Abstract
Introduction
Murine placentation requires trophoblast Notch2, while the Notch ligand, JAGGED1, is reduced in invasive trophoblasts from women with preeclampsia. However, the placental cells with active Notch signaling and expression of other Notch proteins and ligands in placentation have yet to be defined. We sought to identify endothelial cell and trophoblast subtypes with canonical Notch signaling in the decidua and placenta and correlate this to expression of Notch proteins and ligands.
Methods
Notch reporter transgenic mice were used to define canonical Notch activity and immunofluorescence staining performed to characterize expression of Notch1, 2, 3, 4 and ligands, Delta-like 4 (Dll4) and Jagged1 (Jag1) during early placentation and in the mature placenta.
Results
Notch signaling is active in maternal and fetal endothelial cells and trophoblasts during early placentation and in the mature placenta. Dll4, Jag1, Notch1, and Notch4 are expressed in maternal vasculature in the decidua. Dll4, Jag1 and Notch1 are expressed in fetal vasculature in the labyrinth. Dll4, Notch2 and Notch4 are co-expressed in the ectoplacental cone. Notch2 and Notch4 are expressed in parietal-trophoblast giant cells and junctional zone trophoblasts with active canonical Notch signaling and in labyrinthine syncytiotrophoblasts and sinusoidal-trophoblast giant cells.
Discussion
Canonical Notch activity and distinct expression patterns for Notch proteins and ligands was evident in endothelium and trophoblasts, suggesting Notch1, Notch2, Notch4, Dll4, and Jag1 have distinct and overlapping functions in placentation. Characterization of Notch signaling defects in existing mouse models of preeclampsia may shed light on the role of Notch in developing the preeclampsia phenotype.
Keywords: Notch, Decidua, Placenta, Trophoblast, Endothelium, Preeclampsia
1. Introduction
In humans and mice, interactions between maternal decidual cells, maternal vascular cells and embryo-derived trophoblasts (TBs) are necessary for post-implantation embryo development and proper placentation. The molecular signaling pathways that mediate these interactions and cellular functions have yet to be fully characterized. Prior to placentation, the decidua contains spiral arteries that carry maternal blood to the implantation site and implantation-induced decidual capillaries. In mice, the pre-placental structure (Fig.1A) consists of embryo-derived trophoblast giant cells (TGCs), the ectoplacental cone (EPC), which contains glycogen trophoblast cells (GlyTCs) and intermediate TBs, and the chorion [1–3]. Spiral artery associated-TGCs (SpA-TGCs) produce angiogenic factors, vasodilators and anticoagulants, as well as intercalate and replace endothelial cells (ECs) of spiral arteries to create a remodeled, chimeric decidual vasculature composed of ECs and TBs. GlyTCs invade interstitially into the decidua. Parietal-TGCs (P-TGCs) line the implantation chamber and EPC and are in direct contact with maternal decidual and immune cells [4–7].
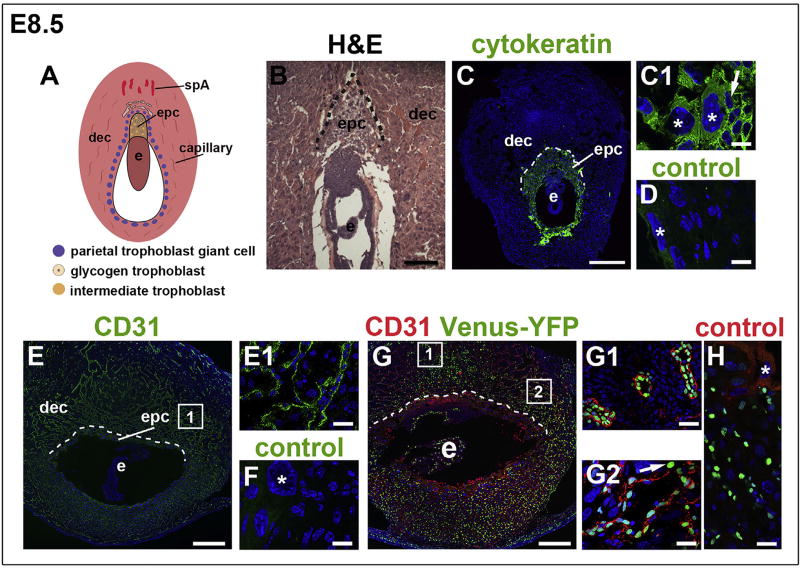
Fig. 1. Notch activity in the decidua at E8.5.
The orientation is the same for all panels; mesometrial at the top and anti-mesometrial at the bottom. (A) Schematic representation of an E8.5 implantation site, highlighting the decidua, maternal blood supply and TB subtypes. (B) H&E highlighting the decidua, EPC and embryo. (C–H) IF was performed on tissue sections and nuclei were stained with DAPI (blue). The dotted line shows the border between the decidua and P-TGC layer. (C) Pan-trophoblast marker, cytokeratin identifies TBs, including P-TGCs (C1, asterisks) and EPC TBs (C1, arrow). (D) Negative control stained with the anti-rabbit secondary used for panels C, C1. Asterisks mark P-TGCs. (E, E1) CD31 identifies ECs of decidual vessels. (F) Negative control stained with the anti-rat secondary used for panels E, E1. Asterisk marks a P-TGC. (G) IF of sections through implantation sites of female CBF:H2B-Venus Notch reporter mice enabled assessment of Notch activity in the decidua. Notch activity (green) was observed in CD31+ ECs (red) of spiral arteries (G1) and decidual capillaries (G2). Notch activity was also observed in non-vascular, decidual cells (G2, arrow). (H) Negative control for the CBF:H2B-Venus Notch reporter stained with the anti-rat secondary used for panels G, G1 and G2. Asterisk marks a P-TGC without Venus-YFP expression. The squares in E and G show the general regions from which the higher magnification areas visualized in E1, G1, and G2 are derived. Dec, decidua; e, embryo; epc, ectoplacental cone; spA, spiral artery. Scale bars B, E, and G = 100 µm. Scale bar C = 500 µm. Scale bars C1, D, E1, F, G1, G2 and H = 25 µm.
The fully developed mouse placenta contains three layers: the maternally-derived decidua basalis, junctional zone and innermost labyrinth. The decidua basalis is primarily comprised of differentiated, decidualized uterine stromal cells and maternal vessels. P-TGCs form the decidua basalis/junctional zone border. The junctional zone, which develops from the EPC, is comprised of many TB cell types, including spongiotrophoblast cells (SpTCs), GlyTCs and TGCs that line arterial and venous maternal blood vessels. Canal-TGCs (C-TGCs) line maternal arterial canals, the coalesced spiral arteries that traverse the junctional zone and deliver maternal blood to the labyrinth for nutrient exchange between mother and fetus. Channel-TGCs (Ch-TGCs) line maternal venous channels that traverse the junctional zone and take blood out of the labyrinth. The labyrinth, which is highly branched with a large surface area for nutrient exchange, is comprised of sinusoidal-TGCs (S-TGCs) that line maternal blood sinusoids and two multinucleated syncytiotrophoblast layers, SynT-I and SynT-II surrounding the endothelium of fetal blood vessels [5,6,8–12]. The process of TB-mediated remodeling of uterine spiral arteries, the presence of fetal blood in EC-lined vessels, and the direct contact of maternal blood with fetal-derived TBs are key features of the placenta that are similar in humans and mice [2,5].
The Notch signaling pathway governs cell fate decisions in many tissues via direct cell-cell contact [13,14]. There are four transmembrane receptors (Notch1, 2, 3, 4) that are activated by four ligands [Delta-like (Dll) 1 and 4 and Jagged (Jag) 1 and 2] [14,15]. Targeted inactivation or overexpression of murine Notch family members results in a variety of vascular defects, commonly causing embryonic lethality [16–21]. In mice, homozygous deletion of Notch2 results in inadequate maternal blood sinus formation in the labyrinth and embryonic lethality before embryonic day (E) 11.5 [22,23]. Conditional deletion of Notch2 in Tpbpa+ TB lineages (SpA-TGCs and GlyTCs) results in decreased size of maternal canals and placental perfusion, suggesting that TBs require Notch2 for proper remodeling of maternal spiral arteries [7,24]. Placentas lacking transcriptional co-repressor Tle3, a downstream effector of Notch signaling that is co-expressed with Notch2 in Ch-TGCs, have small, abnormally shaped venous channels [8]. In humans, expression of JAG1 is reduced in invasive TBs from women with preeclampsia [24] and expression of EGFL7, a modulator of NOTCH signaling, is reduced in preeclamptic placentas [25].
We hypothesized that Notch signaling is active in both ECs and multiple TB subtypes during placentation. We utilized the CBF:H2B-Venus transgenic mouse reporter of canonical Notch signaling activity to identify decidual and placental cell types with canonical Notch activity. Venus-yellow fluorescent protein (YFP) expression provides a quantitative, single-cell resolution read out of Notch signaling [26]. To identify the Notch proteins and ligands that are expressed relative to active Notch signaling during placentation, we characterized protein expression of receptors, Notch1, 2, 3, 4 and angiogenic ligands, Jag1 and Dll4, in ECs and all TB subtypes in the murine decidua and placenta. This analysis lays the foundation for genetic studies that can provide insight into the molecular basis of human placental disorders, such as fetal intrauterine growth restriction and preeclampsia, that are associated with defects in placental vascular development [27,28].
2. Materials and methods
2.1. Animals
Studies were approved by the Columbia University Institutional Animal Care and Use Committee (IACUC #AC-AAAE265). For assessment of Notch signaling activity, we bred CBF:H2B-Venus transgenic virgin females (Jackson Laboratories) [26] with C57BL/6J males (Jackson Laboratories). For assessment of wild type expression patterns, we bred C57BL/6J virgin female mice and C57BL/6J males. Noon on the day a mating plug was observed was designated as E0.5. Uterine implantation sites from pregnant females at E8.5 and placentas from pregnant females at E10.5, E12.5 and E18.5 were analyzed.
2.2. Histology and immunofluorescence
Implantation sites and placenta specimens for histologic examination were fixed in Bouin's solution (Sigma) and paraffin embedded. 10 µm sections were stained with hematoxylin and eosin (H&E). Specimens for immunofluorescence (IF) were fixed in 4% paraformaldehyde at 4 °C, infiltrated with 30% sucrose in PBS, embedded in Tissue-Tek®O.C.T.™ Compound (Sakura Fine Technical Co. Ltd), and cryosectioned at 7 µm. Implantation sites at E8.5, showing inter-embryonic regions and central parts of the decidua were confirmed by H&E staining of every 5th section. Orientation of placenta sections was verified by H&E staining of every 5th section.
IF staining, as previously described [29], was performed at least 3 times and on at least 3 different implantation sites or placentas for analysis of each antibody at each stage. The specificity of Notch protein and ligand primary antibodies was previously determined [30,31]. At all stages, sections were stained with antibodies to Notch1-4, Dll4, Jagged1, and Jagged2. Expression of Jagged2 was inconsistent. Thus, we did not include Jagged2 data. Slides stained with secondary antibody alone served as negative controls for IF staining and are presented in (Figs. 1–3 and Supplementary Fig. 2). Antibodies are listed in the Supplementary Table. Vectashield containing 4′, 6-diamidino-2-phenylindole (DAPI) (Vector) was used for nuclear visualization and mounting.
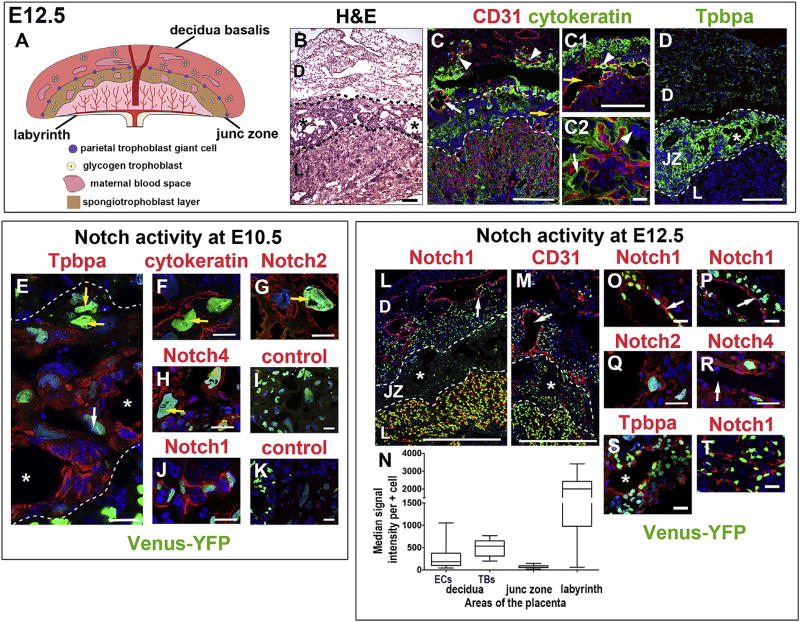
Fig. 3. Notch activity as indicated by Venus-YFP expression in maternal and fetal cells types in the placenta.
(A) Schematic representation of a mature, E12.5 placenta. (B) H&E of a placental section at E12.5 with asterisks in maternal blood spaces. IF was performed on tissue sections and nuclei were stained with DAPI (blue). (C) CD31 identifies maternal ECs in the decidua and fetal ECs in the labyrinth. Cytokeratin identifies TBs in the decidua (white arrow: maternal vessel; arrowheads: TBs surrounding maternal vessels), junctional zone (yellow arrows: maternal canal in C and C1; arrowhead: C-TGC in C1), and labyrinth (C2, arrow: syncytiotrophoblast; arrowhead: S-TGC). In the decidua, chimeric vessels are comprised of CD31+ (red) ECs and cytokeratin+ (green) SpA-TGCs (C, white arrow). (D) Tpbpa identifies SpTCs and GlyTCs around maternal blood spaces (asterisk) in the junctional zone. (E–K) Venus-YFP in placental sections at E10.5 (asterisks are in maternal blood spaces). (E) Notch activity was detected in P-TGCs at the decidua/junctional zone border (yellow arrows), in junctional zone TBs that express Tpbpa (red signal) and in Ch-TGCs (white arrow). Cytokeratin+ P-TGCs with Venus-YFP (F, arrow) express Notch2 (G, arrow) and Notch4 (H, arrow). (I) Negative control stained with the anti-rabbit secondary used for panels E–H. (J) Venus-YFP was detected in ECs of labyrinthine fetal vessels that express Notch1. (K) Negative control stained with the anti-goat secondary used for panel J. (L–T) Venus-YFP in placental sections at E12.5 (asterisks are in maternal blood spaces). Venus-YFP was detected in Notch1+ ECs of maternal decidual vessels (L, O: arrow marks vessel wall) and in TBs surrounding the decidual vessels (M, arrow marks vessel lumen; P, arrow marks vessel wall. Notch2 (Q) and Notch4 (R) expression was detected in decidual TBs with Venus-YFP. Arrow in R marks a vessel lumen. Venus-YFP was detected in junctional zone SpTCs and GlyTCs that express Tpbpa (S) and in ECs of fetal labyrinthine capillaries that express Notch1 (L, T). (N) Notch activity was significantly lower in the junctional zone as compared to labyrinth and decidua. *P < 0.05. Dotted lines separate layers of the placenta. D, decidua; JZ, junctional zone; L, labyrinth. Scale bars B, C, C1, D, L, M = 100 µm. Scale bars C2, E–K and O–T = 25 µm.
2.3. Microscopy
H&E staining was examined with a Nikon MICROPHOT-FXA microscope and images were captured using NIS-Elements D3.10 software. Fluorescent images were captured using a Nikon A1 scanning confocal microscope on an Eclipse Ti microscope stand (Nikon Instruments, Melville, NY). Standard lasers and filters were used. Maximum intensity projections are shown.
2.4. Quantification of Notch intensity
Confocal images of E12.5 placentas expressing the CBF:H2B-Venus transgene were examined with the Fiji package of ImageJ software to measure Notch reporter signal intensity [32]. The intensity of individual positive cells within a measured box was determined in all 3 layers of the placenta. Background signal intensity was subtracted. Median signal intensity per positive cell was compared using Kruskal Wallace (GraphPad Prism 6, Version 6.0a).
3. Results
3.1. Notch activity and expression during early placentation
At E8.5 when placentation begins, P-TGCs line the implantation chamber and EPC (Fig. 1A and B). Cytokeratin identifies TBs and CD31 identifies decidual ECs (Fig. 1C, C1 and Fig. 1E, E1). Notch signaling activity, indicated by Venus-YFP expression, was observed primarily in CD31+ ECs of spiral arteries and capillaries throughout the decidua (Fig. 1G, G1, G2). Notch signaling activity was also observed in non-vascular, decidual cells (Fig. 1G, arrow in G2) but not in P-TGCs or EPC TBs (Fig. 1G and H). These data suggest that Notch signaling functions in the decidual endothelium at the initiation of placentation.
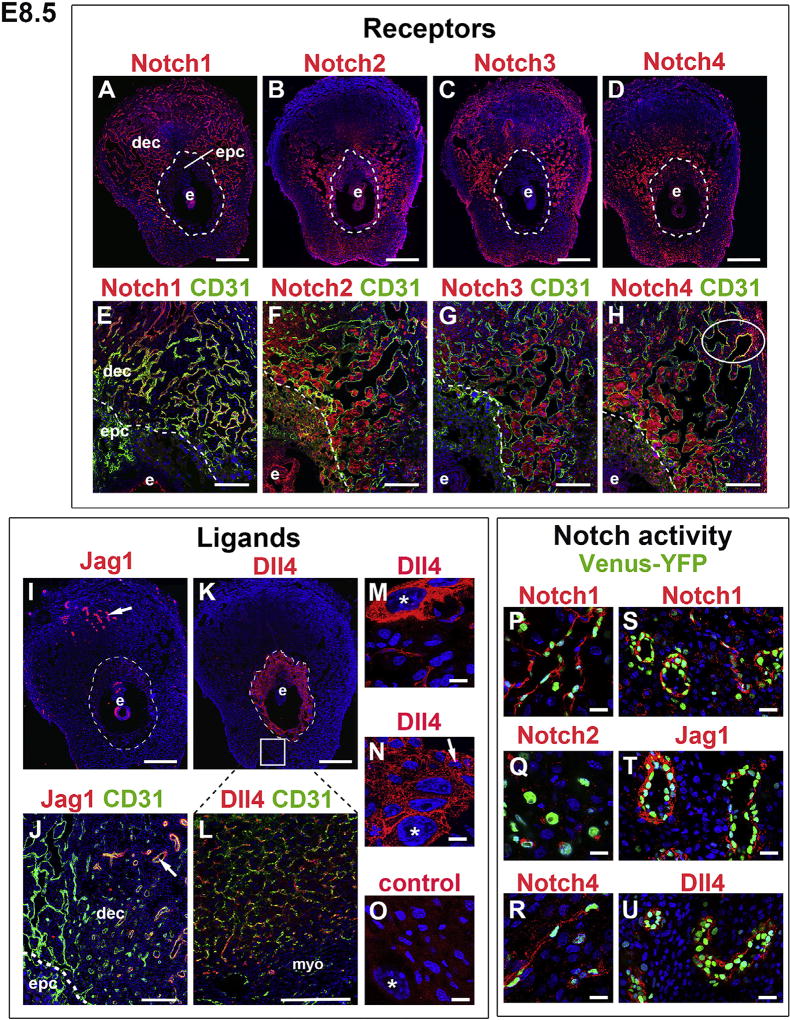
To define the expression of Notch family members at E8.5, sections were co-stained for each Notch protein or ligand and the EC marker, CD31. The adjacent serial section was stained with pantrophoblast marker, cytokeratin (data not shown). Decidual capillary ECs expressed both Notch1 (Fig. 2A and E) and Notch4 (Fig. 2D and H), with Notch4 expression restricted to a subset of peripheral capillary ECs in the decidua (oval in Fig. 2H). Notch2, Notch3, and Notch4 were expressed in decidual cells adjacent to ECs (Fig. 2B–D and 2F-H). Notch2 (Fig. 2B and F) and Notch4 (Fig. 2D and H) were expressed in the EPC and P-TGCs.
Fig. 2. Notch receptors and ligands in the decidua and ectoplacental cone at E8.5.
IF was performed on tissue sections and nuclei were stained with DAPI (blue). The orientation is the same for all panels; mesometrial at the top and anti-mesometrial at the bottom. The dotted line shows the border between the decidua and P-TGC layer. (E–H) Co-staining with CD31 (green) shows Notch expression with respect to ECs. (A, E) Notch1 was expressed in ECs of decidual vessels. (B, F) Notch2 was expressed in decidual cells, P-TGCs and EPC TBs. (C, G) Notch3 was expressed in decidual cells. (D, H) Notch4 was expressed in decidual cells, P-TGCs and EPC TBs. Notch4 was also expressed in ECs of decidual capillaries (oval in H). (I) Jag1 was expressed in spiral arteries (arrow). (J) Co-staining for CD31 (green) and Jag1 (red) showed Jag1 expression in spiral artery ECs (arrow, yellow signal) and perivascular cells (arrow, red signal). Dll4 was expressed in P-TGCs (K, asterisks in M and N) and TBs in the EPC (K, arrow in N). Dll4 was also expressed in capillary ECs in the anti-mesometrial decidua. The rectangle in K shows the general region from which the higher magnification area visualized in L is derived. (O) Negative control stained with the anti-goat secondary used for panels I–N. Asterisk marks a P-TGC. (P–U) Sections through E8.5 implantation sites of female CBF:H2B-Venus Notch reporter mice to correlate Notch protein expression with Notch activity in the decidua. Notch1 (P) and Notch4 (R) expression was detected in decidual capillaries with Notch activity. Notch2 was detected in decidual cells with Notch activity (Q). Spiral arteries with Notch activity express Notch1 (S), Jag1 (T) and Dll4 (U). Dec, decidua; e, embryo; epc, ectoplacental cone; myo, myometrium. Scale bars in A–D, I, K = 500 µm. Scale bars in E–H, K, L = 100 µm. Scale bars in M–U = 25 µm.
Notch ligands, Jag1 and Dll4 were expressed in ECs and TBs (Fig. 2I–N). Jag1 was expressed in spiral arteries (Fig. 2I and J; arrows). Co-staining with CD31 demonstrated that Jag1 was expressed in spiral artery ECs (Fig. 2J, yellow signal) and perivascular cells (Fig. 2J, red signal). Dll4 expression was observed in P-TGCs, EPC TBs and ECs lining the capillaries in the anti-mesometrial decidua (Fig. 2K–N).
We determined the expression of Notch proteins and ligands, Jag1 and Dll4, in CBF:H2B-Venus mice at E8.5 (Fig. 2P–U). Notch1 and Notch4 were co-expressed with Venus-YFP in decidual capillaries and Notch2 was co-expressed with Venus-YFP in decidual cells (Fig. 2P–R). Notch1, Jag1, and Dll4 were expressed in spiral arteries with Notch signaling activity (Fig. 2S–U). In summary, Notch1, Notch4, Dll4 are expressed in uterine endothelium and Jag1 is expressed in both endothelial and perivascular cells of the uterine vasculature at the initiation of placentation, suggesting these Notch proteins have a role in EC Notch signaling in the E8.5 decidua (Table 1).
Table 1.
Notch activity and expression of ligands and receptors at E8.5
| Maternal | Fetal | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Decidua | Capillaries | Spiral arteries | EPC | P-TGCs | |
| CBF:H2B-Venus | + | + | + | ||
| Notch1 | + | + | |||
| Notch2 | + | + | + | ||
| Notch3 | + | ||||
| Notch4 | + | + | + | + | |
| Dll4 | + | + | + | ||
| Jag1 | + | ||||
EPC: ectoplacental cone; P-TGCs: parietal-trophoblast giant cells.
3.2. Notch activity in endothelial cells and trophoblasts in the mature placenta
The three placental layers, decidua basalis, junctional zone, and labyrinth, are established by E10.5 [33] and well developed by E12.5, with all TB subtypes present (Fig. 3A and B). We stained placental sections with EC (CD31) and TB markers (cytokeratin, Tpbpa) (Fig. 3). CD31 identifies ECs of maternal vessels in the decidua (Fig. 3C) and fetal vessels in the labyrinth (Fig. 3C, C2). Cytokeratin labels all TB subtypes in the decidua, junctional zone and labyrinth (Fig. 3C, C1, C2), including C-TGCs (arrowhead in C1), labyrinthine syncytiotrophoblasts (Fig. 3C, C2, arrow) and S-TGCs (arrowhead in C2). Tpbpa labels junctional zone SpTCs and GlyTCs (Fig. 3D) [8]. In the decidua, cytokeratin+ TBs surround maternal vessels (Fig. 3C, arrowheads). At the decidua/junctional zone interface, we found remodeled chimeric vessels comprised of CD31+ ECs and cytokeratin+ TBs, resulting from SpA-TGC invasion of spiral arteries (Fig. 3C, white arrow) and a maternal canal (Fig. 3C, C1, yellow arrows) lined by C-TGCs traversing the junctional zone.
To evaluate Notch signaling, placental sections from CBF:H2B-Venus mice at E10.5, E12.5 and E18.5 were stained with antibodies that identify ECs (CD31), TBs (Tpbpa and cytokeratin) and Notch proteins (Fig. 3E–T and Supplementary Fig. 1). At E10.5, Venus-YFP was detected in P-TGCs at the decidua/junctional zone border (Fig. 3E, yellow arrows), in junctional zone TBs that express Tpbpa and in Ch-TGCs (Fig. 3E, white arrow). Cytokeratin+ P-TGCs with Notch activity also express Notch2 and Notch4 (Fig. 3F–H). Notch activity was detected in ECs of labyrinthine fetal vessels that express Notch1 (Fig. 3J). At E12.5, Notch signaling was observed in ECs of maternal vessels that express Notch1 (Fig. 3L, O) and in invasive TB cells (Fig. 3M, P) that express Notch2 (Fig. 3Q) and Notch4 (Fig. 3R). Notch activity was detected in Tpbpa+ and Tpbpa− TBs in the junctional zone (Fig. 3S, asterisk in maternal blood space) and in ECs of labyrinthine fetal vessels that express Notch1 and Dll4 (Fig. 3L, T and Supplementary Fig. 2). Notch signaling was not detected in labyrinthine syncytiotrophoblasts or S-TGCs at E12.5.
The intensity of Venus-YFP in the different placental layers was variable (Fig. 3L, M). As the CBF:H2B-Venus strain provides a quantitative, readout of Notch signaling [26], Venus-YFP intensity was quantified to determine levels of Notch activity. Median signal intensity per positive cell was significantly lower in junctional zone TBs as compared to that in decidual ECs, decidual TBs or fetal ECs in the labyrinth (P < 0.05) (Fig. 3N).
3.3. Notch expression in endothelial cells and trophoblasts in the mature placenta
To characterize the expression of Notch family members in the mature placenta, we co-stained placental sections with antibodies against Notch proteins and CD31 (ECs), Muc1 (S-TGCs), Mct1 (Syn-TI) or Mct4 (SynT-II) and stained serial sections for Notch protein, cytokeratin or Tpbpa (Fig. 4, Table 2). We found that Notch1 and Notch ligands, Dll4 and Jag1 were expressed in CD31+ ECs of maternal vessels in the decidua (Fig. 4A–C, arrows). Notch2 and Notch4 were expressed in non-EC, non-TB decidual cells (Fig. 4D–E).
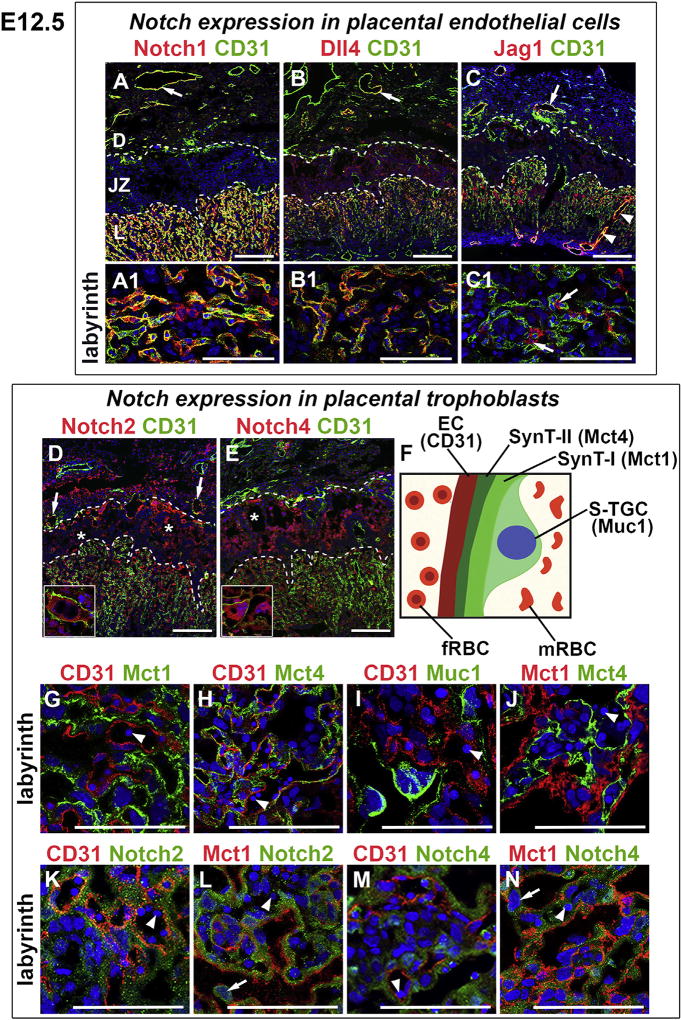
Fig. 4. Notch receptors and ligands in the E12.5 placenta.
IF was performed on tissue sections and nuclei were stained with DAPI (blue). (A–E) CD31 stains ECs of maternal vessels in the decidua and ECs of fetal vessels in the labyrinth. (A, A1) Notch1 was expressed in ECs of maternal vessels (arrow) and fetal labyrinthine vessels (A1). (B, B1) Dll4 was expressed in CD31+ ECs of maternal vessels (arrow) and fetal labyrinthine vessels. (C, C1) Jag1 (red) was expressed in CD31+ ECs of maternal vessels (arrow); in ECs (yellow signal) and perivascular cells of maternal canals (arrowheads); and in labyrinthine perivascular cells (arrows in C1). (D, E) Notch2 (D) and Notch4 (E) were expressed in decidual cells; P-TGCs (insets); and in SpTCs and GlyTCs surrounding maternal blood spaces (asterisks). Notch2 was expressed in TBs of chimeric maternal vessels (remodeled spiral arteries) at the decidua/junctional interface (arrows in D). (F) Schematic of the labyrinthine maternal-fetal interface with the markers used to identify cell types. (G–J) CD31 stains ECs, Mct1 identifies synctiotrophoblast layer SynT-I, Mct4 identifies synctiotrophoblast layer SynT-II, and Muc1 identifies S-TGCs in the labyrinth. Notch2 (K, L) and Notch4 (M, N) were not co-expressed with CD31 or Mct1. Notch2 and Notch4 were expressed in SynT-II, the synctiotrophoblast layer directly adjacent to fetal vessels, which contain nucleated red blood cells (arrowheads). Notch2 (L) and Notch4 (N) were expressed in S-TGCs (arrows). Dotted lines separate the layers of the placenta. fRBC, fetal red blood cell; mRBC, maternal red blood cell, D, decidua; JZ, junctional zone; L, labyrinth. All scale bars = 100 µm.
Table 2.
Notch activity and expression of ligands and receptors in the placenta at E12.5.
| Decidua | Junctional zone | Labyrinth | |
|---|---|---|---|
| CBF:H2B-Venusa | Maternal ECs, TBs | P-TGCs, GlyTCs, SpTCs, Ch-TGCs | Fetal ECs |
| Notch1 | Maternal ECs | nd | Fetal ECs |
| Notch2 | Decidual cells, TBs | P-TGCs, GlyTCs, SpTCs | SynT-II, S-TGCs |
| Notch4 | Maternal ECs,TBs | P-TGCs, GlyTCs, SpTCs | SynT-II, S-TGCs |
| Dll4 | Maternal ECs | nd | Fetal ECs |
| Jag1 | Maternal ECs | nd | Perivascular cells, Maternal canals |
nd: not detected; ECs: endothelial cells; Ch-TGCs: Channel-trophoblast giant cells; GlyTCs: glycogen trophoblast cells; P-TGCs: parietal-trophoblast giant cells; S-TGCs: sinusoidal-trophoblast giant cells; SpTCs: spongiotrophoblasts; SynT-II: syncytiotrophoblast layer II; TBs: trophoblasts.
Venus expression was detected in the same cell types at E10.5, E12.5 and E18.5.
Remodeled chimeric maternal vessels at the decidua/junctional zone interface were comprised of CD31+ ECs and Notch2+ TBs (Fig. 4D, arrows). P-TGCs at the decidua/junctional zone interface and Tpbpa+ SpTCs and GlyTCs surrounding maternal blood spaces in the junctional zone expressed Notch2 and Notch4 (Fig. 4D and E, respectively; insets show P-TGCs; asterisks are in maternal blood spaces). The junctional zone is analogous to the maternal-fetal interface in the human placenta. Thus, our data suggest that Notch2 and Notch4 have a role in EC-TB communications required to ensure proper maternal blood flow to the placenta.
In the labyrinth, ECs (CD31) line fetal vessels and TB subtypes, S-TGCs (Muc1), SynT-I (Mct1) and SynT-II (Mct4) line maternal blood spaces (Fig. 4F, 4G–J). Notch1 (Fig. 4A, A1) and ligand Dll4 (Fig. 4B, B1) were expressed in CD31+ ECs of fetal vessels. Jag1 was expressed in perivascular cells associated with CD31+ fetal ECs (Fig. 4C, C1 arrows); and in ECs and perivascular cells of maternal arterial canals formed from the convergence of spiral arteries (Fig. 4C, arrowheads). Notch2 (Fig. 4K, L) and Notch4 (Fig. 4M, N) were expressed in syncytiotrophoblast layer, SynT-II and S-TGCs (Fig. 4L, N, arrows), suggesting a role in differentiation of TB subtypes that control maternal blood flow in the labyrinth.
In summary, we detected active Notch signaling in all three layers of the mature placenta, with highest levels of activity in invasive decidual TBs and in the decidual and fetal labyrinthine endothelium. Our data suggest that Notch2 and Notch4 mediate canonical Notch signaling in P-TGCs at the decidua/junctional zone interface and in junctional zone TBs. S-TGCs and syncytiotrophoblasts expressed Notch2 and Notch4, but did not display canonical Notch signaling.
4. Discussion
We have previously shown that Notch signaling is active in ECs and pericytes in the uterine decidua prior to placentation, supporting a role for Notch in successful embryo implantation [31]. The present analysis of canonical Notch signaling correlated with expression of Notch proteins in ECs and TB subtypes advances our understanding of the possible roles of Notch signaling in placental vascular development and function. We found that Notch signaling is active in maternal and fetal ECs and many TB subtypes, with distinct Notch protein and ligand expression in the decidua, developing placenta (Table 1) and mature placenta (Table 2). Our findings extend and corroborate previous analyses that focused on Notch mRNA expression [34] and Notch signaling [8,24] in TB subtypes.
Based on these studies, we postulate that unique ligand-receptor pairs mediate Notch signaling in placental capillary formation and vessel remodeling. Our analysis suggests that Dll4 activates Notch1 and Notch4 in decidual capillaries, whereas both Dll4 and Jag1 activate Notch1 in spiral arteries. Expression of Notch2 in TBs of remodeled, chimeric vessels suggests activation of Notch2 in TBs by endothelial Dll4 and/or Jag1. Dll4 is predominantly expressed in vascular endothelium, whereas Jag1 is expressed in both endothelial and perivascular cells [35,36]. We found similar Dll4/Jag1 expression patterns in the decidual and placental vasculature. As Jag1 can act downstream of Dll4/Notch signaling with antagonistic or synergistic effects [37], Jag1 in the same Dll4 expressing cell could inhibit or potentiate Notch signaling.
Multiple TB subtypes use Notch signaling during early placentation [24] and in the mature placenta (Table 2). Previous reports using TNR Notch reporter mice show Notch activity in SpA-TGCs and GlyTCs at E12.5 [24]. We found overlapping expression domains for Dll4, Notch2 and Notch4 in P-TGCs around the implantation chamber and EPC TBs. Although Notch signaling activity was lowest in junctional zone TBs, Notch2 and Notch4 expression was readily observed in these cells. Therefore, Notch4 may mediate formation of maternal canals when Notch2 is deleted in invasive TBs [24]. These data implicate Dll4-mediated activation of Notch2/Notch4 in TB subtype specification and development and TB-mediated formation of maternal arterial canals and venous channels.
Taken together, these data point to a requirement for Notch activity in both ECs and TBs, primary cell types involved in vascular remodeling during placentation. This study forms the basis for developing additional cell type-specific Notch loss-of-function and gain-of-function models to better understand the requirements for Notch in placentation. There are key similarities in murine and human placentation that allow genetic studies in mice to provide insights into the molecular basis of human placenta disorders. TGCs mediate uterine vascular invasion in mice and invasive extravillous cytotrophoblasts mediate uterine vascular invasion in humans. The murine labyrinth is analogous in function to the chorionic villi of the human placenta. We propose that characterizing Notch signaling defects in existing mouse models of preeclampsia may shed light on the role of Notch in developing the preeclampsia phenotype as well as serve as a basis to create Notch-based mouse models of human placentopathies.
Supplementary Material
Acknowledgments
Research was funded by NIH/NICHD grant 5R37 HD033082 (V.E.P.), NIH/NHLBI grant 1R01HL112626 (J.KK.), the Robert Wood Johnson Foundation/Amos Medical Faculty Development Program (N.C.D.) and NIH/NHLBI grant 1R01HL127013-01A1 (N.C.D.). Images were collected in the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by NIH grant P30 CA013696. The confocal microscope was purchased with NIH grant S10 RR025686. The authors thank Theresa Swayne, Ph.D. and Emilia Laura Munteanu, Ph.D. for their technical assistance with confocal microscopy.
Abbreviations
- Dll4
Delta-like4
- Jag1
Jagged1
- ECs
Endothelial cells
- TB
Trophoblast
- EPC
Ectoplacental cone
- TGCs
Trophoblast giant cells
- C-TGC
Canal-TGC
- Ch-TGC
Channel-TGC
- P-TGC
Parietal-TGC
- S-TGC
Sinusoidal-TGC
- SpA-TGC
Spiral artery associated-TGC
- GlyTCs
Glycogen trophoblast cells
- SpTCs
Spongiotrophoblast cells
- SynT-I
Syncytiotrophoblast layer I
- SynT-II
Syncytiotrophoblast layer II
Footnotes
Conflict of interest
No conflicts of interest exist for any of the authors.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.placenta.2017.04.014.
References
- 1.Tesser RB, Scherholz PL, do Nascimento L, Katz SG. Trophoblast glycogen cells differentiate early in the mouse ectoplacental cone: putative role during placentation. Histochem Cell Biol. 2010;134(1):83–92. doi: 10.1007/s00418-010-0714-x. [DOI] [PubMed] [Google Scholar]
- 2.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2001;2(7):538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 3.Hemberger M, Cross JC. Genes governing placental development. Trends Endocrinol. Metab. 2001;12(4):162–168. [Google Scholar]
- 4.Hemberger M, Nozaki T, Masutani M, Cross JC. Differential expression of angiogenic and vasodilatory factors by invasive trophoblast giant cells depending on depth of invasion. Dev. Dyn. 2003;227(2):185–191. doi: 10.1002/dvdy.10291. [DOI] [PubMed] [Google Scholar]
- 5.Rai A, Cross JC. Development of the hemochorial maternal vascular spaces in the placenta through endothelial and vasculogenic mimicry. Dev. Biol. 2014;387(2):131–141. doi: 10.1016/j.ydbio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Hu D, Cross JC. Development and function of trophoblast giant cells in the rodent placenta. Int. J. Dev. Biol. 2010;54(2–3):341–354. doi: 10.1387/ijdb.082768dh. [DOI] [PubMed] [Google Scholar]
- 7.Hu D, Cross JC. Ablation of Tpbpa-positive trophoblast precursors leads to defects in maternal spiral artery remodeling in the mouse placenta. Dev. Biol. 2011;358(1):231–239. doi: 10.1016/j.ydbio.2011.07.036. [DOI] [PubMed] [Google Scholar]
- 8.Gasperowicz M, Surmann-Schmitt C, Hamada Y, Otto F, Cross JC. The transcriptional co-repressor TLE3 regulates development of trophoblast giant cells lining maternal blood spaces in the mouse placenta. Dev. Biol. 2013;382(1):1–14. doi: 10.1016/j.ydbio.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 2007;304(2):567–578. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Cross JC, Hemberger M, Lu Y, Nozaki T, Whiteley K, Masutani M, Adamson SL. Trophoblast functions, angiogenesis and remodeling of the maternal vasculature in the placenta. Mol. Cell Endocrinol. 2002;187(1–2):207–212. doi: 10.1016/s0303-7207(01)00703-1. [DOI] [PubMed] [Google Scholar]
- 11.Simmons DG, Natale DR, Begay V, Hughes M, Leutz A, Cross JC. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development. 2008;135(12):2083–2091. doi: 10.1242/dev.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai A, Takebe K, Nio-Kobayashi J, Takahashi-Iwanaga H, Iwanaga T. Cellular expression of the monocarboxylate transporter (MCT) family in the placenta of mice. Placenta. 2010;31(2):126–133. doi: 10.1016/j.placenta.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 14.Kofler NM, Shawber CJ, Kangsamaksin T, Reed HO, Galatioto J, Kitajewski J. Notch signaling in developmental and tumor angiogenesis. Genes Cancer. 2011;2(12):1106–1116. doi: 10.1177/1947601911423030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shawber CJ, Kitajewski J. Notch function in the vasculature: insights from zebrafish, mouse and man. Bioessays. 2004;26(3):225–234. doi: 10.1002/bies.20004. [DOI] [PubMed] [Google Scholar]
- 16.Uyttendaele H, Ho J, Rossant J, Kitajewski J. Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium. Proc. Natl. Acad. Sci. U. S. A. 2001;98(10):5643–5648. doi: 10.1073/pnas.091584598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK. Essential role of endothelial Notch1 in angiogenesis. Circulation. 2005;111(14):1826–1832. doi: 10.1161/01.CIR.0000160870.93058.DD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs LT, Starling C, Chervonsky AV, Gridley T. Notch1 activation in mice causes arteriovenous malformations phenocopied by ephrinB2 and EphB4 mutants. Genesis. 2010;48(3):146–150. doi: 10.1002/dvg.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa L, Henrique D, Rossant J. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18(20):2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128(4):491–502. doi: 10.1242/dev.128.4.491. [DOI] [PubMed] [Google Scholar]
- 21.McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44(1):29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- 22.Hamada Y, Hiroe T, Suzuki Y, Oda M, Tsujimoto Y, Coleman JR, Tanaka S. Notch2 is required for formation of the placental circulatory system, but not for cell-type specification in the developing mouse placenta. Differentiation. 2007;75(3):268–278. doi: 10.1111/j.1432-0436.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- 23.Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126(15):3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- 24.Hunkapiller NM, Gasperowicz M, Kapidzic M, Plaks V, Maltepe E, Kitajewski J, Cross JC, Fisher SJ. A role for Notch signaling in trophoblast endovascular invasion and in the pathogenesis of pre-eclampsia. Development. 2011;138(14):2987–2998. doi: 10.1242/dev.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacko LA, Massimiani M, Sones JL, Hurtado R, Salvi S, Ferrazzani S, Davisson RL, Campagnolo L, Stuhlmann H. Novel expression of EGFL7 in placental trophoblast and endothelial cells and its implication in preeclampsia. Mech. Dev. 2014;133:163–176. doi: 10.1016/j.mod.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowotschin S, Xenopoulos P, Schrode N, Hadjantonakis AK. A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Dev. Biol. 2013;13:15. doi: 10.1186/1471-213X-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 2012;18(12):1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher SJ. The placental problem: linking abnormal cytotrophoblast differentiation to the maternal symptoms of preeclampsia. Reprod. Biol. Endocrinol. 2004;2:53. doi: 10.1186/1477-7827-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vorontchikhina MA, Zimmermann RC, Shawber CJ, Tang H, Kitajewski J. Unique patterns of Notch1, Notch4 and Jagged1 expression in ovarian vessels during folliculogenesis and corpus luteum formation. Gene Expr. Patterns. 2005;5(5):701–709. doi: 10.1016/j.modgep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Murtomaki A, Uh MK, Choi YK, Kitajewski C, Borisenko V, Kitajewski J, Shawber CJ. Notch1 functions as a negative regulator of lymphatic endothelial cell differentiation in the venous endothelium. Development. 2013;140(11):2365–2376. doi: 10.1242/dev.083865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shawber CJ, Lin L, Gnarra M, Sauer MV, Papaioannou VE, Kitajewski JK, Douglas NC. Vascular Notch proteins and Notch signaling in the periimplantation mouse uterus. Vasc. Cell. 2015;7:9. doi: 10.1186/s13221-015-0034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adamson SL, Lu Y, Whiteley KJ, Holmyard D, Hemberger M, Pfarrer C, Cross JC. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev. Biol. 2002;250(2):358–373. doi: 10.1016/s0012-1606(02)90773-6. [DOI] [PubMed] [Google Scholar]
- 34.Gasperowicz M, Rai A, Cross JC. Spatiotemporal expression of Notch receptors and ligands in developing mouse placenta. Gene Expr. Patterns. 2013;13(7):249–254. doi: 10.1016/j.gep.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Shutter JR, Scully S, Fan W, Richards WG, Kitajewski J, Deblandre GA, Kintner CR, Stark KL. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14(11):1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 36.Villa N, Walker L, Lindsell CE, Gasson J, Iruela-Arispe ML, Weinmaster G. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech. Dev. 2001;108(1–2):161–164. doi: 10.1016/s0925-4773(01)00469-5. [DOI] [PubMed] [Google Scholar]
- 37.Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137(6):1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.