uevld | Kif18a | Anxa4 | |
| LOC690467 | Sash3 | Rrm2 | Mad2l1 | |
| Cdkn2c | Vim | Chchd2 | Il33 | |
| Tubb6 | Plac8 | Cd8a | Gulp1 | |
| S100a4 | Pop4 | Depdc1 | Cdca2 | |
| Mmp14 | Cd86 | Igf1 | Ebpl | |
| LOC679586 | Mrpl46 | S100a10 | LOC100910367 | |
| Emp1 | Rcbtb2 | Ms4a11 | Hemgn | |
| Rpa3 | Pdpn | Ly96 | LOC363337 | |
| Efcab1 | Ect2 | Traf4af1 | Decr1 | |
| Dlgap5 | RGD1562655 | Cdkn3 | Kif11 | |
| Casc5 | Clrn1 | Ska3 | Scrg1 | |
| Eef1b2 | Cd244 | Cdca3 | Ncapg | |
| Tfec | Ccnd1 | RGD1559588 | Casp1 | |
| Anxa2 | Ckap2l | Timp1 | Dhfr | |
| Up → up | Naaa | Lcn2 | ||
Note. Down → up represents that mRNAs are downregulated in the TBI group and upregulated in the A2B5+ iPSC group. Up → down represents that mRNAs are upregulated in the TBI group and downregulated in the A2B5+ iPSC group. Up → up represents that mRNAs are simultaneously upregulated in the TBI group and in the A2B5+ iPSC group. lncRNAs, long noncoding RNAs; mRNA, messenger RNAs; TBI, traumatic brain injury; iPSC, induced pluripotent stem cell.
Table 5.
The lncRNAs Near the Cochanged mRNAs.
| Gene Symbol | TBI versus Sham | A2B5 versus TBI | lncRNA Accession | Location |
|---|---|---|---|---|
| Tpx2 | Up | Down | ENSRNOT00000068979 | Antisense |
| Pld5 | Up | Down | ENSRNOT00000070593 | Antisense |
| Top2a | Up | Down | ENSRNOT00000063054 | Antisense |
| Anxa4 | Up | Down | ENSRNOT00000053564 | Antisense |
| Ect2 | Up | Down | ENSRNOT00000069536 | Antisense |
| Sgol2 | Up | Down | ENSRNOT00000070519 | Up stream |
| Ms4a11 | Up | Down | ENSRNOT00000070035 | Up stream |
| Ska3 | Up | Down | ENSRNOT00000062274 | Up stream |
| Dhfr | Up | Down | ENSRNOT00000054548 | Overlap |
| Dhfr | Up | Down | ENSRNOT00000070189 | Overlap |
| Eef1b2 | Up | Down | ENSRNOT00000052469 | Overlap |
| Eef1b2 | Up | Down | ENSRNOT00000053393 | Overlap |
| Stk32a | Up | Down | ENSRNOT00000063242 | Overlap |
| Cmc1 | Up | Down | ENSRNOT00000069415 | Overlap |
| Ebpl | Up | Down | ENSRNOT00000070634 | Overlap |
| Cenpq | Up | Down | ENSRNOT00000069770 | Down stream |
| Decr1 | Up | Down | ENSRNOT00000070684 | Down stream |
| Kif2c | Up | Down | ENSRNOT00000052577 | Down stream |
| Kif2c | Up | Down | ENSRNOT00000052697 | Down stream |
| Kif2c | Up | Down | ENSRNOT00000053497 | Down stream |
| Vim | Up | Down | ENSRNOT00000063302 | Down stream |
Abbreviation: lncRNAs, long noncoding RNAs; mRNA, messenger RNAs; TBI, traumatic brain injury.
GO and Pathway Analysis of Differentially Expressed mRNAs in the A2B5+ iPSC Group
The function of lncRNAs may be reflected in their associated protein-coding genes. Therefore, GO enrichment analysis of differentially expressed mRNAs may provide insight into the function of these differentially expressed lncRNAs.36 Pathway analysis of differentially expressed mRNAs is designed to provide insight into the cell pathways associated with these genes.
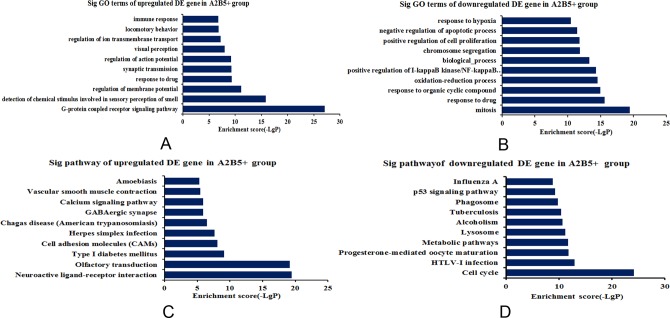
In the A2B5+ iPSC group, the highest enriched GO terms targeted by upregulated mRNAs were “G-protein-coupled receptor signaling pathway,” “regulation of membrane potential,” “synaptic transmission,” “regulation of action potential,” “visual perception,” “regulation of ion transmembrane transport,” and “locomotor behavior” (Fig. 6A). The highest enriched GO terms targeted by downregulated transcripts included “mitosis,” “response to organic cyclic compound,” “oxidation-reduction process,” “positive regulation of I κ B kinase/NFκ B cascade,” “biological process,” “chromosome segregation,” “positive regulation of cell proliferation,” “negative regulation of apoptotic process,” and “response to hypoxia,” which may be associated with the effect of iPSCs for TBI (Fig. 6B).
Fig. 6.
GO enrichment and KEGG pathway analysis of differentially expressed mRNAs in the A2B5+ iPSC group. (A) Top 10 GO terms enriched among upregulated mRNAs according to biological processes. (B) Top 10 GO terms enriched among downregulated mRNAs according to biological processes. (C) The top 10 pathways enriched among the upregulated mRNAs. (D) The top 10 pathways enriched among the downregulated mRNAs. The bar plot depicts the enrichment scores. The P value indicates the significance of the correlation between the pathway and the transplantation of A2B5+ iPSCs in TBI. mRNA, messenger RNAs; TBI, traumatic brain injury; iPSC, induced pluripotent stem cell.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of upregulated mRNAs was “neuroactive ligand-receptor interaction,” “olfactory transduction,” “type 1 diabetes mellitus,” “cell adhesion molecules,” “herpes simplex infection,” “GABAergic synapse,” “calcium signaling pathway,” “vascular smooth muscle contraction,” and “Amoebiasis” (Fig. 6C). However, “cell cycle,” “progesterone-mediated oocyte maturation,” “metabolic pathways,” “lysosome,” “alcoholism,” “tuberculosis,” “phagosome,” “p53 signaling pathway,” and “influenza A” were the top pathway enriched among the downregulated mRNAs (Fig. 6D).
Except for the top 10 GO terms and KEGG pathways, some upregulated genes were enriched for GO terms associated with “positive regulation vascular endothelial growth factor (VEGF) production,” “neuromuscular junction development,” “neurological system process,” “brain development, angiogenesis,” “wound healing,” “memory,” and “behavior,” which were related to recovery of neurological function. The lncRNAs and their target genes were ENSRNOT00000069658 and roar (overlap), ENSRNOT00000062221 and cacnb4 (antisense), NR_032112 and Igf2 (overlap), and so on. However, some downregulated genes were enriched for GO terms associated with “negative regulation of apoptotic process,” “negative regulation of extrinsic apoptotic signaling pathway,” “negative regulation of inflammatory response,” and “negative regulation of I κ B kinase/NF-κ B,” which could participate in the relief of the injury response. The lncRNAs and their target genes were ENSRNOT00000069227 and Birc6 (antisense), ENSRNOT00000070359/ENSRNOT00000052495 and Tex11 (antisense), ENSRNOT00000069658 and roar (overlap). Pathway analysis also indicated that inflammation-associated pathways, such as “Mitogen-activated protein kinase (MAPK) signaling pathway,” “Wnt signaling pathway,” “natural killer cell-mediated cytotoxicity,” and “B cell receptor signaling pathway,” were the enriched pathways targeted by the downregulated genes. The lncRNAs and their target genes were ENSRNOT00000052926 and Aspm (overlap), ENSRNOT00000069536 and Ect2 (antisense), ENSRNOT00000052818 and Nfatc3 (antisense).
The Combined Analysis of lncRNAs and mRNAs in the A2B5+ iPSC Group and TBI Group
In order to find the lncRNA which played the leading role in the action of A2B5+ iPSC transplantation in TBI, we analyzed 37 differentially expressed lncRNAs and 195 differentially expressed mRNAs, which were simultaneously changed in the A2B5+ iPSC group versus the TBI group and the TBI group versus the sham group. Then, through scanning the 10 kb upstream and downstream of the common differentially expressed lncRNAs, the potential target genes of lncRNAs were obtained. Finally, we took an intersection of the scanned target genes and the differentially expressed genes from the microarray; we got the valuable lncRNA (ENSRNOT00000052577) and its target gene (Kif2c; Table 6). The results showed that ENSRNOT00000052577 and kinesin family member 2C (Kif2c) were upregulated in the TBI group versus the sham group (fold change: 3.220 and 3.398, respectively) and downregulated in the A2B5+ iPSC group versus the TBI group (fold change: 2.071 and 2.080). In addition, Kif2c was located downstream of ENSRNOT00000052577.
Table 6.
The Intersected lncRNA and mRNA.
| Gene Symbol | TBI versus sham | A2B5 versus TBI | lncRNA Accession | TBI versus Sham | A2B5 versus TBI | Location |
|---|---|---|---|---|---|---|
| Kif2c | Up | Down | ENSRNOT00000052577 | Up | Down | Down stream |
Abbreviation: lncRNAs, long noncoding RNAs; mRNA, messenger RNAs; TBI, traumatic brain injury.
Discussion
In the present study, we successfully generated a TBI model, evaluated the effect of A2B5+ iPSCs for the treatment of TBI, and explored the expression of lncRNAs and mRNAs in TBI with A2B5+ iPSC transplantation in rats. In order to overcome the challenge of finding a proper stem cell source, we transplanted A2B5+ iPSCs, which were generated from MEFs by the ectopic expression of defined transcription factors, into the brains of TBI rats. As a result, we found that A2B5+ iPSCs could differentiate into oligodendrocytes and promote neurological recovery after TBI. Moreover, 1 wk after transplantation, we evaluated the expression of lncRNAs and mRNAs in the A2B5+ iPSC group, the TBI group, and the sham group, and found that the restoration of neurological function after A2B5+ iPSC transplantation could be associated with the alternations of related lncRNAs and mRNAs such as ENSRNOT00000052577 and Kif2c.
TBI is defined as the damage of brain tissue caused by an external mechanical force with long-term motor and sensory functional deficits.5,37 Saber et al. found that the necrosis, apoptosis, and excessive autophagy of cerebral cells were also related to severe axonal injury due to TBI.38 Meanwhile, the disconnection of neural circuits in the CNS is the chief reason for the neurological impairments including long-term memory problems, emotional disturbances, unconsciousness, and vegetative state.39 However, we transplanted A2B5+ iPSCs which could differentiate into oligodendrocytes to repair the axonal injury in TBI rats in this study.6,40 In order to overcome the challenge of finding a suitable stem cell source, we induced MEFs into iPSCs by the ectopic expression of defined transcription factors in vitro, then, A2B5+ iPSCs which could differentiate into oligodendrocytes were transplanted into the TBI brain for functional improvement. Recent evidence indicated that NPCs, especially OPCs, had been transplanted into the mammalian nervous system9,41,42 and that they were able to facilitate the regeneration of injured axons, differentiate into neurons, and promote functional recovery after TBI.18 Xu et al. found that the transplantation of exogenous OPCs may furnish competent oligodendrocytes, which could assist remyelination/myelin remodeling and prevent axonal degeneration in TBI.15 However, a series of problems, such as ethical issues and limited sources, impeded the application of stem cells in research and in the clinic. Comparatively, iPSCs in our study could largely overcome those problems. A2B5, a cell surface ganglioside, is the best characterized marker of developing oligodendroglial progenitors which could assist in remyelination/myelin remodeling and prevent axonal degeneration in TBI.16–18,40,41 In this study, we used A2B5+ iPSCs and transplanted them into the tissue surrounding the injury epicenter in TBI rats. We found that the transplanted iPSCs could differentiate into oligodendrocytes expressing Olig2, and the level of MBP was largely increased in the A2B5+ cell-transplanted condition, which indicated remyelination. Meanwhile, we found that rats gained significant neurological and functional improvement.40
In NSS evaluation, the higher the scores, the more serious the neurological impairment. So far, NSS is one of the most reliable and widely accepted methods for the evaluation of neurological function, such as movement, sensory, equilibrium, and nervous reflex.10 In our study, the results of NSS confirmed the protective effect of A2B5+ iPSCs in TBI rats. We found that TBI effectively induced the increase in NSS, which meant that a successful TBI model has been produced. However, on the seventh day after transplantation, the NSSs of the A2B5+ iPSC group decreased greatly, when compared to the TBI group. This demonstrated that transplantation of A2B5+ iPSCs significantly promotes the recovery of neurological function in TBI rats. Our results provided new evidence for the protective effect of A2B5+ iPSCs for TBI.
At the molecular level, we evaluated the expression of lncRNAs and mRNAs in the A2B5+ iPSC group, the TBI group, and the sham group, and showed that the restoration of neurological function after A2B5+ iPSC transplantation might be associated with the alternations of related lncRNAs and mRNAs. Recent evidence has shown that lncRNAs exhibit strong expression in developing and adult brains20,27,29 and are related to neural differentiation and synaptic plasticity.27,30 Meanwhile, many researchers have reported considerable progress toward understanding the relationship between lncRNAs and brain physiology and diseases.19,20 In our study, in order to gain insight into the potential role of the differentially expressed lncRNAs in the infarct region, we performed GO and pathway analysis to predict the biological functions and mechanisms of 360 correlated targets in the A2B5+ iPSC group. In the GO analysis, some upregulated genes were related to recovery of neurological function. However, some downregulated genes participated in the reduction of the injury response. Biological analysis also indicated that inflammation-associated pathways were the enriched pathways targeted by the downregulated genes. Moreover, lncRNAs and mRNAs in the injured region may have participated in the A2B5+ iPSC transplantation-mediated protection.
More specifically, we found that 37 lncRNAs and 195 mRNAs were simultaneously changed in the A2B5+ iPSC group versus the TBI group and the TBI group versus the sham group. It has been discovered that lncRNAs can be divided into 5 categories: sense, antisense, intronic, intragenic, and bidirectional42 according to their relative position in the genome to the protein-coding genes. In order to gain insight into the potential roles of the common differentially expressed lncRNAs and mRNAs, we found their potential target genes or lncRNAs through scanning 10 kb upstream and downstream of common differentially expressed lncRNAs or mRNAs. The major location relationship of lncRNAs and their target genes was overlap. Then, we intersected the lncRNA and mRNA analysis and ultimately identified the crucial lncRNA (ENSRNOT00000052577) and its target gene (Kif2c), which functioned as a microtubule-dependent molecular motor43 and could depolymerize microtubules at the plus end, thereby promoting mitotic chromosome segregation.43,44 Importantly, the related pathways of Kif2c are related to the immune system and Aurora B signaling,45 and the GO annotations are ATPase activity and microtubule motor activity.46 We therefore speculated that Kif2c might be related to the ability of iPSCs to differentiate into oligodendrocytes and aid in the recovery of neurological function in TBI rats.
Conclusion
In conclusion, A2B5+ iPSC grafts could promote the recovery of neurological function in the TBI brain. Moreover, the present study is the first to detect the differential expression of lncRNAs and mRNAs in brain tissue of rats subjected to TBI and A2B5+ iPSC grafts, in which the lncRNAs and mRNAs in the injured region may contribute to the neuroprotective effects of A2B5+ iPSC transplantation. These available findings provide a new strategy for the treatment of TBI using A2B5+ cell transplantation and explain the possible molecular mechanisms.
Footnotes
Authors’ Note: Qiang Lyu and Zi-Bin Zhang contributed equally to this work.
Ethical Approval: This study was approved by the guidelines of the Institutional Medical Experimental Animal Care Committee of Kunming Medical University.
Statement of Human and Animal Rights: This study was carried out according to the recommendations of guidelines for laboratory animal care and safety from the United States National Institutes of Health.
Statement of Informed Consent: The Statement of Informed Consent is Not Applicable in this study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. DePalma RG. Combat TBI: history, epidemiology, and injury modes. In: Kobeissy FH, editor. Brain Neurotrauma: molecular, neuropsychological, and rehabilitation aspects, frontiers in neuroengineering Boca Raton (FL): CRC Press, 2015. [Google Scholar]
- 2. Iorio-Morin C, Fortin D, Blanchard J. TBI prognosis calculator: a mobile application to estimate mortality and morbidity following traumatic brain injury. Clin Neurol Neurosurg. 2016;142:48–53. [DOI] [PubMed] [Google Scholar]
- 3. Zhou Y, Lui YW. Changes in brain organization after TBI: evidence from functional MRI findings. Neurology. 2013;80(20):1822–1823. [DOI] [PubMed] [Google Scholar]
- 4. Flygt J, Gumucio A, Ingelsson M, Skoglund K, Holm J, Alafuzoff I, Marklund N. Human traumatic brain injury results in oligodendrocyte death and increases the number of oligodendrocyte progenitor cells. J Neuropathol Exp Neurol. 2016;75(6):503–515. [DOI] [PubMed] [Google Scholar]
- 5. Libin AV, Scholten J, Schladen MM, Danford E, Shara N, Penk W, Grafman J, Resnik L, Bruner D, Cichon S, et al. Executive functioning in TBI from rehabilitation to social reintegration: COMPASS (goal,) a randomized controlled trial (grant: 1I01RX000637-01A3 by the VA ORD RR&D, 2013-2016). Mil Med Res. 2015;2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armstrong RC, Mierzwa AJ, Marion CM, Sullivan GM. White matter involvement after TBI: clues to axon and myelin repair capacity. Exp Neurol. 2016;275(pt 3):328–333. [DOI] [PubMed] [Google Scholar]
- 7. Lajiness-O’Neill R, Erdodi L, Bigler ED. Demographic and injury-related moderators of memory and achievement outcome in pediatric TBI. Appl Neuropsychol. 2011;18(4):298–308. [DOI] [PubMed] [Google Scholar]
- 8. Opydo-Chanek M. Bone marrow stromal cells in traumatic brain injury (TBI) therapy: true perspective or false hope? Acta Neurobiol Exp (Wars). 2007;67(2):187–195. [DOI] [PubMed] [Google Scholar]
- 9. Cahu X, Labopin M, Giebel S, Aljurf M, Kyrcz-Krzemien S, Socie G, Eder M, Bonifazi F, Bunjes D, Vigouroux S, et al. Impact of conditioning with TBI in adult patients with T-cell ALL who receive a myeloablative allogeneic stem cell transplantation: a report from the acute leukemia working party of EBMT. Bone Marrow Transplant. 2016;51(3):351–357. [DOI] [PubMed] [Google Scholar]
- 10. Fu XM, Liu SJ, Dan QQ, Wang YP, Lin N, Lv LY, Zou Y, Liu S, Zhou X, Wang TH. Combined bone mesenchymal stem cell and olfactory ensheathing cell transplantation promotes neural repair associated with CNTF expression in traumatic brain-injured rats. Cell Transplant. 2015;24(8):1533–1544. [DOI] [PubMed] [Google Scholar]
- 11. Choi HW, Kim JS, Choi S, Hong YJ, Kim MJ, Seo HG, Do JT. Neural stem cells differentiated from iPS cells spontaneously regain pluripotency. Stem Cells. 2014;32(10):2596–2604. [DOI] [PubMed] [Google Scholar]
- 12. Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. [DOI] [PubMed] [Google Scholar]
- 13. Aoki J, Seo S, Kanamori H, Tanaka M, Fukuda T, Onizuka M, Kobayashi N, Kondo T, Sawa M, Uchida N, et al. Impact of low-dose TBI on outcomes of reduced intensity conditioning allogeneic hematopoietic stem cell transplantation for AML. Bone Marrow Transplant. 2016;51(4):604–606. [DOI] [PubMed] [Google Scholar]
- 14. Shen WB, Plachez C, Tsymbalyuk O, Tsymbalyuk N, Xu S, Smith A, Michel S, Yarnell D, Mullins R, Gulapalli R, et al. Cell-based therapy in TBI: magnetic retention of neural stem cells in vivo. Cell Transplant. 2016;25(6):1085–1099. [DOI] [PubMed] [Google Scholar]
- 15. Xu L, Ryu J, Hiel H, Menon A, Aggarwal A, Rha E, Mahairaki V, Cummings BJ, Koliatsos VE. Transplantation of human oligodendrocyte progenitor cells in an animal model of diffuse traumatic axonal injury: survival and differentiation. Stem Cell Res Ther. 2015;6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Back SA, Riddle A, McClure MM. Maturation-dependent vulnerability of perinatal white matter in premature birth. Stroke. 2007;38(suppl 2):724–730. [DOI] [PubMed] [Google Scholar]
- 17. Baracskay KL, Kidd GJ, Miller RH, Trapp BD. NG2-positive cells generate A2B5-positive oligodendrocyte precursor cells. Glia. 2007;55(10):1001–1010. [DOI] [PubMed] [Google Scholar]
- 18. Ogawa S, Tokumoto Y, Miyake J, Nagamune T. Immunopanning selection of A2B5-positive cells increased the differentiation efficiency of induced pluripotent stem cells into oligodendrocytes. Neurosci Lett. 2011;489(2):79–83. [DOI] [PubMed] [Google Scholar]
- 19. Liu Y, Li G, Lu H, Li W, Li X, Liu H, Li X, Li T, Yu B. Expression profiling and ontology analysis of long noncoding RNAs in post-ischemic heart and their implied roles in ischemia/reperfusion injury. Gene. 2014;543(1):15–21. [DOI] [PubMed] [Google Scholar]
- 20. Chen R, Liu L, Xiao M, Wang F, Lin X. Microarray expression profile analysis of long noncoding RNAs in premature brain injury: a novel point of view. Neuroscience. 2016;319:123–133. [DOI] [PubMed] [Google Scholar]
- 21. Bohmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25(10):623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lan X, Yan J, Ren J, Zhong B, Li J, Li Y, Liu L, Yi J, Sun Q, Yang X, et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64(1):58–72. [DOI] [PubMed] [Google Scholar]
- 23. Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. [DOI] [PubMed] [Google Scholar]
- 24. Zhao XY, Lin JD. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40(10):586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carmichael GG, Taylor HS, Huang Y, Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Commun. 2016;17(1):47–62. [DOI] [PubMed] [Google Scholar]
- 26. Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res./ 2014;115(7):668–677. [DOI] [PubMed] [Google Scholar]
- 27. Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88(5):861–877. [DOI] [PubMed] [Google Scholar]
- 28. Mills JD, Chen J, Kim WS, Waters PD, Prabowo AS, Aronica E, Halliday GM, Janitz M. Long intervening non-coding RNA 00320 is human brain-specific and highly expressed in the cortical white matter. Neurogenetics. 2015;16(3):201–213. [DOI] [PubMed] [Google Scholar]
- 29. Guennewig B, Cooper AA. The central role of noncoding RNA in the brain. Int Rev Neurobiol. 2014;116:153–194. [DOI] [PubMed] [Google Scholar]
- 30. Gudenas BL, Wang L. Gene coexpression networks in human brain developmental transcriptomes implicate the association of long noncoding RNAs with intellectual disability. Bioinform Biol Insights. 2015;9(suppl 1):21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Welstead GG, Brambrink T, Jaenisch R. Generating iPS cells from MEFS through forced expression of Sox-2, Oct-4, c-Myc, and Klf4. J Vis Exp. 2008;(14). pii:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tat PA, Sumer H, Jones KL, Upton K, Verma PJ. The efficient generation of induced pluripotent stem (iPS) cells from adult mouse adipose tissue-derived and neural stem cells. Cell Transplant. 2010;19(5):525–536. [DOI] [PubMed] [Google Scholar]
- 33. Shapira Y, Lam AM, Eng CC, Laohaprasit V, Michel M. Therapeutic time window and dose response of the beneficial effects of ketamine in experimental head injury. Stroke. 1994;25(8):1637–1643. [DOI] [PubMed] [Google Scholar]
- 34. Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19(18):2448–2455. [DOI] [PubMed] [Google Scholar]
- 35. Clarke R, Ressom HW, Wang A, Xuan J, Liu MC, Gehan EA, Wang Y. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008;8(1):37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao F, Qu Y, Liu J, Liu H, Zhang L, Feng Y, Wang H, Gan J, Lu R, Mu D. Microarray profiling and co-expression network analysis of LncRNAs and mRNAs in neonatal rats following hypoxic-ischemic brain damage. Sci Rep. 2015;5:13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cappa KA, Conger JC, Conger AJ. Injury severity and outcome: a meta-analysis of prospective studies on TBI outcome. Health Psychol. 2011;30(5):542–560. [DOI] [PubMed] [Google Scholar]
- 38. Saber M, Kokiko-Cochran ON, Puntambekar S, Lathia J, Lamb BT. TREM2 deficiency alters acute macrophage distribution and improves recovery after TBI. J Neurotrauma. 2017;34(2):423–435. [DOI] [PubMed] [Google Scholar]
- 39. Ramos-Cejudo J, Gutierrez-Fernandez M, Otero-Ortega L, Rodriguez-Frutos B, Fuentes B, Vallejo-Cremades MT, Hernanz TN, Cerdan S, Diez-Tejedor E. Brain-derived neurotrophic factor administration mediated oligodendrocyte differentiation and myelin formation in subcortical ischemic stroke. Stroke. 2015;46(1):221–228. [DOI] [PubMed] [Google Scholar]
- 40. Asher RA, Morgenstern DA, Shearer MC, Adcock KH, Pesheva P, Fawcett JW. Versican is upregulated in CNS injury and is a product of oligodendrocyte lineage cells. J Neurosci. 2002;22(6):2225–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y, Wang H, Sun T, Chen J, Guo L, Shen H, Du Z, Zhou Y. Biological characteristics of a new human glioma cell line transformed into A2B5(+) stem cells. Mol Cancer. 2015;14:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. [DOI] [PubMed] [Google Scholar]
- 43. Wang W, Jiang Q, Argentini M, Cornu D, Gigant B, Knossow M, Wang C. Kif2C minimal functional domain has unusual nucleotide binding properties that are adapted to microtubule depolymerization. J Biol Chem. 2012;287(18):15143–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang W, Shen T, Guerois R, Zhang F, Kuerban H, Lv Y, Gigant B, Knossow M, Wang C. New insights into the coupling between microtubule depolymerization and ATP hydrolysis by kinesin-13 protein Kif2C. J Biol Chem. 2015;290(30):18721–18731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18(8):2970–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shimizu Y, Shimizu T, Nara M, Kikumoto M, Kojima H, Morii H. Effects of the KIF2C neck peptide on microtubules: lateral disintegration of microtubules and beta-structure formation. FEBS J. 2013;280(7):1681–1692. [DOI] [PubMed] [Google Scholar]