Abstract
The human innate immune protein calprotectin (CP, S100A8/S100A9 oligomer, calgranulin A/calgranulin B oligomer, MRP-8/MRP-14 oligomer) chelates a number of first-row transition metals, including Mn(II), Fe(II), and Zn(II), and can withhold these essential nutrients from microbes. Here we elucidate the Ni(II) coordination chemistry of human CP. We present a 2.6-Å crystal structure of Ni(II)- and Ca(II)-bound CP, which reveals that CP binds Ni(II) ions at both its transition-metal-binding sites: the His3Asp motif (site 1) and the His6 motif (site 2). Further biochemical studies establish that coordination of Ni(II) at the hexahistidine site is thermodynamically preferred over Zn(II). We also demonstrate that CP can sequester Ni(II) from two human pathogens, Staphylococcus aureus and Klebsiella pneumoniae, that utilize this metal nutrient during infection, and inhibits the activity of the Ni(II)-dependent enzyme urease in bacterial cultures. In total, our findings expand the biological coordination chemistry of Ni(II)-chelating proteins in nature and provide a foundation for evaluating putative roles of CP in Ni(II) homeostasis at the host-microbe interface and beyond.
Graphical abstract
Introduction
Nickel is an important trace nutrient for many organisms.1-4 Several decades of investigations have illuminated the regulation and utilization of this metal in bacteria.1,2 A number of microbial enzymes employ nickel as a cofactor for catalytic activity,3 including superoxide dismutase,5 urease,6 [NiFe]-hydrogenase,7 and carbon monoxide dehydrogenase.8 In the context of infectious disease, recent reports have highlighted the importance of Ni(II) import systems for the growth and virulence of the pathogenic bacterium Staphylococcus aureus under metal-depleted conditions in vitro and in animal models of infection.9-11 Furthermore, urease is an enzyme that contributes to the virulence of human pathogens,12,13 including Helicobacter pylori,14 Staphylococcus spp.,15,16 and Klebsiella pneumoniae.17 In contrast, less is known about nickel homeostasis in mammals and other higher organisms, and no mammalian nickel-dependent enzyme has been identified.1
Several mechanisms to withhold nutrient transition metals from microbial invaders are employed by the mammalian host during the early stages of infection in a process termed “nutritional immunity.”18-22 Antimicrobial proteins such as lactoferrin and siderocalin prevent microbial Fe(III) uptake,23,24 and the S100 family proteins S100A7 (psoriasin) and S100A12 (calgranulin C) scavenge Zn(II).25-29 The S100 protein calprotectin (CP, S100A8/S100A9 oligomer) is a versatile metal-sequestering protein that coordinates Mn(II), Fe(II), and Zn(II) with high affinity and is able to withhold these metals from microbial pathogens.28,30-32 Despite the importance of nickel in microbial pathogenesis,12-15,17 to the best of our knowledge, a host- defense strategy that limits the microbial acquisition of this metal is unknown.33
CP is released from neutrophils and epithelial cells during the innate immune response.19-21,28,30 At sites of infection, human CP has been reported to be present at levels up to ≈1 mg/mL (≈40 μM heterodimer).34 As a member of the Ca(II)-binding S100 protein family, human CP is the heterooligomer of S100A8 (α) and S100A9 (β) and exists as an αβ heterodimer or α2β2 heterotetramer.35,36 CP has four EF-hand domains that coordinate Ca(II) ions, including a C-terminal canonical (“calmodulin-like”) site and a N-terminal non-canonical site in each subunit.36-38 Ca(II) chelation causes two αβ heterodimers to self-associate to form an α2β2 heterotetramer.35,36 In addition, Ca(II) binding enhances the transition-metal affinities, antimicrobial activity, and protease stability of CP.39,40 Distinct from the Ca(II)-binding EF-hands, two transition-metal-binding sites form at the S100A8/S100A9 dimer interface.38,41,42 These sites are a His3Asp motif (site 1) and a His6 motif (site 2). Site 1 has high affinity for Zn(II)39 and has been observed to chelate Mn(II)42,43 and Fe(II),44 albeit with relatively low affinity. Site 2 comprises a unique hexahistidine metal-binding motif that coordinates Mn(II)32,41-43, 45 Fe(II),31,44 and Zn(II)39,46 with high affinity. Our metal-substitution studies demonstrated that site 2 exhibits thermodynamic preference for these divalent cations (i.e., Kd,Zn < Kd,Fe < Kd,Mn)31,43 consistent with the Irving-Williams series.47 Moreover, our prior work revealed that CP treatment of bacterial growth medium also reduces the concentrations of nickel.31 In these experiments, the metal-binding-site variants of CP (e.g. ΔHis3Asp, ΔHis4; Table S1, Supporting Information) afforded metal-depletion profiles indicating that site 2 is a high-affinity site for nickel.31 Based on these observations, we reasoned that CP may also contribute to the sequestration of Ni(II) ions from microbial pathogens and thereby play an as-yet unidentified role in mammalian Ni homeostasis and host defense. To evaluate this notion, we sought to explore the Ni(II) coordination chemistry of CP and to understand the role of Ni within the broader context of its metal-withholding function.
In this work, we report structural, biochemical, and functional evaluation of Ni(II) coordination by CP. We present a crystal structure of Ni(II)- and Ca(II)-bound CP that reveals Ni(II) chelation at both sites 1 and 2. We demonstrate that CP coordinates two equivalents of Ni(II) in solution, and that the His6 site coordinates Ni(II) with greater affinity than Zn(II). In addition, we show that CP can limit Ni uptake into bacterial pathogens, and inhibit bacterial urease activity. These discoveries underscore the functional versatility of CP in sequestering essential metal nutrients. Moreover, this work on Ni(II) coordination provides the foundation for examining CP in a broad context of Ni homeostasis.
Results
Crystal Structure of Ni(II)- and Ca(II)-bound CP-Ser
CP-Ser is the heterooligomer of S100A8(C42S) and S100A9(C3S) (Table S1). We routinely use this variant in biochemical, biophysical, and functional studies of CP, and under all conditions evaluated to date, it displays comparable metal-binding properties and antimicrobial activity to native CP.31,39,43 To build upon our preliminary observations from metal-depletion studies (vide supra) indicating that the His6 site of CP-Ser binds Ni(II),31 we sought to obtain a crystal structure of Ni(II)- and Ca(II)-bound CP-Ser. Guided by our prior crystallographic study of Mn(II)- and Ca(II)-bound CP-Ser in which we obtained crystals following incubation of the protein with 1 equiv Mn(II) and observed Mn(II) bound only at the His6 site,42 we screened crystallization conditions where CP-Ser (αβ) was incubated with ≈1 equiv Ni(II). We anticipated that we would obtain Ni(II)-bound CP-Ser where the Ni(II) ion populates the His6 site. We solved the structure of Ni(II)- and Ca(II)-bound CP-Ser to 2.6-Å resolution by molecular replacement with two α2β2 heterotetramers in the asymmetric unit (Figure 1, Table S2). Formation of α2β2 heterotetramers under these conditions is consistent with prior work establishing that Ca(II) binding to the EF-hand domains and transition-metal binding at the His6 site promote formation of CP heterotetramers.35,36,40
Figure 1.
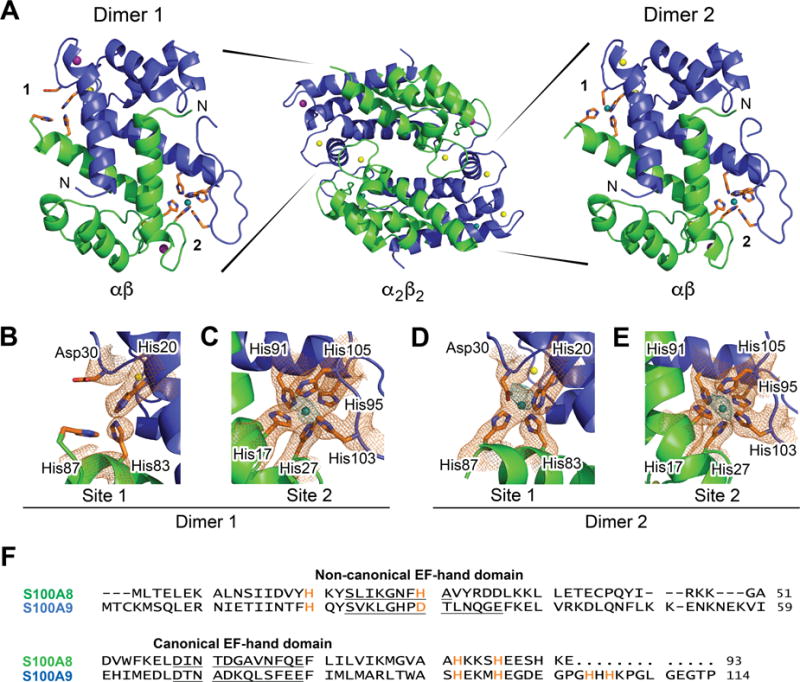
X-ray crystallographic analysis of Ni(II)- and Ca(II)-bound CP. (A) Dimer (αβ) and tetramer (α2β2) models of CP-Ser coordinated to Ni(II) (teal), Ca(II) (yellow), and Na(I) (purple). The S100A8 subunit is green, and the S100A9 subunit is blue. The two dimers, denoted dimers 1 and 2, are depicted in 90° rotation to the tetramer and exhibit different metal binding. Dimer 1 (left) contains a Ni(II) ion at site 2 only with apparent 100% occupancy. Dimer 2 (right) contains a Ni(II) ion at site 1 refined at 75% occupancy and a Ni(II) ion at site 2 refined at 100% occupancy. The N-terminus of each subunit is labeled. (B) Site 1 of dimer 1. (C) Site 2 of dimer 1. (D) Site 1 of dimer 2. (E) Site 2 of dimer 2. A 2Fo-Fc composite omit electron density map (orange mesh) to 2.6-Å resolution is contoured at 1σ around the metal sites. A 3.6-Å resolution nickel anomalous difference map, calculated using data collected at a wavelength of 1.4831 Å, is contoured at 3σ and shown in teal. (F) Amino acid sequence alignment of human S100A8 and S100A9. The metal-binding residues are orange. The residues of the EF-hand domains are underlined. The metal speciation of each subunit is described in Table 1
For each heterotetramer, all eight EF-hand domains exhibited electron density consistent with the presence of metals, and we were able to assign Ca(II) ions refined at 100% occupancy to five of the EF-hands (Table 1, Figure S1). The four canonical sites contained Ca(II) ions, similar to the three published crystal structures where CP-Ser has bound Ca(II) ions.38,41,42 In addition, we modeled a Ca(II) ion at 100% occupancy at the non-canonical EF-hand of S100A9 of dimer 2 and Na(I) ions at 100% occupancy for the other three non-canonical EF-hand domains in the tetramer. These results differ from the tetramers of the two reported Mn(II)- and Ca(II)-bound CP-Ser structures that exhibit (i) Ca(II) ions or no metal at the non-canonical domains,41 or (ii) Na(I) ions at all four non-canonical EF-hands instead of only three of these sites (see Supporting Discussion).42
Table 1.
Occupancy of Metal Ions in Ni(II)-, Ca(II)- and Na(I)-bound CP-Ser Crystal Structure
| Dimer | Subunit | N-EFa | C-EFb | His3Aspc | His6 |
|---|---|---|---|---|---|
| 1 | S100A8 | Na | Ca | – | Ni |
| S100A9 | Na | Ca | |||
| 2 | S100A8 | Na | Ca | Nid | Ni |
| S100A9 | Ca | Ca |
Non-canonical N-terminal EF-hand domain.
Canonical C-terminal EF-hand domain.
Dashes denote binding sites without a metal modeled in.
Ni is modeled in at 75% occupancy at the His3Asp site. All other metals are modeled at 100% occupancy.
To interrogate the relative affinities of site 1 and 2 for Ni(II) ions, we added less Ni(II) to our crystals than would be necessary to completely fill both sites on each heterodimer (≈1.0 equiv Ni(II) relative to the CP-Ser heterodimer instead of 2.0 equiv), and then used X-ray data at λ = 1.4831 Å to generate a Ni anomalous difference map at 3.6-Å resolution and to identify where the Ni(II) ions were bound (Table S2). This map reveals that Ni(II) ions are coordinated at both His6 sites and at one of the two His3Asp sites of the tetramer (Figure 1B–E, Table 1). Our refinement indicates that the Ni(II) ion at the one occupied His3Asp site is not at full occupancy (approximately 75%), compared to the two His6 sites of the heterotetramer, which do appear to bind Ni(II) at full occupancy. This differential binding of Ni(II) at the two sites suggests that CP has higher affinity for Ni(II) at site 2 over site 1. These results are reminiscent of our prior observation that Ni depletion in bacterial growth medium was dependent on the presence of site2.31
Site 2 coordinates Ni(II) using the hexahistidine motif where the metal ion is coordinated by the Nε2 atoms of residues His17 and His27 of S100A8 and His91, His95, His103, and His105 of S100A9 (Figure 1C,E). The bond distances and angles indicate distorted octahedral geometry (Tables S3–S4). These results are consistent with previous studies of Mn(II),41-43, 45 Fe(II),31, 44 and Zn(II)39, 46 bound at this site. The two Mn(II)- and Ca(II)-bound CP structures comprise the Mn(II)-His6 site,41, 42 and structural alignments of the metal-bound His6 motifs indicate little difference between Mn(II) and Ni(II) coordination at this site (Figure S2).
Site 1 coordinates Ni(II) in a tetrahedral fashion by residues His83 (Nε2) and His87 (Nε2) of S100A8 and His20 (Nε2) and a monodentate Asp30 (Oδ1) of S100A9 (Figure 1E, Tables S5–S6). In contrast, one of the Mn(II)-bound crystal structures was refined with a Mn(II) ion at 50% occupancy at site 1 and shows that the His3Asp motif harbors a five-coordinate Mn(II) center, where the Asp residue provides bidentate coordination.41
CP Binds Two Equivalents of Ni(II) in Solution
To confirm that CP coordinates two equiv Ni(II) per heterodimer in solution under conditions where Ni(II) is in excess, we employed size-exclusion chromatography (SEC) (Figures 2, S3). Samples of CP-Ser, ΔHis3Asp, and ΔHis4 (Table S1) prepared in the absence and presence 5.0 equiv Ni(II) were incubated and analyzed by SEC, and the metal content of the eluent fractions was measured by inductively coupled plasma- mass spectrometry (ICP-MS). Following incubation with Ni(II), CP-Ser retains ≈2 equiv Ni(II) per heterodimer in the eluent fractions, indicating that both the His3Asp and His6 sites coordinate this metal ion with sufficient affinity for the metal to be retained over the course of elution from the SEC column (Figure 2A). This experiment also indicates that only ≈1 equiv Ni(II) is retained during the SEC elution of ΔHis3Asp or ΔHis4, variants that lack metal-binding residues of site 1 or site 2 (Table S1), and further confirms that Ni(II) coordination to CP is dependent on the presence of the transition-metal sites (Figure 2B,C). These results demonstrate that both the His3Asp and His6 sites coordinate Ni(II) in solution, and that a 2:1 Ni(II):CP complex forms.
Figure 2.
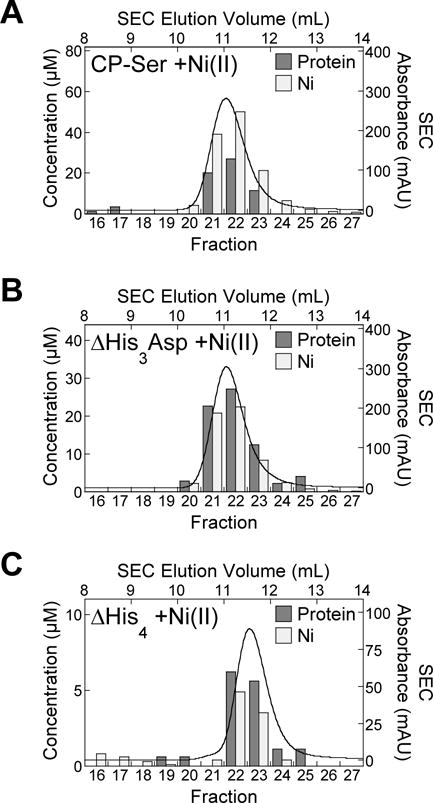
The CP-Ser heterodimer binds two equivalents of Ni(II) in solution whereas the ΔHis3Asp and ΔHis4 variants coordinate only one equivalent of Ni(II) under the same conditions. Samples of 300 μM (A) CP-Ser, (B) ΔHis3Asp, and (C) ΔHis4 preincubated with 5.0 equiv Ni(II) were monitored by analytical SEC in 75 mM HEPES, 100 mM NaCl, pH 7.0. The SEC chromatograms are shown as absorbance (right y-axis) as a function of elution volume (top x-axis). The protein and Ni concentrations (left y-axis) of the eluent fractions (bottom x-axis) were measured by absorbance at 280 nm and by ICP-MS, respectively, and these data are shown as bar plots. Protein concentration is shown as dark gray bars, and the Ni concentration is shown as light gray bars. Data from one representative experiment for each condition is shown.
CP Prefers to Coordinate Ni(II) over Other First-Row Transition Metals at Site 2
To further evaluate the N(II)-binding properties of CP, we investigated the relative affinities for Ni(II) and Zn(II) of site 2 by performing metal-substitution assays. We focused on site 2 because we observed full occupancy of this site in both heterodimers in the current crystal structure (Figure 1), and this site was required for CP to deplete Ni from bacterial growth medium in our prior work.33 We designed and prepared the biotinylated CP variant B-ΔHis3Asp (Table S1, S4–S6), established a biotin-streptavidin pull-down assay where B-ΔHis3Asp is removed from aqueous solution using streptavidin agarose resin (Figure S7), and employed this assay to investigate the metal selectivity of the His6 site (Figure 3). In this experiment, B-ΔHis3Asp was preincubated with 1.0 equiv of either Ni(II) or Zn(II) in the presence of excess Ca(II) to form the Ni(II)- and Zn(II)-bound proteins. Subsequently, 1.0 equiv of Zn(II) or Ni(II) were added to the solutions of Ni(II)-bound or Zn(II)-bound B-ΔHis3Asp, respectively. After 72 h of incubation at 37 °C, the samples were treated with streptavidin resin, and the unbound metal content in the supernatant was measured by ICP-MS. A comparison of the unbound metal concentration for the two metals in each sample provides an assessment of relative metal affinities because the metal ion that exhibits a higher unbound concentration is the one for which CP has a lower affinity. The order of metal addition was reversed to ensure that sufficient incubation time was given to reach equilibrium. Samples incubated with only one metal were also analyzed as controls (Figure 3).
Figure 3.
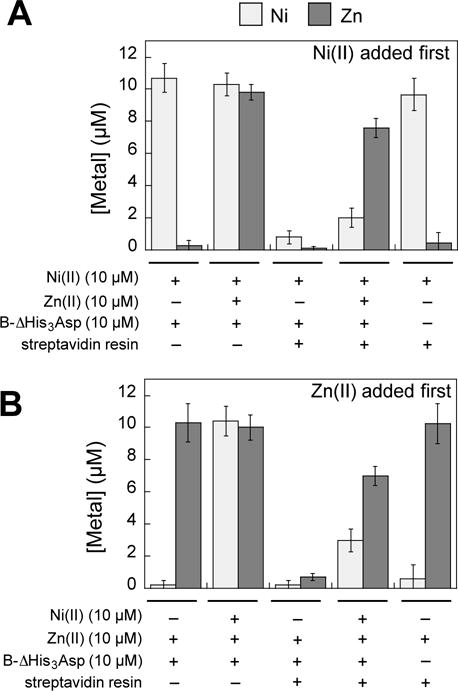
Metal selectivity of the His6 site ascertained by the B-ΔHis3Asp pull-down assay. The concentrations of Ni(II) and Zn(II) in the supernatant of each sample were determined by ICP- MS. B-ΔHis3Asp (10 μM) was incubated with 10 μM Ni(II), and/or Zn(II) for 72 h at 37 °C in 75 mM HEPES, 100 mM NaCl, 2 mM CaCl2, pH 7.0 and the mixture was treated with streptavidin agarose resin. (A) Ni(II) was added first and the Ni(II) + B-ΔHis3Asp mixture was incubated for 30 min at room temperature prior to addition of Zn(II). (B) Zn(II) was added first. The mean and SDM are reported (n = 4 for samples with B-ΔHis3Asp n added, n = 2 for samples without B-ΔHis3Asp).
The results from this metal substitution experiment using Ca(II)-bound B-ΔHis3Asp demonstrate that, regardless of the order of metal addition, the quantity of unbound Zn(II) is greater than that of unbound Ni(II) (Figure 3). Thus, the hexahistidine site of CP has a thermodynamic preference for Ni(II) over Zn(II). Taken together with our prior metal substitution studies,31,43 we conclude that the thermodynamic preference of site 2 for divalent first-row metals is Kd,Ni < Kd,Zn < Kd,Fe < Kd,Mn. This trend is consistent with the Irving-Williams series.47
Ni(II) Preincubation Blocks Metal Sequestration Associated with Site 2
We performed antibacterial activity assays employing CP-Ser, ΔHis3Asp, and ΔHis4 preincubated with 0, 1 and/or 2 equiv Ni(II) or Zn(II) against Escherichia coli (Ni and Zn) and S. aureus (Ni) to evaluate the effect of preloading Ni(II) and Zn(II) on the site-dependent metal-sequestering antibacterial action of CP (Figure S8). We observed that the antibacterial activity of CP-Ser is partially attenuated in the presence of 1 equiv Ni(II) (Figure S8A,B). Following a 20-h incubation, E. coli cultures treated with CP-Ser (1.0 mg/mL) preincubated with 1 equiv Ni(II) exhibited OD600 ≈0.1, whereas treatment with apo CP-Ser (1.0 mg/mL) afforded negligible E. coli growth (OD600 < 0.02; Figure S8A). Moreover, CP-Ser preincubated with 2 equiv Ni(II) provided growth inhibition comparable to that of CP-Ser preincubated with only 1 equiv Ni(II), suggesting that Ni(II) prevents only one site of CP from sequestering nutrient metals in the growth medium. Preincubation of ΔHis3Asp with 1 equiv Ni(II) resulted in full E. coli growth, whereas Ni(II) preloading did not affect the antibacterial activity of ΔHis4 variant. We observed similar trends after preincubating CP-Ser with Ni(II) when the assay was conducted with S. aureus (Figure S8B). These bacterial growth studies indicate that Ni(II) only blocks site 2. In contrast, Zn(II) effectively blocks the antibacterial activity associated with both sites 1 and 2 (Figure S8C).
Our crystallographic and biochemical analyses of CP demonstrate that both sites 1 and 2 chelate Ni(II). However, the antibacterial activity assays suggest that only site 2 has the capacity to sequester Ni(II) because preincubation with Ni(II) only blocks the growth inhibitory activity attributed to this site. We reason that metal exchange occurs at site 1 under our assay conditions. Preincubation of CP with 2 equiv Ni(II) affords a Ni(II)-bound His3Asp motif, but because Ni(II) binding at this site is relatively labile, site 1 still contributes to the antibacterial activity of CP by binding and sequestering Zn(II) after the Ni(II) ion dissociates. In contrast, our data indicate that metal exchange occurs less readily at site 2. As a result, Ni(II) remains coordinated at site 2 following pre-incubation, which prevents this site from capturing and withholding other nutrient metals.
CP Sequesters Ni(II) from Bacterial Pathogens
To probe the functional relevance of Ni(II) coordination by CP, we evaluated whether CP has the capacity to withhold Ni(II) from microbes. First, we measured the intracellular Ni(II) content of S. aureus suspensions (OD600 = 6, ≈109 CFU/mL) that were obtained from cultures treated with CP or the ΔΔ variant (a variant that lacks both transition-metal-binding sites; Table S1) by acid digestion and ICP-MS (Figures 4, S9A,B). Because CP coordinates other nutrient transition metals at the Ni(II)-binding sites, we selected a metal-depleted chemically-defined staphylococcal growth medium (dCDM) to which no transition metals were added (Table S7), modified from an earlier protocol.48 This medium contained less than 200 nM Mn, Fe, or Zn (Table S8). Bacteria were grown overnight in dCDM with 1 μM Ni(II) in the absence and presence of 1 μM CP or the ΔΔ variant. In this assay, we employed the methicillin-resistant S. aureus (MRSA) strains USA300 JE2 and M2, and the methicillin-sensitive strain S. aureus ATCC 29213 (Table S9). In addition, we evaluated the ΔcntA mutant strain of S. aureus USA300 JE2.49 The cntA gene encodes the extracellular solute- binding protein of the broad-spectrum metallophore staphylopine.11 Bacterial strains lacking genes of the cnt system are deficient in Ni uptake and exhibit reduced urease activity under metal-limiting conditions.10
Figure 4.
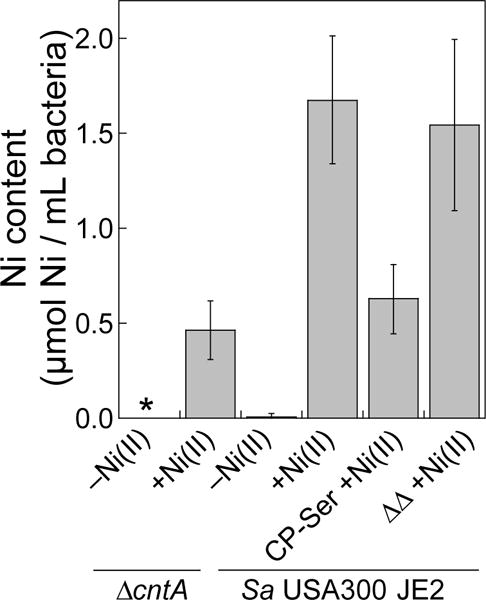
CP treatment results in decreased intracellular Ni in S. aureus USA300 JE2 as measured by ICP-MS. The mean Ni content of the bacterial cells (OD600 = 6, ≈109 CFU/mL) and SDM are reported (n = 6). The asterisk denotes Ni levels below the detection limit. Data for S. aureus M2 and ATCC 29213 are provided in Figures S9.
We observed intracellular Ni levels in the 1.0–2.0 μM range when S. aureus was grown in the presence of Ni(II) or with the ΔΔ variant (Figures 4, S9A,B). In contrast, cultures grown in the presence of CP-Ser exhibited markedly lower intracellular Ni content (Figures 4, S9A,B). These data indicate that CP-Ser coordinated Ni(II) present in the growth medium and thereby prevented microbial Ni(II) uptake under these growth conditions. In addition, S. aureus USA300 JE2 ΔcntA exhibited decreased levels of Ni compared to the parent strain (Figure 4), supporting that the Cnt system is an active Ni(II) acquisition system under the metal-depleted conditions used in this work, as previously observed in metal-deplete media.9-11
CP Attenuates the Activity of a Bacterial Ni(II) Enzyme
Next, we examined the impact of reduced intracellular Ni(II) uptake on the activity of a staphylococcal Ni(II) enzyme. Ni(II) is a cofactor for urease, a bacterial enzyme that catalyzes the hydrolysis of urea to ammonium and carbon dioxide.13 Bacterial pathogens are thought to utilize ammonium as a local pH buffer in acidic host environments, such as in the gastrointestinal and urinary tracts.12 CP is capable of inhibiting the growth of S. aureus (Figure S10A), and this organism utilizes urease during infection.14,15 On the basis of the Ni(II)-uptake study (Figure 4), we hypothesized that Ni(II) sequestration by CP in the extracellular space would perturb intracellular Ni(II) homeostasis and diminish the urease activity of S. aureus.
To test this notion, we first designed and validated a whole cell urease activity assay employing the metal-depleted chemically defined medium dCDM (Figures 5, S11). This assay is based on a standard urease test, employed in medical microbiology for strain identification, where microbes are cultured in a urea broth containing phenol red. Similar to the Ni(II)-uptake study, bacteria were grown overnight in dCDM with 1 μM Ni(II) in the absence and presence of 1 μM CP-Ser or the ΔΔ variant. The bacteria were then incubated in chemically defined urea broth (dCDMU) that contained the colorimetric pH indicator phenol red, which exhibits a color change between pH 6.8 and 8.2 in aqueous solutions (pKa = 7.5, 25 °C).50 The color and pH of the supernatant of the bacterial suspension were monitored over time to determine relative levels of urease activity between growth conditions. Bacteria cultured in dCDM without the 1 μM Ni(II) supplement were tested as negative controls. In this assay, we employed S. aureus strains USA300 JE2 and the ΔcntA mutant, M2, and ATCC 29213. We also utilized the ΔureC mutant strain of S. aureus USA300 JE249 as a negative control. The ureC gene encodes the Ni(II)-binding α subunit of urease; thus, the ΔureC strain lacks a functional urease.
Figure 5.
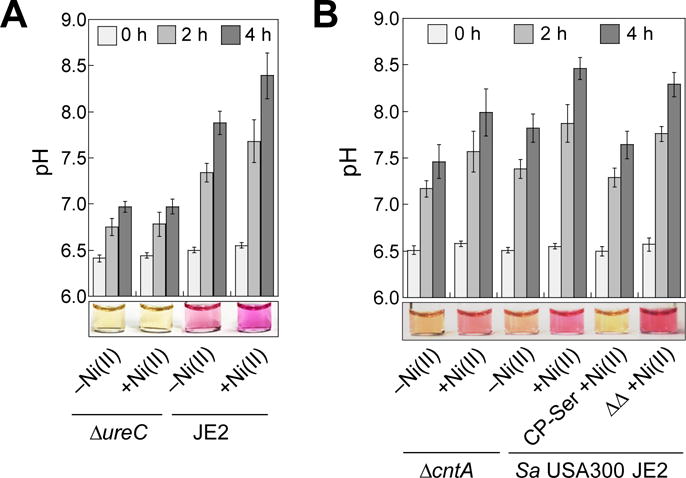
CP attenuates urease activity of S. aureus USA300 JE2 as indicated by directly measuring pH (bar plots) and visual detection using the colorimetric pH indicator phenol red (photographs below bar plots, pKa = 7.5 at 25 °C, ref. 50), which turns from yellow to purple with increasing pH. (A) The pH profile and a representative image of dCDMU from bacterial cultures of S. aureus USA300 JE2 and ΔureC grown in the absence and presence of a 1-μM Ni(II) supplement (14-18 h, 37 °C). The mean pH values and SDM are reported (n = 3). The image was taken at t = 4 h. (B) The pH profile and a representative image of dCDMU from bacterial cultures of S. aureus USA300 JE2 and ΔcntA grown in the absence and presence of 1 μM Ni(II) and CP variants (14-18 h, 37 °C). The image was taken at t = 2 h. The mean pH values and SDM are reported (n = 6).
For S. aureus USA300 JE2 incubated with Ni(II), we observed that the pH values of dCDMU increased indicative of urease activity (Figure 5A,B). In contrast, negligible change in pH was observed for ΔureC, which indicates the observed increase in pH results from urease activity (Figure 5A,B). As expected, ΔcntA exhibited reduced urease activity compared to the parent strain under +Ni(II) growth conditions (Figure 5B). S. aureus USA300 JE2 grown in the presence of CP-Ser and Ni(II) showed lower pH levels over the time course compared to bacteria cultured in the presence of Ni(II) only (Figure 5B). Moreover, cultures of S. aureus USA300 JE2 grown in the presence of 1 μM Ni(II) and the ΔΔ variant exhibited comparable pH levels to those grown under +Ni(II) only conditions (Figure 5B). Urease assays conducted with the M2 and ATCC 29213 strains afforded similar trends (Figure S9C–F). Taken together, these results suggest that attenuated urease activity is a consequence of Ni(II) sequestration by CP. To confirm that the variations in urease activity were not caused by differences in the number of bacteria, we quantified the bacterial cells in select dCDMU samples at the beginning and end of each assay by colony counting, and each suspension tested exhibited comparable numbers of bacterial cells (Table S10).
Next, to directly monitor urease activity in CP-treated cultures and to validate our whole-cell urease assay results, we performed a phenol-hypochlorite assay (also termed indophenol assay)51 to quantify ammonia production in cell lysates supplemented with urea (Figure 6). This assay employs the Berthelot reaction, a reaction of ammonium ions with phenols under oxidizing conditions that results in formation of an indophenol dye.51 Thus, the generation of indophenol provides a colorimetric and quantitative readout for ammonia levels in cell lysates, and this assay is employed to detect urease activity.17,52 To minimize background ammonia production in these assays, dCDM prepared without ammonium sulfate was utilized for culture growth. In this medium, S. aureus USA300 JE2 or ΔureC was grown with 1 μM Ni(II) in the presence or absence of 1 μM CP-Ser or ΔΔ. Cultures were lysed enzymatically and the soluble lysate was incubated in urea-containing HEPES buffer for 20 min before assaying for ammonia content. As expected, the lysates of the ΔureC strain yielded negligible ammonia production. We observed that lysates of cultures treated with Ni(II) only catalyzed more ammonia production than a negative control where no Ni(II) was added. Moreover, the lysates from cultures treated with CP-Ser and Ni(II) exhibited attenuated ammonia production compared to cultures treated with ΔΔ and Ni(II). These data agree with our findings from the whole cell urease assays, further supporting the role of CP in withholding Ni(II) and inhibiting urease activity in S. aureus.
Figure 6.
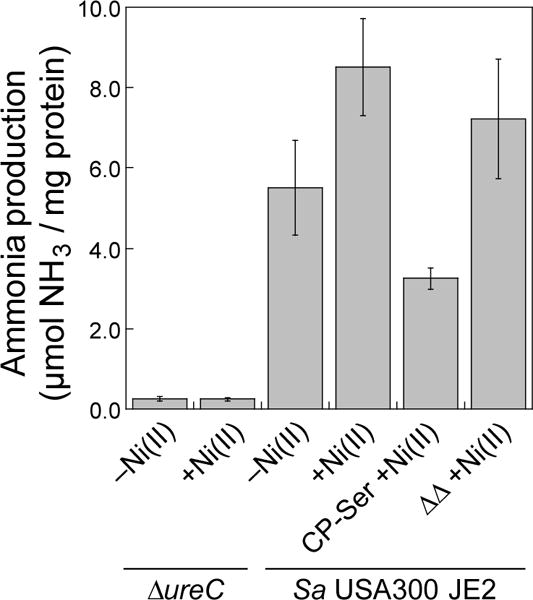
Urease activity of S. aureus USA300 JE2 and ΔureC cell lysates monitored by the direct detection of ammonium ions using the phenol-hypochlorite assay. Prior to the assay, bacteria were cultured in dCDM without ammonium in the absence or presence of 1 μM Ni(II) and CP variants as indicated (8 h, 37 °C). Mean μmol ammonia/mg protein and SDM are reported (n = 12).
K. pneumoniae also utilizes urease during infection,17 and CP exhibits growth inhibitory activity against this Gram-negative pathogen (Figure S10B). We therefore examined the Ni(II) uptake and urease activity of K. pneumoniae ATCC 13883 cultured in dCDM in the absence and presence of CP-Ser (Figure S12, Table S10). Analysis of the Ni content in K. pneumoniae revealed markedly lower intracellular levels of this metal (≈0.01 μM Ni) under +Ni(II) conditions compared to S. aureus USA300 JE2, M2 and ATCC 29213. Despite differences in intracellular Ni, the urease activity profiles are similar between these organisms over the time course. As observed for S. aureus, the presence of CP-Ser in the growth medium resulted in decreased urease activity for K. pneumoniae. Taken together, the results from these microbiology studies demonstrate that CP can perturb intracellular Ni(II) homeostasis and attenuate urease activity in two different human pathogens under laboratory conditions.
Discussion
In this work, we report structural and biochemical studies of Ni(II) complexation by human CP and demonstrate that this host-defense protein can sequester this metal from microbes. We present a crystal structure of Ni(II)- and Ca(II)-bound CP, which shows that CP chelates Ni(II) at both transition-metal-binding sites and expands the biological coordination chemistry of Ni(II) centers in proteins. To the best of our knowledge, the Ni(II)-His6 motif of CP is unique amongst structurally characterized nickel proteins (Table S11).53 Proteins that contain a Ni(II)- His6 site identified in the Protein Data Bank54 include: (i) the metallochaperone SlyD from Thermus thermophilus where a conserved His3 motif, two residues of a His6-tag incorporated for protein purification, and one residue of a neighboring monomer in the crystal complete the coordination sphere (PDB: 3CGM);55 (ii) the homotrimeric engineered four-helix bundle protein construct MBPC-1 that undergoes metal-templated oligomerization where each monomer contributes two His residues around the metal center (PDB: 3DE9);56 and (iii) the N-terminal domain of a Na+/K+ ATPase crystallized with Ni(II) ions bound to polyhistidine tags that were employed during protein purification (PDB: 1Q3I).57 The tetrahedral Ni(II)-His3Asp site is also noteworthy. Metalloproteins that contain a native four-coordinate Ni(II) site include NikR of E.coli (PDB: 2HZA),58 NikM of Thermoanaerobacter tengcongensis (PDB: 4M58),59 and LarA of Lactobacillus plantarum (PDB: 4YNS).60 In contrast to the Ni(II)-His3Asp site of CP, these three metalloproteins exhibit square planar geometries at each Ni(II) center.
Our current findings demonstrate that CP prefers to coordinate Ni(II) over Zn(II) at the His6 site. This result is in agreement with the Irving-Williams series,47 which shows that Ni(II) is thermodynamically favored over Mn(II), Fe(II), and Zn(II) for an octahedral coordination site. Moreover, the established stability constants of Ni(II) and Zn(II) centers coordinated by small- molecule imidazole-containing ligands (Table S12)61-63 support the relative affinities of CP for these two metal ions. We have previously reported lower limits to the Kd values of CP for Zn(II) in the presence of Ca(II) (Kd1,Zn ≤ 90 fM and Kd2,Zn ≈ 0.9 pM), and we did not assign these values to the His3Asp and His6 sites of CP.46 Based on the relative Ni(II)/Zn(II) affinities determined in this work from qualitative metal selectivity studies (Figure 3), we conclude that CP coordinates Ni(II) at site 2 with Kd,Ni ≤ 0.9 pM in the presence of Ca(II).
To the best of our knowledge, a study to examine the metal content and speciation of metal-bound CP in a biological sample has not been reported. However, based on the fact that the His6 site binds Ni(II) with greater affinity than Zn(II), we posit that CP can function as a Ni(II)-chelating protein in vivo. Because the His6 site sequesters multiple first-row metals, the speciation and relative concentrations of bioavailable metals at a given site will influence the speciation of metal-bound CP. Moreover, in prior work on the coordination chemistry of the His6 site, we reasoned that the His6 site of CP will likely withhold the transition metal ion that it encounters first.31 CP can be released at levels that are expected to be in excess of the bioavailable metal concentrations found at sites of infection.21,34 For instance, extracellular CP has found in concentrations up to 1 mg/mL (≈40 μM heterodimer).34 The concentration of nickel in human blood is ≈0.5 nM, and subnanomolar to low nanomolar concentrations have been measured in human serum, plasma, and urine.64,65 Provided that CP is adequately abundant in an environment where Ni(II) is available, the current work indicates that CP has the ability to function as a nickel-sequestering protein.
In the context of infectious disease, a number of human pathogens such as S. aureus and K. pneumoniae that can infect the gastrointestinal and urinary tracts require Ni(II) for successful colonization of the host.3,9,12,15,17 To the best of our knowledge, no mammalian Ni(II)-sequestering antimicrobial host-defense mechanism has been identified. Our current work shows that CP has the remarkable capacity to prevent Ni(II) uptake by these organisms. CP is abundant in the gastrointestinal tract and is a biomarker for irritable bowel diseases.66 In addition, elevated levels of CP have been associated with urinary tract infection67 and other urinary diseases such as bladder cancer and kidney injury.68,69 Given that CP is present in these physiological locales where urea is abundant and where microbial pathogens are known to colonize, our work affords a new hypothesis that human CP may be involved in the homeostasis of Ni(II) at the host- microbe interface. In addition to human pathogens such as S. aureus and K. pneumoniae, the role of Ni(II) is established for the virulence of H. pylori,14,70 and whether CP influences Ni(II) trafficking and utilization in H. pylori and other gastrointestinal pathogens during infection is an avenue for future research.
Lastly, the ability of CP to bind Ni(II) with high affinity may have other physiological implications. A recent report indicates that human Toll-like receptor 4 (TLR4) is involved in the proinflammatory response to Ni contact dermatitis, and the S100A9 subunit of CP was employed as a marker for leukocyte infiltration at sites of Ni(II) exposure.71 In addition, an abstract of an ongoing clinical study noted that elevated levels of fecal CP were found in patients with systemic Ni allergy syndrome.72 These studies suggest that CP may be involved in the immune response to metal contact allergy. Future work is required to understand Ni(II) regulation and utilization in higher organisms, and our discovery of Ni(II) coordination by CP provides a foundation and motivation for investigating this protein broadly in the context of mammalian nickel homeostasis.
Supplementary Material
Acknowledgments
We gratefully acknowledge the MIT Center for Environmental Health Sciences (NIH Grant P30-ES002109), MIT Center for Environmental Health Sciences Theron Randolph Gift (E.M.N.), the MIT Research Support Committee Wade Award (E.M.N), and the Camille and Henry Dreyfus Foundation (E.M.N.) and NIH grant GM069857 (C.L.D). C.L.D. is a Howard Hughes Medical Institute Investigator. T.G.N. is a recipient of a NSF Graduate Research Fellowship. The crystallographic work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The S. aureus M2 strain was obtained from the Oglesby-Sherrouse Laboratory at the University of Maryland School of Pharmacy. We thank Ms. E. C. Wittenborn, Ms. T. A. J. Grell, and Dr. S. E. J. Bowman for assistance in X-ray diffraction data collection and refinement. We thank Professor A. E. Keating for use of her laboratory’s CD spectrometer. We acknowledge the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for providing the S. aureus USA300 JE2 parent strain as well as the ΔcntA and ΔureC mutant strains of the Nebraska Transposon Mutant Library (NTML) that is supported by NIH NIAID grant HHSN272200700055C.49
Footnotes
Supporting Information
This material is available free of charge via the Internet at http://pubs.acs.org
Complete experimental methods, Tables S1-S12, Figures S1-S12.
References
- 1.Ragsdale SW. J Biol Chem. 2009;284:18571–18575. doi: 10.1074/jbc.R900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Zamble DB. Chem Rev. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- 3.Mulrooney SB, Hausinger RP. FEMS Microbiol Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 4.Brown PH, Welch RM, Cary EE. Plant Physiol. 1987;85:801–803. doi: 10.1104/pp.85.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 6.Pearson MA, Michel LO, Hausinger RP, Karplus PA. Biochemistry. 1997;36:8164–8172. doi: 10.1021/bi970514j. [DOI] [PubMed] [Google Scholar]
- 7.Volbeda A, Garcin E, Piras C, de Lacey AL, Fernandez VM, Hatchikian EC, Frey M, Fontecilla-Camps JC. J Am Chem Soc. 1996;118:12989–12996. [Google Scholar]
- 8.Doukov TI, Iverson TM, Seravalli J, Ragsdale SW, Drennan CL. Science. 2002;298:567–572. doi: 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- 9.Hiron A, Posteraro B, Carrière M, Remy L, Delporte C, La Sorda M, Sanguinetti M, Juillard V, Borezée-Durant E. Mol Microbiol. 2010;77:1246–1260. doi: 10.1111/j.1365-2958.2010.07287.x. [DOI] [PubMed] [Google Scholar]
- 10.Remy L, Carrière M, Derré-Bobillot A, Martini C, Sanguinetti M, Borezée-Durant E. Mol Microbiol. 2013;87:730–743. doi: 10.1111/mmi.12126. [DOI] [PubMed] [Google Scholar]
- 11.Ghssein G, Brutesco C, Ouerdane L, Fojcik C, Izaute A, Wang S, Hajjar C, Lobinski R, Lemaire D, Richaud P, Voulhoux R, Espaillat A, Cava F, Pignol D, Borezée-Durant E, Arnoux P. Science. 2016;352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 12.Konieczna I, Żarnowiec P, Kwinkowski M, Kolesińska B, Frączyk J, Kamiński Z, Kaca W. Curr Protein Pept Sci. 2012;13:789–806. doi: 10.2174/138920312804871094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutherford JC. PLoS Pathog. 2014;10:e1004062. doi: 10.1371/journal.ppat.1004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Reuse H, Vinella D, Cavazza C. Front Cell Infect Microbiol. 2013;3:94. doi: 10.3389/fcimb.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatermann S, John J, Marre R. Infect Immun. 1989;57:110–116. doi: 10.1128/iai.57.1.110-116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedelin H. Int J Antimicrob Agents. 2002;19:484–487. doi: 10.1016/s0924-8579(02)00095-x. [DOI] [PubMed] [Google Scholar]
- 17.Maroncle N, Rich C, Forestier C. Res Microbiol. 2006;157:184–193. doi: 10.1016/j.resmic.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Weinberg ED. J Am Med Assoc. 1975;231:39–41. [Google Scholar]
- 19.Kehl-Fie TE, Skaar EP. Curr Opin Chem Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood MI, Skaar EP. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Ochoa VE, Jellbauer S, Klaus S, Raffatellu M. Front Cell Infect Microbiol. 2014;4:2. doi: 10.3389/fcimb.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerasi M, Ammendola S, Battistoni A. Front Cell Infect Microbiol. 2013;3:108. doi: 10.3389/fcimb.2013.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 24.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 25.Gläser R, Harder J, Lange H, Bartels J, Christophers E, Schroder J-M. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 26.Haley KP, Delgado AG, Piazuelo MB, Mortensen BL, Correa P, Damo SM, Chazin WJ, Skaar EP, Gaddy JA. Infect Immun. 2015;83:2944–2956. doi: 10.1128/IAI.00544-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunden LS, Gaillard A, Nolan EM. Chem Sci. 2016;7:1338–1348. doi: 10.1039/c5sc03655k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohnle PG, Collins-Lech C, Wiessner JH. J Infect Dis. 1991;164:137–142. doi: 10.1093/infdis/164.1.137. [DOI] [PubMed] [Google Scholar]
- 29.Clohessy P, Golden BE. Scand J Immunol. 1995;42:551–556. doi: 10.1111/j.1365-3083.1995.tb03695.x. [DOI] [PubMed] [Google Scholar]
- 30.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 31.Nakashige TG, Zhang B, Krebs C, Nolan EM. Nat Chem Biol. 2015;11:765–771. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro Kim A, Chazin Walter J, Skaar Eric P. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer LD, Skaar EP. Annu Rev Genet. 2016;50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johne B, Fagerhol MK, Lyberg T, Prydz H, Brandtzaeg P, Naess-Andresen CF, Dale I. J Clin Pathol: Mol Pathol. 1997;50:113–123. doi: 10.1136/mp.50.3.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogl T, Leukert N, Barczyk K, Strupat K, Roth J. Biochim Biophys Acta. 2006;1763:1298–1306. doi: 10.1016/j.bbamcr.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 36.Gifford JL, Walsh MP, Vogel HJ. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 37.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korndörfer IP, Brueckner F, Skerra A. J Mol Biol. 2007;370:887–898. doi: 10.1016/j.jmb.2007.04.065. [DOI] [PubMed] [Google Scholar]
- 39.Brophy MB, Hayden JA, Nolan EM. J Am Chem Soc. 2012;134:18089–18100. doi: 10.1021/ja307974e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephan JR, Nolan EM. Chem Sci. 2016;7:1962–1975. doi: 10.1039/c5sc03287c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. Proc Natl Acad Sci US A. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gagnon DM, Brophy MB, Bowman SEJ, Stich TA, Drennan CL, Britt RD, Nolan EM. J Am Chem Soc. 2015;137:3004–3016. doi: 10.1021/ja512204s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayden JA, Brophy MB, Cunden LS, Nolan EM. J Am Chem Soc. 2013;135:775–787. doi: 10.1021/ja3096416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker TM, Nakashige TG, Nolan EM, Neidig ML. Chem Sci. 2017;8:1369–1377. doi: 10.1039/c6sc03487j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brophy MB, Nakashige TG, Gaillard A, Nolan EM. J Am Chem Soc. 2013;135:17804–17817. doi: 10.1021/ja407147d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakashige TG, Stephan JR, Cunden LS, Brophy MB, Wommack AJ, Keegan BC, Shearer JM, Nolan EM. J Am Chem Soc. 2016;138:12243–12251. doi: 10.1021/jacs.6b06845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irving H, Williams RJP. J Chem Soc. 1953;138:3192–3210. [Google Scholar]
- 48.Taylor D, Holland KT. J Appl Bacteriol. 1989;66:319–329. doi: 10.1111/j.1365-2672.1989.tb02485.x. [DOI] [PubMed] [Google Scholar]
- 49.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. MBio. 2013;4:e00537–12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robert-Baldo GL, Morris MJ, Byrne RH. Anal Chem. 1985;57:2564–2567. [Google Scholar]
- 51.Weatherburn MW. Anal Chem. 1967;39:971–974. [Google Scholar]
- 52.Benoit S, Maier RJ. J Bacteriol. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boer JL, Hausinger RP. Nickel-binding sites in proteins. In: Kretsinger RH, Uversky VN, Permyakov EA, editors. Encyclopedia of Metalloproteins. Springer; New York: New York, NY: 2013. pp. 1528–1534. [Google Scholar]
- 54.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Löw C, Neumann P, Tidow H, Weininger U, Haupt C, Friedrich-Epler B, Scholz C, Stubbs MT, Balbach J. J Mol Biol. 2010;398:375–390. doi: 10.1016/j.jmb.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 56.Salgado EN, Lewis RA, Mossin S, Rheingold AL, Tezcan FA. Inorg Chem. 2009;48:2726–2728. doi: 10.1021/ic9001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Håkansson KO. J Mol Biol. 2003;332:1175–1182. doi: 10.1016/j.jmb.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 58.Schreiter ER, Wang SC, Zamble DB, Drennan CL. Proc Natl Acad Sci U S A. 2006;103:13676–13781. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Y, Zhou M, Kirsch F, Xu C, Zhang L, Wang Y, Jiang Z, Wang N, Li J, Eitinger T, Yang M. Cell Res. 2014;24:267–277. doi: 10.1038/cr.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desguin B, Zhang T, Soumillion P, Hols P, Hu J, Hausinger RP. Science. 2015;349:66–69. doi: 10.1126/science.aab2272. [DOI] [PubMed] [Google Scholar]
- 61.Smith RM, Martell AE. Critical Stability Constants Volume 2: Amines. Vol. 2 Plenum Press; New York: 1975. [Google Scholar]
- 62.Smith RM, Martell AE. Critical Stability Constants First Supplement. Vol. 5 Springer Science+Business Media; New York: 1982. [Google Scholar]
- 63.Martell AE, Smith RM. Critical Stability Constants Second Supplement. Vol. 6 Springer Science+Business Media; New York: 1989. [Google Scholar]
- 64.Nriagu JO. The global cycle of nickel. In: Nriagu JO, editor. Nickel in the Environment. John Wiley & Sons; New York: 1980. p. 1. [Google Scholar]
- 65.Herber RFM. Int Arch Occup Environ Health. 1999;72:279–283. doi: 10.1007/s004200050374. [DOI] [PubMed] [Google Scholar]
- 66.Smith LA, Gaya DR. World J Gastroenterol. 2012;18:6782–6789. doi: 10.3748/wjg.v18.i46.6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reyes L, Allam AB, Canales BK, Brown MB. The role of calgranulins in urinary tract infection. In: Nikibakhsh A, editor. Clinical Management of Complicated Urinary Tract Infection. InTech; Rijeka: 2011. pp. 249–266. [Google Scholar]
- 68.Ebbing J, Mathia S, Seibert FS, Pagonas N, Bauer F, Erber B, Günzel K, Kilic E, Kempkensteffen C, Miller K, Bachmann A, Rosenberger C, Zidek W, Westhoff TH. World J Urol. 2014;32:1485–1492. doi: 10.1007/s00345-013-1227-8. [DOI] [PubMed] [Google Scholar]
- 69.Ebbing J, Seibert FS, Pagonas N, Bauer F, Miller K, Kempkensteffen C, Günzel K, Bachmann A, Seifert HH, Rentsch CA, Ardelt P, Wetterauer C, Amico P, Babel N, Westhoff TH. PLoS ONE. 2016;11:e0146395. doi: 10.1371/journal.pone.0146395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benoit SL, Miller EF, Maier RJ. Infect Immun. 2013;81:580–584. doi: 10.1128/IAI.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmidt M, Raghavan B, Muller V, Vogl T, Fejer G, Tchaptchet S, Keck S, Kalis C, Nielsen PJ, Galanos C, Roth J, Skerra A, Martin SF, Freudenberg MA, Goebeler M. Nat Immunol. 2010;11:814–819. doi: 10.1038/ni.1919. [DOI] [PubMed] [Google Scholar]
- 72.Cazzato IA, Vadrucci E, Ainora ME, Gasbarrini G, Gasbarrini A, Minelli M. Gastroenterology. 2011;140:S-283. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


