Figure 1. Epitope cluster, alignments, and comparative 3D structures of the V1C7 family of VHHs.
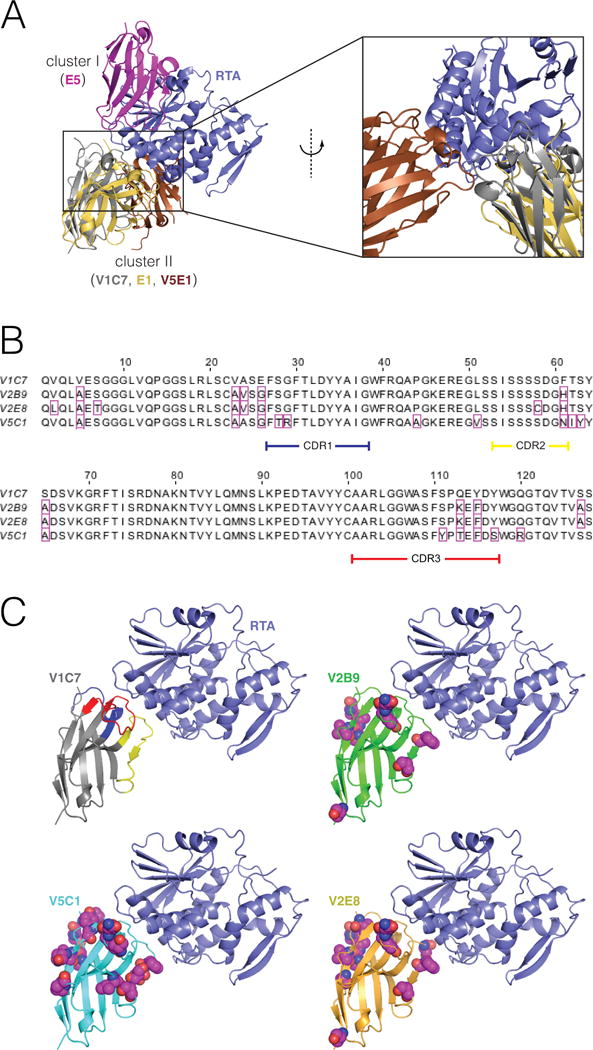
(Panel A) Representative binding modes for epitope clusters I and II on the surface of RTA. VHHs were assigned to cluster I or II based on whether they were competitively inhibited from binding to ricin by the mouse mAbs PB10 (which is similar to UNIVAX R70) or SyH7, respectively. These are the only two clusters for which our group has published X-ray structures of bound VHHs. VHH E5 binds to a cluster-I epitope. V1C7 and E1 bind to a cluster-II epitopes; these two VHHs have a 69% overall sequence identity 25% over the CDRs. (Panel B) Primary amino acid sequence alignment of the V1C7 family of VHHs. Residues that differ from V1C7 are boxed in magenta. V1C7 has a sequence identity of >87% overall and of >81% over the CDRs with any other member of the family. (Panel C) Proposed structures of the four VHHs in complex with RTA. The X-ray structure of V1C7 in complex with RTA was solved at 1.8-Å resolution (PDB: 5J56). The structures of V2B9, V2E8, and V5C1 are computational models having the same backbone structure as V1C7. Residues that differ from their V1C7 counterparts are represented as spheres (carbon: magenta; oxygen: red; nitrogen: blue; sulfur: yellow).
