SUMMARY
Targeting immune checkpoints such as the one mediated by programmed cell death protein 1 (PD-1) and its ligand PD-L1 have been approved for treating human cancers with durable clinical benefit 1,2. However, many cancer patients fail to respond to anti-PD-1/PD-L1 treatment, and the underlying mechanism(s) is not well understood 3–5. Recent studies revealed that response to PD-1/PD-L1 blockade might correlate with PD-L1 expression levels in tumor cells 6,7. Hence, it is important to mechanistically understand the pathways controlling PD-L1 protein expression and stability, which can offer a molecular basis to improve the clinical response rate and efficacy of PD-1/PD-L1 blockade in cancer patients. Here, we report that PD-L1 protein abundance is regulated by cyclin D-CDK4 and the Cullin 3SPOP E3 ligase via proteasome-mediated degradation. Inhibition of CDK4/6 in vivo elevates PD-L1 protein levels largely through inhibiting cyclin D-CDK4-mediated phosphorylation of SPOP, thereby promoting SPOP degradation by APC/CCdh1. Loss-of-function mutations in SPOP compromise ubiquitination-mediated PD-L1 degradation, leading to increased PD-L1 levels and reduced numbers of tumor-infiltrating lymphocytes (TILs) in mouse tumors and in primary human prostate cancer specimens. Notably, combining CDK4/6 inhibitor treatment with anti-PD-1 immunotherapy enhances tumor regression and dramatically improves overall survival rates in mouse tumor models. Our study uncovers a novel molecular mechanism for regulating PD-L1 protein stability by a cell cycle kinase and reveals the potential for using combination treatment with CDK4/6 inhibitors and PD-1/PD-L1 immune checkpoint blockade to enhance therapeutic efficacy for human cancers.
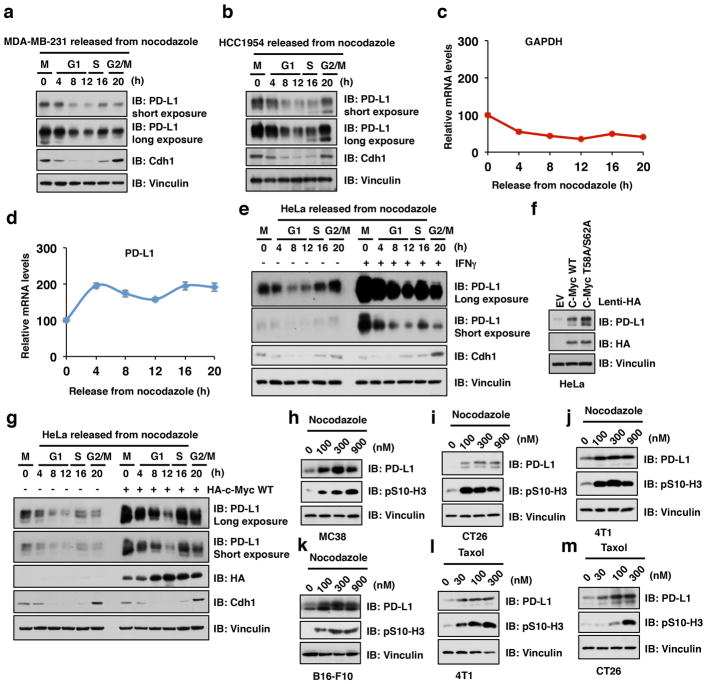
Deregulated cell cycle progression is a hallmark of human cancer, and targeting cyclin-dependent kinases (CDKs) to block cell proliferation has been validated as an effective anti-cancer therapy 8. Although it has been reported that PD-L1 expression can be regulated at both transcriptional 9,10 and post-translational levels 11,12, it remains unclear whether PD-L1 stability is regulated under physiological conditions such as during cell cycle progression. We found that PD-L1 protein abundance fluctuated during cell cycle in multiple human cancer cell lines, peaking in M/early G1 phases, followed by a sharp reduction in late G1/S phases (Fig. 1a–d; Extended Data Fig. 1a–g). Elevated PD-L1 protein abundance was also observed in multiple mouse tumor-derived cell lines arrested in M phase by nocodazole or taxol 13 (Extended Data Fig. 1h–m).
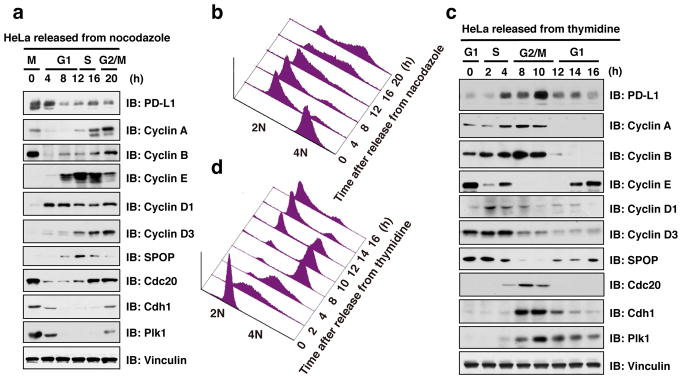
Figure 1. The protein abundance of PD-L1 fluctuates during cell cycle progression.
a, c, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from HeLa cells synchronized in M phase by nocodazole (a) or in late G1/S phase by double thymidine (b) following by releasing back into the cell cycle.
b, d, The cell-cycle profiles in (a) or (c) were monitored by fluorescence-activated cell sorting (FACS).
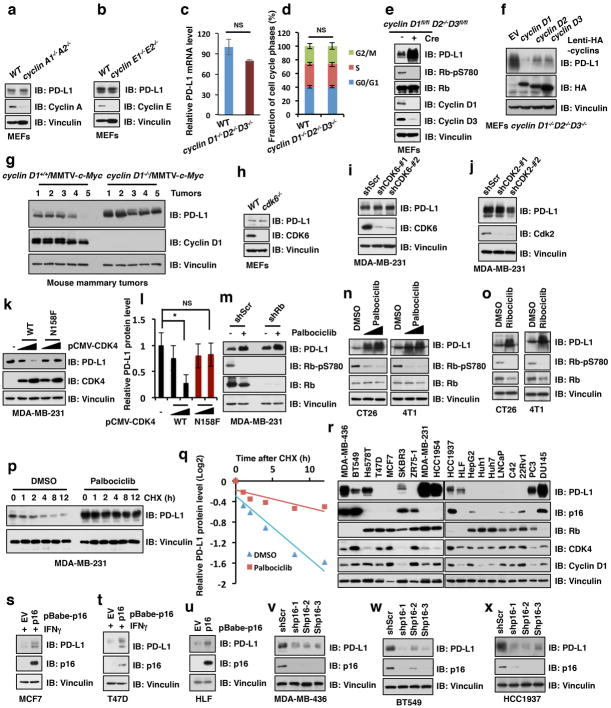
Cyclin-dependent kinases play crucial roles in regulating the stability of cell cycle-related proteins during cell cycle progression 14,15. Therefore, we adopted a genetic method to ablate each major cyclin and found that ablating all three D-type cyclins (D1, D2 and D3), but not cyclin A (A1 and A2) nor cyclin E (E1 and E2), strongly elevated PD-L1 protein abundance in mouse embryonic fibroblasts (MEFs) (Fig. 2a and Extended Data Fig. 2a–e). Using MEFs lacking individual D-type cyclins, we observed that depletion of cyclin D1, and to a lesser extent cyclin D2 or D3, upregulated PD-L1 protein levels (Fig. 2b, c). Conversely, reintroduction of cyclin D1, and to a lesser extent cyclin D2 or D3, suppressed PD-L1 protein abundance in cyclin D1−/−D2−/−D3−/− MEFs (Extended Data Fig. 2f). In further support of a physiological role for cyclin D1 in negatively regulating PD-L1 protein level in vivo, mammary tumors arising in cyclin D1−/− MMTV-Wnt-1 or MMTV-c-Myc mice displayed elevated PD-L1 protein levels, as compared to tumors arising in cyclin D1+/+ animals (Fig. 2d and Extended Data Fig. 2g).
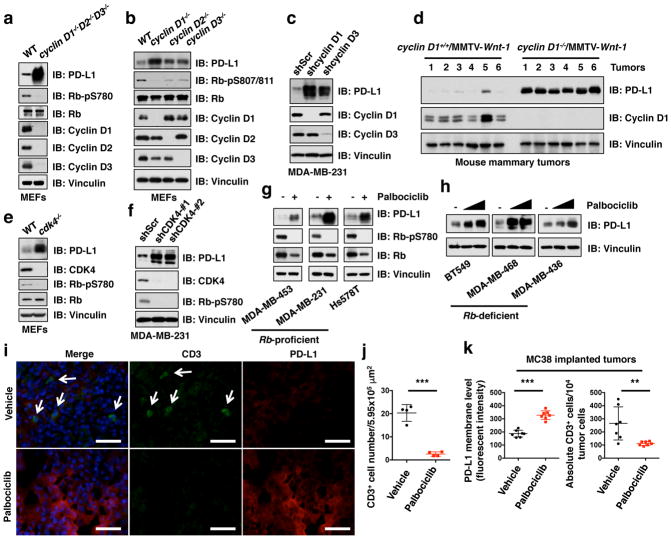
Figure 2. Cyclin D-CDK4 negatively regulates PD-L1 protein stability.
a–d, IB analysis of WCL derived from wild type versus combinational (cyclin D1−/−D2−/−D3−/−) (a) or single isoform cyclin D knockout MEFs (b), MDA-MB-231 cells depleted cyclin D1 or cyclin D3 using shRNAs (c), or MMTV-Wnt1 induced mouse mammary tumors with/without genetic depletion of cyclin D1 (d).
e–h, IB analysis of WCL derived from wild type versus cdk4−/− MEFs (e), MDA-MB-231 cells depleted CDK4 using shRNAs (f), or multiple breast cancer cell lines treated with palbociclib (0.5, 1 μM) for 48 hours (g, h).
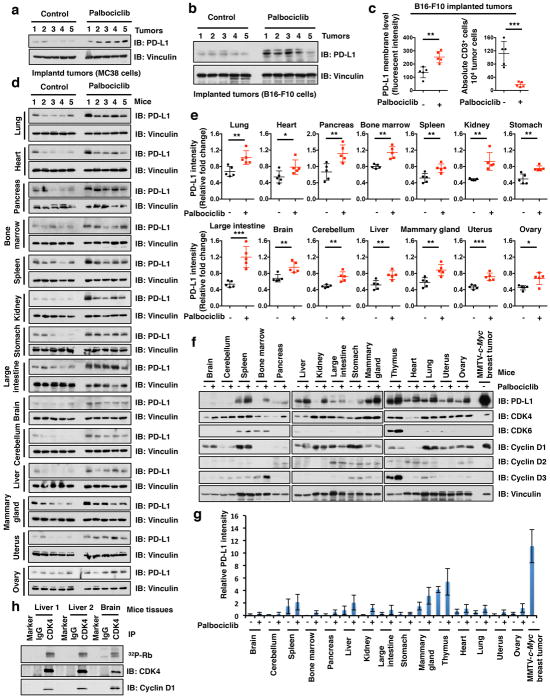
i, j, Immunofluorescence staining of PD-L1 and CD3 in mouse mammary tumors induced by MMTV-ErbB2 treated with vehicle or palbociclib as described in Method (i) and the quantification of CD3+ T cell population (j). The scale bar: 50 μm.
k, FACS analysis for PD-L1 or CD3+ T-cell populations from MC38 implanted tumors treated with vehicle or palbociclib for 7 days. Vehicle, n = 4 for (i, j) or 7 mice for (k); palbociclib, n = 4 for (i, j) or 7 mice for (k). Error bars, ± s.d., two-tailed t-test, **P < 0.01, ***P < 0.001 (two-tailed t-test).
Depletion of cyclin D catalytic partner, the cyclin-dependent kinase 4 (CDK4) 16, but not CDK6 16 nor the cyclin A and cyclin E binding-partner, CDK2 17, also increased PD-L1 protein abundance in cells (Fig. 2e, f; Extended Data Fig. 2h–j). Conversely, ectopic expression of wild-type CDK4, but not kinase-dead N158F mutant, decreased PD-L1 levels (Extended Data Fig. 2k, l). Furthermore, treatment of multiple cancer cell lines with two different selective inhibitors of CDK4/6 kinase, palbociclib or ribociclib 8, upregulated PD-L1 protein abundance and stability even in pRB knock-down cells (Fig. 2g, h; Extended Data Fig. 2m–q).
Rb is frequently inactivated in human cancers 18,19. In agreement with previous reports 20,21, we found that Rb-deficient cancer cells often displayed high levels of cyclin D-CDK4/6 inhibitor, p16INK4a. Consistent with the notion that cyclin D1-CDK4 kinase suppresses PD-L1 levels, we observed that upregulation of 16INK4 correlated with elevated PD-L1 levels. Moreover, in Rb-proficient/p16-low cancer cell lines, higher PD-L1 levels correlated with relatively low CDK4 expression (Extended Data Fig. 2r). In addition, ectopic expression of p16INK4 in Rb-proficient/p16-low cell lines (MCF7 and T47D) or Rb-deficient/p16-low cell line (HLF) elevated PD-L1 protein abundance (Extended Data Fig. 2s–u), while depletion of p16INK4 in Rb-deficient/p16-high cell lines (MDA-MB-436, BT549, and HCC1937) had an opposite effect (Extended Data Fig. 2v–x), further documenting an inverse correlation between the CDK4 activity and PD-L1 expression.
To extend these observations to an in vivo setting, we treated MMTV-ErbB2 mice bearing autochthonous breast cancers, or mice carrying allografts of murine MC38 or B16-F10 cancer cell lines with palbociclib, and monitored PD-L1 levels. Inhibition to CDK4/6 led a significant upregulation of PD-L1 in all these cancer models, which was accompanied by a reduction in the number of infiltrating CD3+ TILs (Fig. 2i–k; Extended Data Fig. 3a–c). We also observed that palbociclib treatment significantly elevated PD-L1 protein levels in various organs of normal mice (Extended Data Fig. 3d–h). Collectively, these results demonstrate that cyclin D-CDK4 kinase plays a rate-limiting role in regulating PD-L1 levels in vivo.
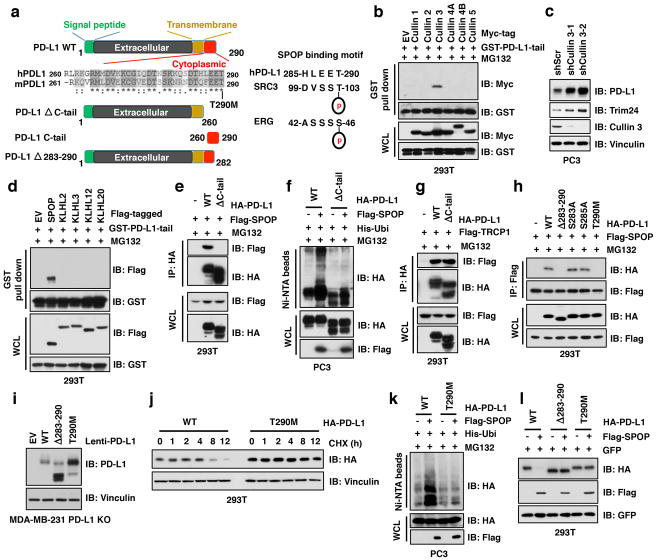
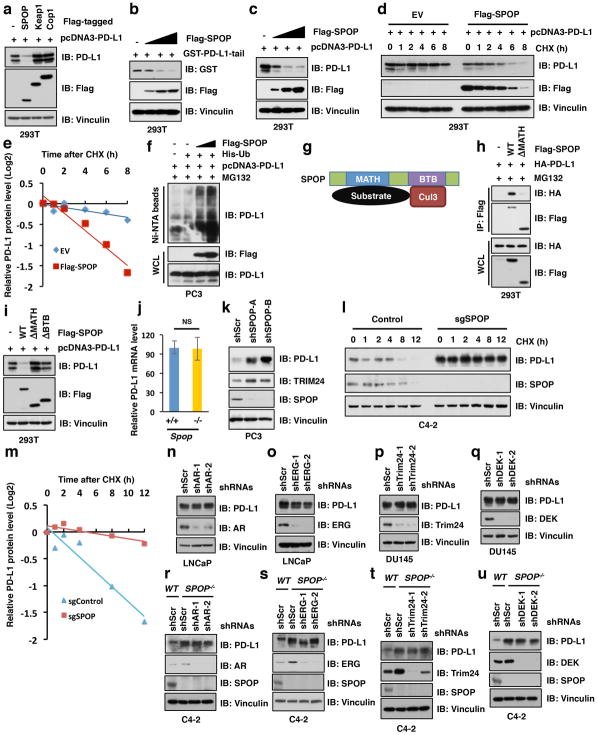
To understand how cyclin D-CDK4 regulates PD-L1 levels, we first determined that treatment of cells with proteasome inhibitor MG132, or with cullin-based ubiquitin E3 ligase inhibitor, MLN4924 22 elevated PD-L1 protein levels (Fig. 3a). To identify which cullin family E3 ligase(s) regulates PD-L1, we screened the potential interaction of PD-L1 with each cullin family proteins and found that Cullin 3, and to a lesser extent, Cullin 1 interacted with PD-L1 in cells (Fig. 3b, Extended Data Fig. 4a, b). These results indicate that in addition to Cullin 1/β-TRCP 11, Cullin 3-based E3 ligase(s) might play a role in regulating PD-L1 stability. Consistent with this notion, depletion of Cullin 3 elevated the protein abundance of endogenous PD-L1 (Extended Data Fig. 4c).
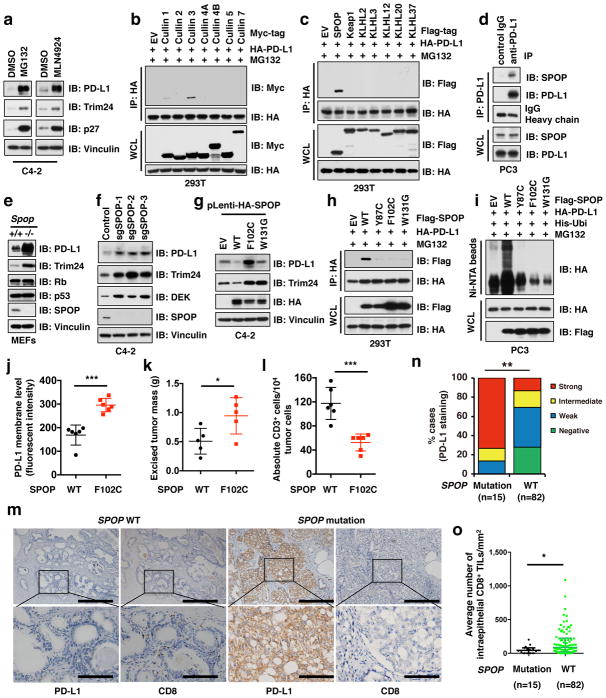
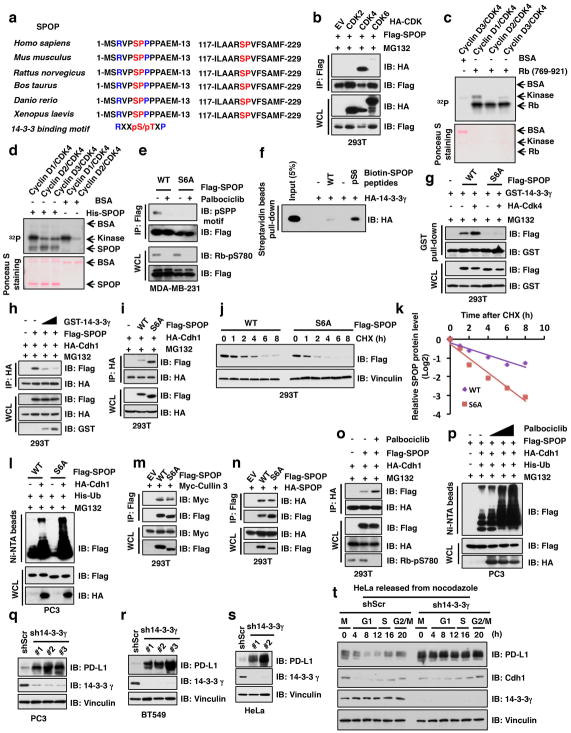
Figure 3. Cullin 3SPOP is the physiological E3 ubiquitin ligase for PD-L1.
a–d, IB analysis of WCL derived from C4-2 cells treated with MG132 (10 μM) or MLN4924 (1 μM) for 12 hours (a), immunoprecipitates (IP) and WCL derived from 293T cells transfected with indicated constructs (b, d), or anti-PD-L1 IP and WCL derived from PC3 cells (e). Cells were treated with MG132 (10 μM) for 12 hours in b, c.
e–g, IB analysis of WCL derived from Spop+/+ versus Spop−/− MEFs (e), C4-2 cells depleted SPOP with sgRNAs (f), or C4-2 cells stably expressing indicated SPOP WT and mutants (g).
h, i, IB analysis of IP and WCL derived from 293T cells (h), or Ni-NTA pull-down products derived from PC3 cells transfected with indicated constructs and treated with 30 μM MG132 for 6 hours.
j–l, FACS analysis for PD-L1 (j) or CD3+ T-cell population (l) of the B16-F10 implanted tumors ectopically expressing SPOP-WT or F102C mutant (n = 6 mice each group). Tumor weight were recorded at the time of sacrifice (k) (n = 5 mice each group).
m, Representative images of PD-L1 and CD8 immunohistochemistry (IHC) staining in SPOP wild-type or mutant primary human prostate cancer samples. The scale bar: 400 μm or 100 μm.
n, o Quantification of IHC analysis for PD-L1 (n) and CD8+ T cells (o) in SPOP wild-type versus mutant human prostate tumor specimens. (n =15 for SPOP mutant, n = 82 for SPOP WT). Error bars, ± s.d., two-tailed t-test, except (n) Mann-Whitney test, *P < 0.05, **P < 0.01, ***P < 0.001.
Cullin 3-based E3 ubiquitin ligases recognize their downstream substrates through substrate-recruiting adaptor proteins 23. We found that SPOP, but not other adaptor proteins examined interacted with PD-L1 in cells (Fig. 3c, d). We further determined that deletion of the C-tail, or the last eight amino acids of PD-L1 (283-290), disrupted binding of PD-L1 to SPOP, and rendered PD-L1 resistant to SPOP-meditated poly-ubiquitination (Extended Data Fig. 4d–h), indicating that the 283-290 region of PD-L1 might represent the potential binding motif for SPOP. Importantly, the cancer-derived PD-L1 T290M mutant (cBioPortal) located within the SPOP-binding motif also lost its ability to interact with SPOP and became more stable through decreased SPOP-mediated poly-ubiquitination and degradation (Extended Data Fig. 4i–l). Furthermore, depleting SPOP or deleting its substrate-interacting MATH domain elevated and stabilized PD-L1 in cells (Fig. 3e, f; Extended Data Fig. 5a–m). However, depleting known SPOP substrates including AR, ERG, Trim24, or DEK in SPOP-WT or SPOP−/− cells did not lead to obvious changes in PD-L1 levels (Extended Data Fig. 5n–u), arguing against a possibility of secondary effects for the observed elevation of PD-L1 upon SPOP depletion.
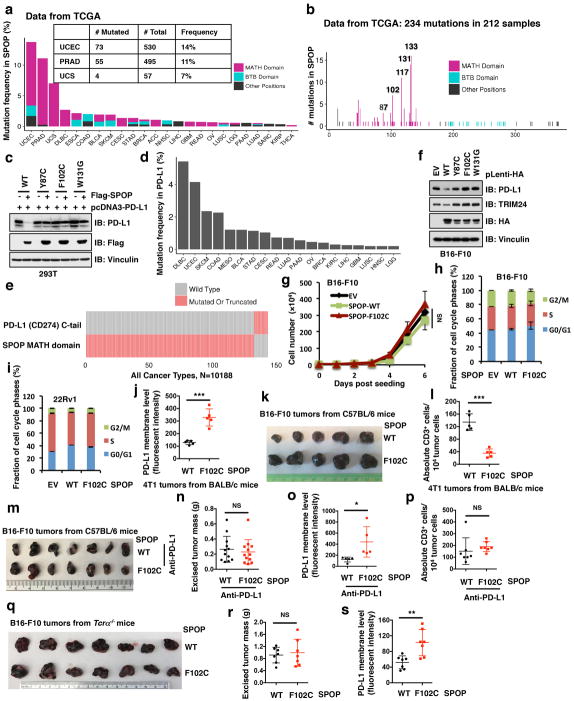
SPOP mutations occur in 10–15% of human prostate cancers, and are largely clustered within the MATH domain 24,25 (Extended Data Fig. 6a). Notably, these cancer-derived SPOP mutants failed to promote PD-L1 degradation due to their deficiency in binding to PD-L1 and promoting PD-L1 poly-ubiquitination (Fig. 3g–i and Extended Data Fig. 6b, c), which resembles the Elongin C-encoding TCEB1 hotspot mutants in clear cell renal carcinoma, resulting in deficiencies in the ability of Cullin 2/Elongin B/C/VHL E3 ligase complex to promote HIF1α degradation 26. We also observed that mutations in the PD-L1 C-tail (degron) are mutually exclusive with mutations in the substrate-interacting MATH domain of SPOP (Extended Data Fig. 6d, e).
To further explore the impact of SPOP mutations on tumorigenesis, we generated tumor cell lines expressing SPOP-WT or cancer-derived mutants. We found that cells expressing cancer-derived SPOP mutants displayed elevated levels of endogenous PD-L1 protein, as compared to cells expressing SPOP-WT. (Fig. 3j and Extended Data Fig. 6f–j). Upon inoculation into immunoproficient mice, the growth of implanted tumors expressing cancer-derived SPOP-F102C was faster than tumors expressing SPOP-WT (Fig. 3k and Extended Data Fig. 6k). Tumors expressing cancer-derived SPOP-F102C mutant displayed elevated PD-L1 levels and significantly reduced numbers of CD3+ TIL (Fig 3l and Extended Data Fig. 6l). Strikingly, the difference in tumor weights between SPOP-WT and SPOP-F102C groups was largely alleviated after treatment with anti-PD-L1 antibody (Extended Data Fig. 6m–p), or when tumor cells were inoculated into T cell-deficient Tcrα−/− mice (Extended Data Fig. 6q–s). Hence, enhanced tumorigenic potential of SPOP-mutant cells is largely caused by elevated PD-L1 levels resulting in increased immune evasion.
We next explored whether loss-of-function SPOP mutations regulate PD-L1 levels or TILs in primary human prostate cancers. To this end, we identified 15 SPOP-mutant and 82 SPOP-wild type tumors through large-scale sequencing as described 27,28. IHC staining results revealed that approximately 80% of SPOP-mutant tumors exhibited strong PD-L1 staining, while only approximately 10% of SPOP-WT tumors exhibited strong staining for PD-L1 and 70% of SPOP-WT cases displayed weak or no PD-L1 staining (Fig. 3m, n; Extended Data Fig. 7a–d). Moreover, the numbers of CD8+ TILs were reduced in samples harboring SPOP mutations, as compared to SPOP-WT tumors (Fig. 3o; Extended Data Fig. 7e–h). These results indicate that SPOP-deficiency correlates with elevated PD-L1 protein abundance and decreased numbers of TILs in primary human prostate cancers.
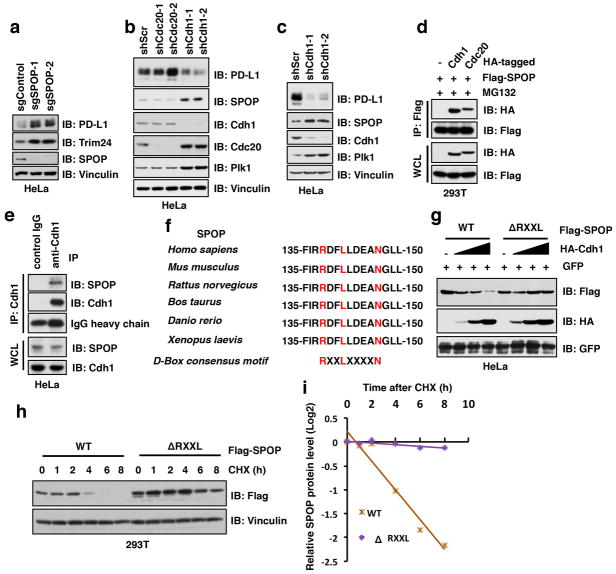
We further found that SPOP protein abundance fluctuated during the cell cycle and displayed an inverse correlation with PD-L1 protein levels (Fig. 1a and Fig. 4a); depleting SPOP resulted in stabilization of PD-L1 across the cell cycle (Fig. 4a and Extended Data Fig. 8a). We noted that the Anaphase-Promoting Complex/Cyclosome (APC/C) E3 ligase adaptor protein Cdh1 displayed an inverse correlation with SPOP protein levels during cell cycle (Fig. 1a, 1c and Fig. 4a). Furthermore, depletion of Cdh1, but not Cdc20, elevated SPOP protein abundance, which was accompanied by a simultaneous reduction in PD-L1 protein levels (Extended Data Fig. 8b, c). Consistent with these results, we detected a physical interaction between the endogenous SPOP and Cdh1 proteins (Fig. 4b and Extended Data Fig. 8d, e), and identified an evolutionarily conserved destruction-box motif (D-box: RxxLxxxxN) 29 in SPOP (Extended Data Fig. 8f). Deleting the D-box motif in SPOP disrupted its binding to Cdh1 and rendered SPOP resistant to Cdh1-mediated poly-ubiquitination and degradation (Fig. 4c, d; Extended Data Fig. 8g–i). Moreover, depletion of Cdh1 led to SPOP stabilization, which subsequently resulted in a reduction in PD-L1 protein level during cell cycle progression (Fig. 4e). Taken together, these results indicate that Cdh1 is a physiologically important upstream E3 ligase responsible for negatively regulating SPOP protein stability.
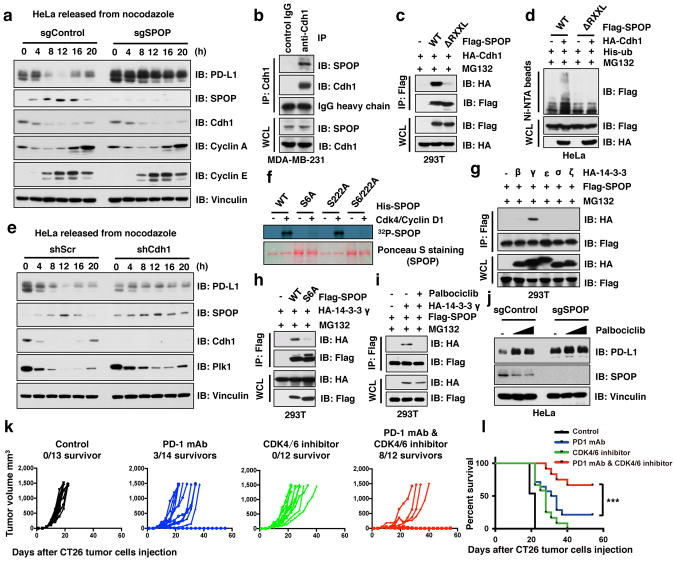
Figure 4. Cyclin D-CDK4-mediated phosphorylation of SPOP stabilizes SPOP largely through recruiting 14-3-3γ to disrupt its binding with Cdh1.
a–e, IB of WCL derived from HeLa cells with/without depletion of SPOP (a) or Cdh1 (e) synchronized in M phase by nocodazole treatment prior to releasing for the indicated times, IP and WCL derived from MDA-MB-231 (b) or 293T (c) cells, or Ni-NTA pull-down products derived from HeLa cells transfected with the indicated constructs (d). Cells were treated with MG132 (30 μM) for 6 hours in b–d.
f, In vitro kinase assays showing that cyclin D1/CDK4 phosphorylates recombinant SPOP at Ser6, not Ser222.
g–j, IB analysis of IP and WCL derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) or with/without palbociclib (1 μM) for 12 hours (g–i), or HeLa cells with/without depletion of SPOP treated with palbociclib (0.5, 1 μM) for 48 hours (j).
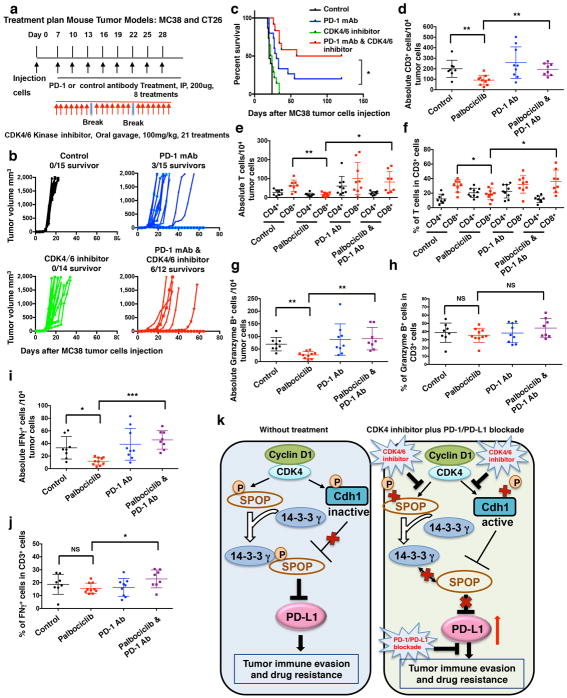
k, CT26 implanted tumor-bearing mice were enrolled in different treatment groups as indicated. Tumor volumes of mice treated with control antibody (n = 13), anti-PD-1 mAb (n = 14), the CDK4/6 inhibitor, palbociclib (n = 12) or combined therapy (n = 12) were measured every three days and plotted individually. We repeated this experiment twice.
l, Kaplan-Meier survival curves for each treatment group demonstrate the improved efficacy of combining PD-1 mAb with the CDK4/6 inhibitor, palbociclib. ***P < 0.001. (Gehan-Breslow-Wilcoxo test). We repeated this experiment twice.
To elucidate how the cyclin D-CDK4 kinase affects this mechanism, we established that cyclin D1-CDK4 directly phosphorylates SPOP at Ser6, but not Ser222, the only two conserved serine-proline sites in SPOP (Fig. 4f and Extended Data Fig. 9a–d). Conversely, treatment of cells with CDK4/6 inhibitor, palbociclib, reduced the phosphorylation of SPOP in cells (Extended Data Fig. 9e). We observed that 14-3-3γ protein physically interacted with SPOP in a pSer6-dependent manner and disrupted the interaction of SPOP with Cdh1 in cells (Fig. 4g, h; Extended Data Fig. 9f–h). Inhibition of SPOP-pSer6 decreased the interaction of SPOP with 14-3-3γ and increased its binding to Cdh1, leading to elevated SPOP poly-ubiquitination (Fig. 4i; Extended Data Fig. 9i–p). Consequently, palbociclib treatment decreased SPOP protein abundance and elevated PD-L1 levels in SPOP-WT, but not SPOP-deficient cells (Fig. 4j). Moreover, depletion of 14-3-3γ dramatically upregulated PD-L1 levels and stabilized PD-L1 during cell cycle progression (Extended Data Fig. 9q–t).
Recent clinical studies revealed that the success of PD1/PD-L1 blockade correlates with PD-L1 expression levels in tumor cells 6,7. Given our observation that inhibition of CDK4/6 elevated PD-L1 levels, we hypothesized that inhibitors of CDK4/6 kinase might synergize with anti-PD-1/PD-L1 therapy to elicit an enhanced therapeutic effect. Notably, we observed that treatment of immunoproficient mice bearing CT26 tumors with palbociclib plus anti-PD-1 antibody dramatically retarded tumor progression and resulted in 8 complete responses out of 12 treated mice (Fig. 4k; Extended Data Fig. 10a). Moreover, combining CDK4/6 inhibitor with anti-PD-1 therapy resulted in a significant improvement of overall survival compared to single-agent treated group (Fig. 4l). Similar results were obtained using mice bearing tumors derived from MC38 cells. (Extended Data Fig. 10b, c). As expected from our earlier observations, treatment of tumor-bearing mice with palbociclib decreased the absolute numbers of TILs, including CD3+, CD4+, CD8+, Granzyme B+ and IFNγ+ cells. Importantly, addition of anti-PD-1 antibody to palbociclib treatment restored essentially normal numbers of TILs (Extended Data Fig. 10d–j).
A recent study revealed that another inhibitor of CDK4/6, abemaciclib, increased immunogenicity of cancer cells via an Rb-dependent mechanism, which activates tumor cell expression of endogenous retroviral elements, thereby stimulating production of type III interferons and antigen presentation by tumor cells 30. Together with our demonstration that cyclin D-CDK4 regulates PD-L1 stability through Cullin 3SPOP (Extended Data Fig. 10k), these studies provide complementary molecular rationale for combining CDK4/6 inhibitor treatment with anti-PD-1/PD-L1 immunotherapy to enhance tumor regression.
Methods
Cell culture, transfections and viral infections
HEK293T, HEK293, HeLa, MDA-MB-231, MCF7, Hs578T, WT MEFs, cyclin D1−/− MEFs, cyclin D2−/− MEFs, cyclin D3−/− MEFs, cyclin D1−/−D2−/−D3−/− MEFs, cyclin D1fl/flD2−/−D3fl/fl, Cdk4+/+ and Cdk4−/− MEFs, cyclin A1+/+A2+/+ and cyclin A1−/−A2−/− MEFs, cyclin E1+/+E2+/+ and cyclin E1−/−E2−/−, Spop+/+ and Spop−/− MEFs (a kind gift of Dr. Nicholas Mitsiadesa, Baylor College of Medicine, Houston, TX) were cultured in DMEM medium supplemented with 10% FBS (Gibco), 100 units of penicillin and 100 μg/ml streptomycin (Gibco). HLF, HepG2, Huh1 and Huh7 were cultured in RPMI medium supplemented with 10% FBS. MDA-MB-231 PD-L1 WT and PD-L1 KO cells are kind gift from Dr. Mien-Chie Hung. BT549, T47D, ZR75-1, HCC1954, HCC1937, MDA-MB436, MDA-MB468 and SKBR3 cells were from Dr. Alex Toker laboratory at BIDMC, Harvard Medical School, and cultured in RPMI medium or McCoy’s5A (Corning, NY) medium supplemented with 10% FBS. PC3, DU145, 22RV1, LNCaP and C42 were kind gifts from Dr. Pier Paolo Pandolfi group at BIDMC, Harvard Medical School, and cultured in RPMI medium (Corning, NY) with 10% FBS. Mouse tumor derived MC38 cell line was a kind gift from Dr. Arlene Sharpe at Harvard Medical School. Mouse tumor derived 4T1 and B16-F10 cell lines were routinely cultured in Gordon Freeman’s laboratory in DMEM medium supplemented with 10% FBS (Gibco), 100 units of penicillin and 100 μg/ml streptomycin (Gibco). All cell lines were routinely tested to be negative for mycoplasma contamination.
Cells with 80% confluence were transfected using lipofectamine plus reagents in Opti-MEM medium (Invitrogen). 293FT cells were used for packaging of lentiviral and retroviral cDNA expressing viruses, as well as subsequent infection of various cell lines were performed. Briefly, medium with secreted viruses were collected twice at 48 hours and 72 hours after transfection. After filtering through 0.45 μM filters, viruses were used to infect cells in the presence of 4 μg/mL polybrene (Sigma-Aldrich). 48 hours post-infection, cells were split and selected using hygromycin B (200 μg/mL) or puromycin (1 μg/mL) for 3 days. Cells were harvested and lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP40) supplemented with protease inhibitors (Roche) and phosphatase inhibitors (Calbiochem) for immunoblot analysis.
Reagents
Nocodazole (M1404) and Taxol (T7402) were purchased from Sigma. Thymidine (CAS: 50-89-5) and cycloheximide (66-81-9) were purchased from Acros organics. Palbociclib (PD0332991, S1116) and ribociclib (LEE011, S7440) were purchased from Selleckchem. MG132 (BML-PI102-0005) was purchased from Enzo life science. MLN4924 was a kind gift from Dr. William Kaelin (Dana-Farber cancer institute).
Plasmids
Myc-tagged Cullin 1, Cullin 2, Cullin 3, Cullin 4A, Cullin 4B, Cullin 5, Flag-tagged SPOP WT, Y87C, F102C, W131G, delta MATH, delta BTB, pLenti-HA-SPOP WT, Y87C, F102C, W131G, pGEX-4T-1-SPOP, Flag-Keap1, Flag-Cop1, shScramble, shCullin 3, shSPOP, and His-ubiquitin constructs were described previously 31. shAR, shERG, shTrim24, shDEK and sgSPOP constructs were described previously 31, 32. Myc-Cullin 7 construct was kindly offered by Dr. James A. DeCaprio (Dana-Farber Cancer Institute). KLHL2 and KLHL3 constructs were generous gifts from Dr. Shinichi Uchida (Tokyo Medical and Dental University). KLHL12 and KLHL37 constructs were purchased from Addgene. KLHL20 construct was offered by Dr. Ruey-Hwa Chen (Institute of Biological Chemistry, Academia Sinica, Taiwan). The construct of HA-PD-L1 (HA tag in the N-terminus of PD-L1) was kindly provided by Dr. Mien-Chie Hung (The University of Texas MD Anderson Cancer Center). HA-Cdh1, HA-Cdc20, shCdh1, hCdc20 and HA-14-3-3 isoform constructs were described previously 33, 34. pCMV-CDK4 WT, pCMV-CDK4 N158F and shcyclin D3 were described previously 35, 36. pBabe-p16 was a kindly gift from Dr. Charles J. Sherr laboratory. pLKO-shCDK4 (Plasmid #78153 and #78154) and pMLP-shCDK6 (Plasmid #73552 and #73553) were purchased from Addgene. pLKO-sh14-3-3γ (TRCN0000078160, TRCN0000078161, TRCN0000078162), pLKO-shp16 (TRCN0000039748, TRCN0000039751, TRCN0000039782) and pLKO-shCD8a (TRCN0000057583, TRCN0000057587) were purchased from Open Biosystems. pcDNA3-PD-L1, pCMV-GST-PD-L1-tail (cytoplasmic amino acids), HA-PD-L1-ΔC-tail, HA-PD-L1-Δ283-290, HA-PD-L1-S283A, HA-PD-L1-S285A, HA-PD-L1-T290M, pLenti-PD-L1 WT, pLenti-PD-L1-Δ283-290, pLenti-PD-L1 T290M, pET-28a-His-SPOP WT, pET-28a-His-SPOP S6A, pET-28a-His-SPOP S22A, Flag-SPOP with delta D-Box (RxxL), pLenti-HA-c-Myc WT, pLenti-HA-c-Myc T58A/S62A, pLenti-HA-cyclin D1, pLenti-HA-cyclin D2, pLenti-HA-cyclin D3, Flag-SPOP S6A, HA-tagged CDK2, CDK4 and CDK6 were generated in this study.
Antibodies
Anti-PD-L1 (E1L3N) rabbit mAb (13684), anti-pS10-H3 (3377), anti-pS780-Rb (8180), anti-pS807/811-Rb (8516), anti-Rb (9309), anti-cyclin D1 (2978), anti-cyclin D2 (3741), anti-CDK6 (3136), anti-cullin 3 (2759), anti-GST (2625), rabbit polyclonal anti-Myc-Tag antibody (2278) and mouse monoclonal anti-Myc-Tag (2276) antibodies were purchased from Cell Signaling Technology. Mouse PD-L1 antibody (MAB90781-100) was purchased from R&D systems. Anti-mPD-L1 for immunoblotting (clone 298B.8E2), anti-mPD-L1 (clone 298B.3G6) for immunohistochemistry, and anti-human PD-L1 for immunoprecipitation (clone 29E.12B1) were generated in the laboratory of Dr. Gordon J. Freeman. Anti-CDK4 (MS-616-P1) was purchased from Thermo Scientific. Anti-SPOP (16750-1-AP) was purchased from Proteintech. Anti-cyclin A (sc-751), anti-cyclin B (sc-245), anti-cyclin E (SC-247), anti-cyclin D3 (sc-182), anti-Cdh1 (sc56312), anti-Cdc20 (sc-8358), anti-Cdc20 (sc-13162), anti-Plk1 (sc-17783), anti-TRIM24 (TIF1α, SC-271266), anti-HA (sc-805, Y-11), anti-PD-L1 (sc-50298) and anti-GST (sc-459) were obtained from Santa Cruz. Anti-GFP (8371-2) was purchased from Clontech. Anti-Flag (F-2425), anti-Flag (F-3165, clone M2), anti-Vinculin (V9131), anti-Flag agarose beads (A-2220), anti-HA agarose beads (A-2095), peroxidase-conjugated anti-mouse secondary antibody (A-4416) and peroxidase-conjugated anti-rabbit secondary antibody (A-4914) were purchased from Sigma. Anti-HA (MMS-101P) was obtained from BioLegend.
Immunoblot and immunoprecipitation analyses
Cells were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). Protein concentrations were measured by the Beckman Coulter DU-800 spectrophotometer using the Bio-Rad protein assay reagent. Equal amounts of protein were resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitations analysis, 1000 μg total cell lysates were incubated with the primary antibody-conjugated beads for 4 hours at 4 °C. The recovered immunocomplexes were washed four times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with indicated antibodies.
Immunohistochemistry (IHC) for cell pellets, xenografted tumors or human prostate tumor specimens
The cultured cells (MDA-MB-231 PD-L1 WT and KO cells; HBP-ALL shScr and shCD8 cells; KE37 shScr and shCD8 cells) were washed and fixed in 4% paraformaldehyde for 20 minutes. Cells pellets or xenografted (MDA-MB-231 PD-L1 WT or KO) tumors were embedded into TFM and frozen. After cryostat sections (10 μM) were placed on Superfrost Plus Stain slides, samples were then permibilized in 0.1% Triton X-100/PBS for 10 minutes. For IHC analysis, we used UltraSensitive™ SP (Mouse) IHC Kit (KIT-9701, Fuzhou Maixin Biotech) following the manufacturer’s instructions with minor modification. The sections were incubated with 3% H2O2 for 15 min at room temperature to block endogenous peroxidase activity. After incubating in normal goat serum for 1 hour to block non-specific binding of IgG, sections were treated with primary antibody (PD-L1, 298B.3G6, 18 μg/ml; CD8α, sc-53212, clone C8/144B, dilution 1:40) at 4°C overnight. Sections were then incubated for 30 minutes with biotinylated goat-anti-mouse IgG secondary antibodies (Fuzhou Maixin Biotech), followed by incubation with streptavidin-conjugated HRP (Fuzhou Maixin Biotech). Specific samples were developed with 3′3-diaminobenzidine (DAB-2031, Fuzhou Maixin Biotech). Images were taken using an Olympus microscopic camera and matched software.
The prostate tumor specimens were obtained from Shanghai Changhai Hospital in China. Usage of these specimens was approved by the Institute Review Board of Shanghai Changhai Hospital. For IHC, the paraformaldehyde fixed paraffin embedded prostate tumor samples were deparaffinized in xylene (3 × 10 min), rehydrated through a series of graded alcohols (100%, 95%, 85%, and 75%) to water. Samples were then subjected to heat-mediated antigen retrieval at 95°C for 20 min. The following IHC steps were the same as described above.
The expression level of PD-L1 in prostate cancer tumor samples was determined according to the intensity of the staining as 0, negative; 1, weak expression; 2, intermediate expression and 3, strong expression. The numbers of intraepithelial CD8+ tumor-infiltrating T lymphocytes (TILs) was counted as described in Hamanishi et al 37. Briefly, three independent areas with the most abundant infiltration were selected under a microscopic field at 200 × magnification (0.0625mm2). The number of intraepithelial CD8+ TILs was counted manually and calculated as cells per mm2. The Mann-Whitney test was used to compare the difference in PD-L1 expression between SPOP mutated and wide type cases. The Student’s t test was used to determine P values of the difference in CD8+ TILs between SPOP mutated and wide type cases. P < 0.05 was considered as significant.
In vitro cyclin D/CDK4 kinase assays
Kinase assays were performed in a final volume of 30 μl of a kinase buffer as described previously 38: 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM DTT, 1 mM EGTA, 0.1 mM NaF, containing 10 μM ATP and 0.4 mCi [32P]γATP (Perkin Elmer). 0.2 μg of CDK4/cyclin D1 (0142-0143-1, Pro-Qinase), CDK4/cyclin D2 (0142-0375-1, Pro-Qinase), or CDK4/cyclin D3 (0142-0373-1, Pro-Qinase) were used as kinases. 2 μg of His-SPOP, His-SPOP-S6A, His-SPOP-S222A, or His-SPOP-S6A/S222A mutant proteins immobilized on Ni-NTA beads were used as kinase substrates. 0.1 μg of Rb1 C-terminal recombinant protein (Cat. SC-4112, Santa Cruz) was used as a positive control for kinase assays. 2 μg of BSA was used as a negative control. After 60 min incubation at 30°C, proteins were denatured, resolved on SDS-PAGE, transferred to nitrocellulose membranes and exposed to X-ray films.
Kinase assay for Rb by immune-purified endogenous CDK4/cyclin D1 from mice tissues
For endogenous kinase assays, the endogenous CDK4 was immunoprecipitated by 6 μg of anti-CDK4 Ab (Santa Cruz, sc-23896 AC) from 2.5 mg of lysates (buffer: 20 mM Tris-HCl pH 8.0, 0.1 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween-20, 0.1% NP40) of livers or brains isolated from C57BL6 mice. The association of cyclin D1 was also confirmed by cyclin D1 antibodies (abcam, ab134175). The immunopurified endogenous CDK4/cyclin D1 was used as kinase, and 0.5 μg of Rb1 C-terminal recombinant protein (SC-4112) was used as the kinase substrate.
In vivo ubiquitination assays
PC3 or HeLa cells with 80% confluence were transfected with His-ubiquitin and the indicated constructs. 36 hours post-transfection, cells were treated with 30 μM MG132 for 6 hours and lysed in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 10 mM imidazole [pH 8.0]). After sonication, the lysates were incubated with nickel-nitrilotriacetic acid (Ni-NTA) beads (QIAGEN) for 3 hours at room temperature. Subsequently, the His pull-down products were washed twice with buffer A, twice with buffer A/TI (1 volume buffer A and 3 volumes buffer TI), and one time with buffer TI (25 mM Tris-HCl and 20 mM imidazole [pH 6.8]). The pull-down proteins were resolved by 2 × SDS-PAGE for immunoblotting.
Protein half-life assays
Cells were transfected or treated under indicated conditions. For half-life studies, cycloheximide (20 μg/ml, Sigma) was added to the medium. At indicated time points thereafter, cells were harvested and protein abundances were measured by immunoblot analysis.
Cell synchronization and FACS analyses
Cells synchronized with nocodazole arrest and double thymidine treatment as described previously 39. Cells synchronized with nocodazole or double thymidine-arrest and release were collected at the indicated time points and stained with propidium iodide (Roche) according to the manufacturer’s instructions. Cells were fixed by 70% ethanol at −20°C overnight and washed 3 times using cold PBS. The samples were digested with RNase for 30 minutes at 37°C and stained with propidium iodide (Roche) according to the manufacturer’s instructions. Stained cells were sorted with BD FACSCanto™ II Flow Cytometer. The results were analyzed by ModFit LT 4.1 and FSC express 5 softwares.
BrdU/PI labelling and FACS analyses
Cells were incubated with/without BrdU (75 μM, Sigma) containing medium for 1 hour. Cells were harvested and washed once with cold PBS for centrifuge 5 min at 1200 rpm. Cells were re-suspended in 200 μl cold PBS and added in 5 ml of cold 90% ethanol for fixation overnight. After centrifuge 5 min at 1200 rpm, cells were washed once using 5 ml PBS and added in 0.5 ml 2N HCl-0.5% Triton X-100 for 30 min at room temperature (RT). After adding in 5 ml PBS, samples were centrifuged for 5 min at 1200 rpm and re-suspended in 1 ml Na2B4O7 (pH 8.5). Samples were re-suspended in 200 μl of anti-BrdU diluted (1: 40) in PBS with 0.5 % tween 20 and 1% BSA and were incubated 30 min at room temperature. After adding in 5 ml 20 mM Hepes-PBS (pH7.4) with 0.5% tween 20, samples were centrifuged for 5 min at 1200 rpm and were re-suspended in 0.5 ml PBS with propidium iodide (PI, 5 μg/ml, Sigma) and RNAse A (200 μg/ml, Roche). After incubating 30 min at RT, samples were transfered into FACS tube and analyzed by flow cytometry.
Real-Time RT-PCR analyses
Total RNAs were extracted using the QIAGEN RNeasy mini kit, and reverse transcription reactions were performed using the ABI Taqman Reverse Transcription Reagents (N808-0234). After mixing the generated cDNA templates with primers/probes and ABI Taqman Fast Universal PCR Master Mix (4352042), reactions were performed with the ABI-7500 Fast Real-time PCR system and SYBR green qPCR Mastermix (600828) from Agilent Technologies Stratagene.
Human GAPDH: Forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′,
Reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′;
Mouse GAPDH: Forward, 5′-AGGTCGGTGTGAACGGATTTG-3′,
Reverse, 5′-GGGGTCGTTGATGGCAACA-3′;
Human PD-L1: Forward, 5′-TGGCATTTGCTGAACGCATTT-3′,
Reverse, 5′-TGCAGCCAGGTCTAATTGTTTT-3′;
Mouse PD-L1: Forward, 5′-GCTCCAAAGGACTTGTACGTG-3′,
Reverse, 5′-TGATCTGAAGGGCAGCATTTC-3′;
Generation of cyclin D-deficient MEFs
Cyclin D1−/−, D2−/−, D3−/− and D1F/FD2−/−D3F/FMEFs were derived from E13.5 mouse embryos as described previously 40, 41.
Generation of mouse tumors
Cyclin D1−/− mice 42 were mated with MMTV-c-Myc or MMTV-Wnt1 mice (from the Jackson Laboratory) yielding cyclin D1−/−/MMTV-c-Myc or cyclin D1−/−/MMTV-Wnt1, as well as control cyclin D1+/+/MMTV-c-Myc or D1+/+/MMTV-Wnt1 mice. Mammary tumors were dissected from multiparous females and snap-frozen.
MMTV-ErbB2 female mice (from the Jackson Laboratory), bred into a mixed C57BL/6 and 129Sv background, were treated with palbociclib or vehicle only for 6 weeks after detection of palpable tumors. Palbociclib was administered daily by gastric gavage (150 mg/kg of body weight); every two weeks the daily dose was lowered to 100mg/kg for 2–3 days. Control mice were treated with vehicle (10% 0.1N HCl, 10% Cremaphor EL, 20% PEG300, 60% 50 mM citrate buffer pH 4.5) 10 ml/kg by gastric gavage. After 6 weeks, tumors were collected and snap-frozen in OCT.
Treatment of wild-type mice with palbociclib
6-weeks old C57BL/6 female mice (from the Jackson Laboratory) were treated with palbociclib (150 mg/kg body weight, by gastric gavage) or vehicle only for 7 days. Subsequently, organs were collected and analyzed by immunoblotting.
Mouse tumor implantation
1 × 105 B16-F10 or 2 × 105 MC38 cells were injected subcutaneously into 6-weeks old C57BL/6 female mice (from the Jackson Laboratory). Starting one week later, mice were treated daily with palbociclib (150 mg/kg body weight, by gastric gavage) or vehicle only, for 7 days. Subsequently, tumors were collected and analyzed by FACS or immunoblotting.
1 × 105 B16-F10 cells stably expressing SPOP WT or F102C mutant were injected subcutaneously into 6-weeks old C57BL/6 female mice (from the Jackson Laboratory). On day 3 after tumor cells were injected, control and PD-L1 mAb treatments were conducted by intra-peritoneal injection (200 μg/mouse in 200 μl HBSS saline buffer) every three days for a total of 3 injections. Subsequently, tumors were collected and analyzed by FACS.
1 × 105 B16-F10 cells stably expressing SPOP WT or F102C mutant were injected subcutaneously into 6-weeks old Tcrα−/− female mice (from the Jackson Laboratory). After 10 days, tumors were collected and analyzed by FACS.
Immunofluorescence staining of cells or tumor tissues
MDA-MB-231 PD-L1 WT and KO cells were seeded in chambers (154534, Thermo Fisher Scientific). Cells were fixed with 4% paraformaldehyde for 20 minutes, followed with 0.1% Triton X-100 in PBS for 10 minutes. Cells were pre-blocked with 2% BSA/PBS for 45 minutes, then incubated with primary antibodies against PD-L1 (PD-L1, 298B.3G6, 1:200), for 2.5 hours at room temperature and followed with secondary anti-mouse antibodies conjugated with Alexa-fluor-568 (Invitrogen, 1:250). Hoechst (life technology, 1:10,000) was used to stain nuclei.
TFM-embedded 10 μM-thick tumor tissue sections were fixed with 2% paraformaldehyde/PBS for 30 min, and permeabilized in 0.1% Triton X-100/PBS for 10 min. Tumor tissue sections were pre-blocked with 2% BSA/PBS for 45 min, then incubated with primary antibodies against PD-L1 (1:200), CD3 (Abcam, 1:250) for 2.5 hours at room temperature and followed with secondary anti-mouse antibodies conjugated with Alexa-fluor-568 (Invitrogen, 1:250) and anti-rabbit antibodies conjugated with Alexa-fluor-488 (Invitrogen, 1:250). Hoechst (life technology, 1:10,000) was used to stain nucleus. Tumor tissues were mounted with fluoromount-G® (SouthernBiotech) at 4°C overnight. Tissue sections were examined with fluorescent microscope under a 20 × objective lens. CD3+ cell numbers were counted in an area of 5.95 × 105 μm2.
Single cell generation from tumor tissue and flow cytometry analysis
Tumor tissues were minced and digested with 5 ml of 2 mg/ml collagenase (Sigma) in DMEM for 1 hour at 37°C. Cells were then collected by centrifuge and filtered through a 70 μm strainer in DMEM. Cell pellets were suspended and lysed in red blood cell lysis buffer for 5 min. The cells were then filtered through a 40 μm strainer in 1 x PBS with 2% BSA. 1 million cells were incubated with antibodies against PD-L1 (564715, BD Biosciences, 1:100) conjugated with APC or antibodies against CD3 (Biolegend, 1:100) conjugated with APC or corresponding isotype IgG1 control at room temperature for 30 min. Cells were washed by 1 x PBS with 2% BSA and analyzed by flow cytometry.
In vivo experimental therapy in MC38 and CT26 mice tumor models
Animal studies were approved by Dana-Farber Cancer Institute Institutional Animal Care and Use Committee (IACUC; protocol number 04-047), and performed in accordance with guidelines established by NIH Guide for the care and use of laboratory animals. MC38 or CT26 tumors were established by subcutaneously injecting 1 × 105 MC38 or CT26 tumor cells in 100 μl HBSS into the right flank of 6-week old C57BL/6 or BALB/c female mice (Jackson Lab, ME). Tumor sizes were measured every three days by caliper after implantation and tumor volume was calculated by length × width2 × 0.5. On day 7 after tumor cells were injected, animals were pooled and randomly divided into four groups with comparable average tumor size. Moreover, the lab members who measured the mice were blinded to the treatment groups. Mice were grouped into control antibody treatment, PD-1 mAb treatment, CDK4/6 inhibitor treatment, and PD-1 mAb plus CDK4/6 inhibitor treatment. As illustrated in Extended Data Fig. 10a, control and PD-1 mAb treatments were conducted by intraperitoneal injection (200 μg/mouse in 200 μl HBSS saline buffer) every three days for a total of 8 injections. The CDK4/6 inhibitor treatment was given by oral gavage once a day with a dosage of 100 mg/kg for three weeks with a break every week for one day. For survival studies, animals were monitored for tumor volumes every three days for 120 days after initial treatment, until tumor volume exceeded 2000 mm3, or until tumor had ulcer with diameter reached 1 cm. Statistical analysis was conducted using the GraphPad Prism software (GraphPad Software, Inc., San Diego, CA). Kaplan-Meier curves and corresponding Gehan-Breslow-Wilcoxo tests were used to evaluate the statistical differences between groups in survival studies. P < 0.05 was considered to be significant.
T cell analysis for MC38 implanted tumors
MC38 implanted tumors were established by subcutaneously injecting 1 × 105 of MC38 cells were injected into the right flank of 6 week old C57BL/6 female mice (Jackson Lab). On the day of the tumor cells injected, mice were randomly divided into four groups: control antibody treatment, PD-1 mAb treatment, CDK4/6 inhibitor treatment, and PD-1 mAb plus CDK4/6 inhibitor treatment. Control and PD-1 mAb treatments were conducted by intraperitoneal injection (200 μg/mouse in 200 μl HBSS saline buffer) every three days for a total of 4 injections. The treatment of palbociclib was given by oral gavage with the dosage of 200 mg/kg for 9 days, with a break after 7 days. Tumors were then collected and single cell was generated from tumor tissues as described in section “Single Cell Generation from Tumor Tissue and Flow Cytometry analysis”. After cells were filtered through 40 μm strainer, cells were fixed in 0.5 ml/tube Fixation buffer (420801, Biolegend) in the dark for 20 minutes at room temperature. Cells were then washed with 1 × PBS with 2% BSA. The fixed cells were suspended in Intracellular Staining Perm Wash Buffer (421002, Biolegend) after centrifuge for two times to permeabilize the cells. Cells were then co-stained with antibodies against CD3 (100236, APC conjugated, Biolegend), Granzyme B (515403, FITC conjugated, Biolegend), IFN-γ (505808, PE conjugated, Biolegend) to check the activities of T cells. Or cells were co-stained with antibodies against CD3 (100236, APC conjugated, Biolegend), CD4 (100510, FITC conjugated, Biolegend), CD8 (100708, PE conjugated, Biolegend). The corresponding isotype IgG1 controls were used for controls. The cells were incubated with corresponding antibodies for 30 minutes at room temperature. Cells were washed by 1 × PBS with 2% BSA and analyzed by flow cytometry.
Data Availability Statement (DAS)
Source data for gels in Figs 1–4 and Extended Data Figs. 1–9 are available in Supplementary Fig. 1. Source data for Figs 2j and 2k are available in Table 1. Source data for Figs 3j–n, 3o are available in Table 2. Source data for Figs 4k, l are available in Table 3. Source data for Extended Data Figs 3c, d are available in Table 4. Source data for Extended Data Figs 6j, l, o, p, r, s are available in Table 5. Source data for Extended Data Figs 10b–j are available in Table 6. All other data supporting the findings of this study are available from the corresponding author upon a reasonable request.
Extended Data
Extended Data Figure 1. PD-L1 fluctuates during cell cycle progression.
a, b, Immunoblot (IB) of whole cell lysates (WCL) derived from MDA-MB-231 or HCC1954 cells synchronized in M phase by nocodazole treatment prior to releasing back into the cell cycle for the indicated times.
c, d, Quantitative real-time PCR (qRT-PCR) analyses of relative mRNA levels of PD-L1 and GAPDH from samples derived from HeLa cells synchronized in M phase by nocodazole treatment prior to releasing back to the cell cycle for the indicated time points.
e, IB of WCL derived from HeLa cells pre-treated with/without IFNγ (10 ng/ml) for 12 hours and then synchronized in M phase by nocodazole treatment prior to releasing back into the cell cycle for the indicated times.
f, IB of WCL derived from HeLa cells stably expressing HA-c-Myc WT, or HA-T58A/S62A-c-Myc as well as empty vector (EV) as a negative control.
g, IB of WCL derived from HeLa cells with/without stably expressing HA-c-Myc WT synchronized in M phase by nocodazole treatment prior to releasing back into the cell cycle for the indicated times.
h–j, IB of WCL derived from MC38, CT26, 4T1, or B16-F10 mouse tumor cells treated with the indicated concentration of nocodazole for 20 hours before harvesting. (k–m) IB of WCL derived from B16-F10, 4T1, or CT26 mouse tumor cells treated with the indicated concentration of taxol for 20 hours before harvesting.
Extended Data Figure 2. Cyclin D/CDK4 negatively regulates PD-L1 protein stability.
a, b, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from wild type (WT), cyclin A1−/−A2−/− or WT, cyclin E1−/−E2−/− MEFs.
c, Quantitative real-time PCR (qRT-PCR) analysis of relative mRNA levels of PD-L1 from wild type MEFs and cyclin D1−/−D2−/−D3−/− MEFs. Data were represented as mean ± s.d, n = 5.
d, Cell cycle profiles for WT and cyclin D1−/−D2−/−D3−/− MEFs, which were labeled with BrdU and analyzed by FACS.
e, IB analysis of WCL derived from cyclin D1fl/flD2−/−D3fl/fl MEFs with or without depleting cyclin D1 and cyclin D3 by pLenti-Cre via viral infection (pLenti-EGFP as a negative control), selected with puromycin (1 μg/ml) for 72 hours before harvesting.
f, IB analysis of WCL derived from cyclin D1−/−D2−/−D3−/− MEFs stably reintroducing cyclin D1, cyclin D2, or cyclin D3, respectively, with empty vector (EV) as a negative control.
g, IB analysis of WCL derived from mouse mammary tumors induced by MMTV-c-Myc with/without genetic depletion of cyclin D1. n = 5 mice per experimental group.
h, IB analysis of WCL derived from WCL derived from wild type and cdk6−/− MEFs.
i, j, IB analysis of WCL derived from MDA-MB-231 cells stably expressing shCDK6 or shCDK2 as well as shScr as a negative control, respectively.
k, l, IB analysis of WCL derived from MDA-MB-231 cells transfected with indicated constructs (k) and the intensity of PD-L1 band was quantified by the ImageJ software (l).
m, IB analysis of WCL derived from MDA-MB-231 cells depleted of Rb (with shScr as a negative control) treated with the CDK4/6 inhibitor, palbociclib, where indicated.
n, o, IB analysis of WCL derived from mouse CT26 or 4T1 tumor cell lines treated with or without the CDK4/6 inhibitor, palbociclib or ribociclib, respectively.
p, q, IB analysis of WCL derived from MDA-MB-231 cells pre-treated with palbociclib (1 μM) for 36 hours before treatment with cycloheximide (CHX) for the indicated time points (p) and PD-L1 protein abundance was quantified by the ImageJ and plotted as indicated (q).
r, IB analysis of WCL derived from 19 different cancer cell lines with indicated antibodies.
s–u, IB analysis of WCL derived from MCF7, T47D or HLF stably expressing p16 as well as EV as a negative control.
v–x, IB analysis of WCL derived from MDA-MB-436, BT549 or HCC1937 stably expressing three independent shRNAs against p16 as well as shScr as a negative control.
Extended Data Figure 3. CDK4/6 inhibitor, palbociclib, treatment elevated PD-L1 levels in vivo.
a, b, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from MC38 or B16-F10 mouse tumor cell line implanted tumors treated with palbociclib (150 mg/kg body weight, by gastric gavage) or vehicle for 7 days. n = 5 mice per experimental group.
c, FACS analysis for PD-L1 or CD3+ T-cell populations from MC38 implanted tumors treated with vehicle or palbociclib for 7 days. n = 5 mice per experimental group.
d, IB analysis of WCL derived from multiple organs in mice treated with palbociclib (150 mg/kg body weight, by gastric gavage) or vehicle for 7 days. n = 5 mice per experimental group.
e, Quantification of PD-L1 protein bands intensity in Extended Data Fig. 3d by using the ImageJ software. n = 5 mice per experimental group.
f, IB analysis of WCL derived from 15 different tissues with/without palbociclib treatment and MMTV-c-Myc induced breast tumors.
g, Quantification of PD-L1 protein bands intensity in Extended Data Fig. 3f by using the ImageJ software. n = 3 biological replicates
h, In vitro kinase assay for Rb through using immunoprecipitated CDK4/cyclin D kinase complex from liver or brain by anti-CDK4 antibody IP. Note that cyclin D-CDK4 complex in non-dividing organs (livers and brains) displayed kinase activity, which might explain why CDK4/6 inhibitor elevated PD-L1 in these organs. Error bars, ± s.d., two-tailed t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Figure 4. Cullin 3SPOP promotes PD-L1 ubiquitination and subsequent degradation largely through interaction with the cytoplasmic tail of PD-L1.
a, A schematic illustration of PD-L1 with N-terminal signal peptide, extracellular domain, trans-membrane domain, cytoplasmic tail and the potential SPOP-binding motif in PD-L1.
b, d, Immunoblot (IB) analysis of whole cell lysates (WCL) and GST pull-down precipitates derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
c, IB analysis of WCL derived from PC3 stably expressing shCullin 3.
e, g, IB analysis of WCL and immunoprecipitation (IP) derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
f, IB of WCL and Ni-NTA pull-down products derived from the lysates of PC3 cells transfected with the indicated constructs. Cells were treated with MG132 (30 μM) for 6 hours before harvesting and lysed in the denature buffer.
h, IB analysis of WCL and IP derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
i, IB of WCL derived from MDA-MB-231 PD-L1 KO cells stably expressing PD-L1 WT, delta 283-290, T290M as well as EV as a negative control.
j, IB analysis of WCL derived from 293T cells transfected with HA-PD-L1 WT and the T290M mutant, which were treated with cycloheximide (CHX) for indicated time points before harvesting.
k, IB of WCL and Ni-NTA pull-down products derived from the lysates of PC3 cells transfected with the indicated constructs. Cells were treated with MG132 (30 μM) for 6 hours before harvesting and lysed in the denaturing buffer.
l, IB of WCL derived from 293T cells transfected with indicated constructs.
Extended Data Figure 5. SPOP negatively regulates PD-L1 protein stability in a poly-ubiquitination dependent manner.
a–c, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with indicated constructs.
d, e, IB analysis of WCL derived from 293T cells transfected with indicated constructs. 36 h post transfection, cells were treated with 20 μg/ml cycloheximide (CHX) at indicated time points (d). The PD-L1 protein abundance were quantified by the ImageJ software and plotted (e).
f, IB of WCL and Ni-NTA pull-down products derived from the lysates of PC3 cells transfected with the indicated constructs. Cells were treated with MG132 (30 μM) for 6 hours before harvesting and lysed in the denaturing buffer.
g, A schematic illustration of SPOP with MATH and BTB domain to interact with substrate and Cullin 3, respectively.
h, IB analysis of WCL and IP derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
i IB analysis of WCL derived from 293T cells transfected with indicated constructs.
j, qRT-PCR analysis of relative mRNA levels of PD-L1 from Spop+/+ and Spop−/− MEFs. Data were represented as mean ± s.d, n=5.
k, IB analysis of WCL derived from PC3 cells infected with indicated lentiviral shRNAs against SPOP and selected with puromycin (1 μg/ml) for 72 hours before harvesting.
l–m, IB analysis of WCL derived from C42 cells with depletion of SPOP using sgRNA and treated with cycloheximide (CHX) for indicated time points before harvesting (l). The PD-L1 protein abundance were quantified by the ImageJ software and plotted (m).
n, o, IB analysis of WCL derived from LNCaP cells stably expressing shAR or shERG as well as shScr as a negative control.
p, q, IB analysis of WCL derived from DU145 cells stably expressing shTrim24 or shDEK as well as shScr as a negative control.
r–u, IB analysis of WCL derived from C42 SPOP WT and SPOP−/− cells that stably expressed shAR, shERG, shTrim24, or shDEK as well as shScr, respectively.
Extended Data Figure 6. Cancer-derived SPOP mutations fail to promote PD-L1 degradation.
a, The mutation frequency (mutated cases/total cases) of SPOP across 24 cancer types from the TCGA database. Mutations are categorized as happening in the MATH domain, in the BTB domain or at any other position of the gene, including UTRs. Because some patient cases contain mutations of two or three categories, the proportion of three colors are allocated mutation-wise, instead of case-wise.
b, The distribution of mutation positions of SPOP in 24 cancer types from the TCGA database. Mutations with low translational consequences have been discarded.
c, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with indicated constructs.
d, The mutation frequency (mutated cases/total cases) of PD-L1 (CD274) across 19 cancer types from the TCGA database.
e, Oncoplot of PD-L1 (CD274) and SPOP across all 39 cancer types in the TCGA database. Only mutations or truncations in the C terminal tail of PD-L1 or in the MATH domain of SPOP are counted.
f, IB of WCL derived from B16-F10 mouse tumor cell line stably expressing the indicated SPOP constructs.
g, h, Growth curve and cell cycle profile of B16-F10 cells stably expressing SPOP WT and the F102C mutant as well as EV as a negative control.
i, Cell cycle profile of 22Rv1 cells stably expressing SPOP WT and the F102C mutant as well as EV as a negative control.
j, Relative cell surface PD-L1 expression of 4T1 implanted tumors ectopically expressing SPOP-WT or the SPOP-F102C mutant were subjected to FACS analysis. n = 5 mice per experimental group.
k, B16-F10 cells stably expressing SPOP-WT or the SPOP-F102C mutant implanted tumors from C57BL/6 mice were dissected and taken a picture after euthanizing the mice.
l, The number of CD3+ T-cell populations from the isolated tumor-infiltrating lymphocytes in 4T1 cells stably expressing SPOP-WT or the SPOP-F102C mutants implanted tumors were subjected to FACS analysis. n = 5 mice per experimental group.
m, B16-F10 cells stably expressing SPOP-WT or the SPOP-F102C mutant implanted tumors from C57BL/6 mice treated with anti-PD-L1 antibody were dissected and taken a picture after euthanizing the mice. n = 7 mice per experimental group.
n, The weight of B16-F10 cells implanted tumors from C57BL/6 mice treated with anti-PD-L1 antibody. 12 mice per experimental group.
o, Relative cell surface PD-L1 expression of B16-F10 cells implanted tumors ectopically expressing SPOP-WT or the SPOP-F102C mutant treated with anti-PD-L1 antibody were subjected to FACS analysis. n = 5 mice per experimental group.
p, The number of CD3+ T-cell populations from the isolated tumor-infiltrating lymphocytes in B16-F10 cells implanted tumors ectopically expressing SPOP-WT or the SPOP-F102C mutant treated with control IgG or anti-PD-L1 antibody were subjected to FACS analysis. n = 7 mice per experimental group.
q, B16-F10 cells stably expressing SPOP-WT or the SPOP-F102C mutant implanted tumors from Tcrα−/− mice were dissected and taken a picture after euthanizing the mice. n = 7 mice per experimental group.
r, Relative cell surface PD-L1 expression of B16-F10 cells stably ectopically expressing SPOP-WT or the SPOP-F102C mutant implanted tumors from Tcrα−/− mice were subjected to FACS analysis. n = 7 mice per experimental group.
s, The number of CD3+ T-cell populations from the isolated tumor-infiltrating lymphocytes in B16-F10 cells stably ectopically expressing SPOP-WT or the SPOP-F102C mutant implanted tumors from Tcrα−/− mice were subjected to FACS analysis. n = 7 mice per experimental group. Error bars, ± s.d., two-tailed t-test, *P < 0.05, **P < 0.01, ***P < 0.001, NS: no significance.
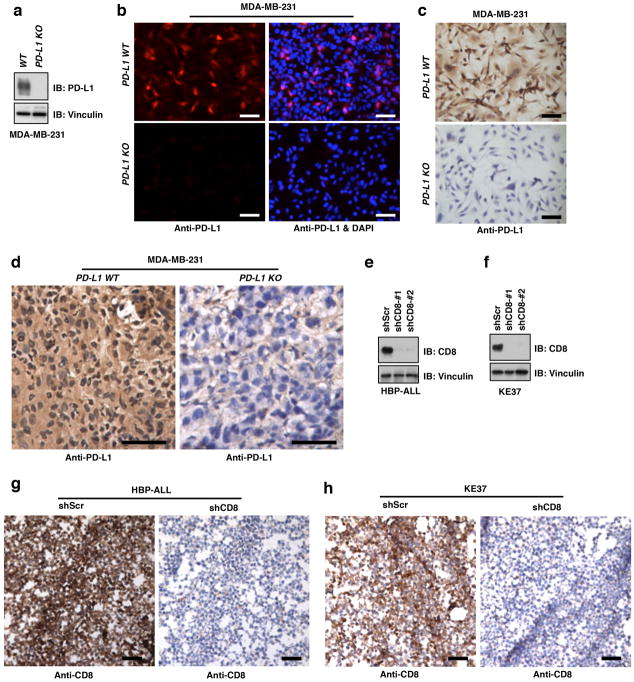
Extended Data Figure 7. Validation of anti-PD-L1 and anti-CD8 antibodies through using PD-L1 KO or shCD8 cells.
a, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from MDA-MB-231 cells depleted PD-L1 through the CRISPR-Cas9 system.
b, Immunofluorescence (IF) for MDA-MB-231 PD-L1 WT and KO cells using the anti-PD-L1 antibody. The scale bar represents 50 μm.
c, d, Immunochemistry (IHC) for MDA-MB-231 PD-L1 WT and KO cells from cultured on glass slides (c) or implanted tumors (d) using the anti-PD-L1 antibody. The scale bar represents 50 μm.
e, f, IB analysis of WCL derived from HBP-ALL (e) or KE37 (f) cells stably expressing shCD8 as well as shScr as a negative control using the anti-CD8 antibody.
g, h, IHC for HBP-ALL (g) or KE37 (h) cell pellets stably expressing shCD8 as well as shScr as a negative control using the anti-CD8 antibody. The scale bar represents 50 μm.
Extended Data Figure 8. Depletion of Cdh1, but not Cdc20, prolongs SPOP proteins stability, which is simultaneously coupled with a decrease in PD-L1 protein level.
a–c, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from HeLa depleted SPOP through the CRISPR-Cas9 system (a) or depleted Cdc20 or Cdh1 through multiple independent shRNAs (b, c).
d, IB analysis of WCL and immunoprecipitation (IP) derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
e, IB analysis of WCL and IP derived from HeLa cells treated with MG132 (10 μM) for 12 hours before harvesting.
f, A sequence comparison of D-box motif (RxxLxxxxN) in SPOP derived from different species.
g, IB analysis of WCL derived from HeLa cells transfected with indicated constructs.
h, i, IB analysis of WCL derived from 293T cells transfected with indicated constructs. 36 h post transfection, cells were treated with cycloheximide (CHX) as indicated time points before harvesting (h). The protein abundance of SPOP-WT and deletion of RxxL mutant were quantified by the ImageJ software (i).
Extended Data Figure 9. Cyclin D/CDK4-mediated phosphorylation of SPOP at the Ser6 residue promotes its binding with 14-3-3γ to reduce its poly-ubiquitination and subsequent degradation by APC/Cdh1.
a, A sequence comparison of conserved SP sites and putative 14-3-3γ binding motif in SPOP.
b, Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitation (IP) derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
c, d, In vitro kinase assays with recombinant Rb and SPOP as substrates and cyclin D1/CDK4, cyclin D2/CDK4 and cyclin D3/CDK4 as kinase complex were performed. BSA was used as a negative control where indicated.
e, IB analysis of WCL and immunoprecipitation (IP) derived from MDA-MB-231 cells transfected with indicated constructs, which were treated with/without palbociclib (1 μM) for 12 hours.
f, Streptavidin beads pull-down assay for biotin-labeled SPOP peptide with/without phosphorylation at the Ser6 residue to examine its in vitro association with 14-3-3γ.
g, IB analysis of WCL and GST pull-down precipitates derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
h, i, IB analysis of WCL and IP derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) for 12 hours before harvesting.
j, k, IB analysis of WCL derived from 293T cells transfected with indicated constructs. 36 h post transfection, cells were treated with 20 μg/ml cycloheximide (CHX) as indicated time points (j). The protein abundance of SPOP-WT and S6A mutant were quantified by the ImageJ software and plotted accordingly (k).
l, p, IB of WCL and Ni-NTA pull-down products derived from the lysates of PC3 cells transfected with the indicated constructs. Cells were treated with MG132 (30 μM) for 6 hours before harvesting and lysed in the denaturing buffer for following assay.
m–o, IB analysis of WCL and IP derived from 293T cells transfected with indicated constructs and treated with MG132 (10 μM) and with/without palbociclib (1 μM) for 12 hours before harvesting.
q–s, IB of WCLs derived from PC3, BT549 and HeLa cells stably expressing sh14-3-3γ as well as shScr as a negative control.
t, IB of WCL derived from HeLa cells stably expressing shScr or sh14-3-3γ synchronized in M phase by nocodazole treatment prior to releasing back into the cell cycle for the indicated times.
Extended Data Figure 10. Combination therapy of anti-PD-1 mAb and CDK4/6 inhibitor in MC38 colon cancer mouse model.
a, A schematic model that illustrates the treatment plan for mice bearing subcutaneous MC38 tumors. Female C57BL/6 mice were implanted with 0.1 × 106 MC38 cells subcutaneously and treated with four arms: control antibody treatment, anti-PD-1 mAb treatment, CDK4/6 inhibitor treatment, anti-PD-1 mAb plus CDK4/6 inhibitor combination treatment.
b, MC38 implanted tumor-bearing mice were enrolled in different treatment groups as indicated. Tumor volumes of mice treated with control antibody (n = 15), anti-PD-1 mAb (n = 15), the CDK4/6 inhibitor, palbociclib (n = 14) or combined therapy (n = 12) were measured every three days and plotted individually. We repeated this experiment twice.
c, Kaplan-Meier survival curves for each treatment group demonstrate the improved efficacy of combining PD-1 mAb with the CDK4/6 inhibitor, palbociclib. *P < 0.05. (Gehan-Breslow-Wilcoxo test). We repeated this experiment twice.
d, e, g, i, The absolute number of CD3+, CD4+, CD8+, Granzyme B+, or IFNγ+ TILs cells of implanted MC38 tumors treated with indicated agents was analyzed by FACS. Control: n = 8, palbociclib: n = 10, PD-1 Ab: n = 9, Palbociclib & PD-1 Ab: n = 8.
f, h, j, The percentage of CD4+, CD8+ in CD3+ TILs cells of implanted MC38 tumors treated with indicated agents was analyzed by FACS. Control: n = 8, palbociclib: n = 10, PD-1 Ab: n = 9, Palbociclib & PD-1 Ab: n = 8.
k, A proposed working model to illustrate how PD-L1 protein stability is regulated by the cyclin D/CDK4-SPOP-Cdh1 signaling pathway. The cyclin D/CDK4 negatively regulates PD-L1 protein stability largely through phosphorylating its upstream physiological E3 ligase SPOP to promote SPOP binding with 14-3-3γ, which subsequently disrupts Cdh1-mediated destruction of SPOP. As such, CDK4/6 inhibitor treatment could unexpectedly elevate PD-L1 protein levels largely through inhibiting cyclin D/CDK4-mediated phosphorylation of SPOP to promote its degradation by APC/CCdh1. The unexpected rise of PD-L1 could present a severe clinical problem for patients receiving CDK4 inhibitor treatment and could be one of the underlying mechanisms accounting for CDK4 inhibitor resistance via evading immune surveillance checkpoint. Hence, our work provides a novel molecular mechanism as well as the rationale for the combinational treatment of PD-L1 blockage treatment and the CDK4/6 inhibitors as a more efficient anti-cancer clinical option. Error bars, ± s.d., two-tailed t-test, *P < 0.05, **P < 0.01, ***P < 0.001, NS: no significance.
Supplementary Material
Acknowledgments
We thank members of the Wei, Freeman, Sicinski, and Pandolfi laboratories for discussions. J.Z. is supported by the career transition award (1K99CA212292-01). W.W. is a Leukemia & Lymphoma Society (LLS) research scholar. This work was supported in part by the NIH grants GM094777 and CA177910 (W.W.), R01 CA083688, P01 CA080111 and R01 CA202634 (to P.S.), and P50CA101942 (G.J.F).
Footnotes
Supplementary Information is linked to the online version of the paper.
Disclosure of Potential Conflicts of Interest
GF has patents/pending royalties on the PD-1 pathway from Roche, Merck, Bristol-Myers-Squibb, EMD-Serono, Boehringer-Ingelheim, AstraZeneca, Dako and Novartis. GF has served on advisory boards for CoStim, Novartis, Roche, Eli Lilly, Bristol-Myers-Squibb, Seattle Genetics, Bethyl Laboratories, Xios, and Quiet. PS in a consultant and a recipient of a research grant from Novartis. No potential conflicts of interests were disclosed by other authors.
Author Contributions
J.Z., X.B. and H.W. performed most of the experiments with assistance from Y.Z. Y.G., N.T.N, Y.T., Y.C., F.W., X.D., J.G., Y.H., C.F., S.R., and Y.S.. Y.Z, S.R., and Y.S performed IHC staining for human prostate cancer samples. Y.G., Y.T., and Y.C. helped mice work. J.Z., X.B., H.W. P.S. and W.W. designed the experiments. G.F., P.S., and W.W. supervised the study. J.Z. and W.W. wrote the manuscript with help from X.B, H.W., P.S. and G.F. All authors commented on the manuscript.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv324. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boussiotis VA. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med. 2016;375:1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotwals P, et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer. 2017;17:286–301. doi: 10.1038/nrc.2017.17. [DOI] [PubMed] [Google Scholar]
- 4.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 5.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 6.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwai Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A. 2002;99:12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorand RD, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim SO, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci U S A. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bates S, et al. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene. 1994;9:71–79. [PubMed] [Google Scholar]
- 17.Lees E, Faha B, Dulic V, Reed SI, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 18.Takaki T, et al. Preferences for phosphorylation sites in the retinoblastoma protein of D-type cyclin-dependent kinases, Cdk4 and Cdk6, in vitro. J Biochem. 2005;137:381–386. doi: 10.1093/jb/mvi050. [DOI] [PubMed] [Google Scholar]
- 19.Fry DW, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 20.Parry D, Bates S, Mann DJ, Peters G. Lack of cyclin D-Cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumour suppressor gene product. EMBO J. 1995;14:503–511. doi: 10.1002/j.1460-2075.1995.tb07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukas J, et al. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 22.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 23.Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–2320. doi: 10.1038/emboj.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163 doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44:1231–1235. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gan W, et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol Cell. 2015;59:917–930. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goel S, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548:471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan W, et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Molecular cell. 2015;59:917–930. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai X, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23:1063–1071. doi: 10.1038/nm.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan L, et al. The APC/C E3 Ligase Complex Activator FZR1 Restricts BRAF Oncogenic Function. Cancer Discovery. 2017;7:424–441. doi: 10.1158/2159-8290.CD-16-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao D, et al. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009;11:397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Q, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23–32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Sicinska E, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–61. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 37.Hamanishi J, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci U S A. 2007;104:3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546:426–430. doi: 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan L, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Developmental cell. 2014;29:377–391. doi: 10.1016/j.devcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozar K, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 41.Choi YJ, et al. The requirement for cyclin D function in tumor maintenance. Cancer cell. 2012;22:438–451. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sicinski P, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for gels in Figs 1–4 and Extended Data Figs. 1–9 are available in Supplementary Fig. 1. Source data for Figs 2j and 2k are available in Table 1. Source data for Figs 3j–n, 3o are available in Table 2. Source data for Figs 4k, l are available in Table 3. Source data for Extended Data Figs 3c, d are available in Table 4. Source data for Extended Data Figs 6j, l, o, p, r, s are available in Table 5. Source data for Extended Data Figs 10b–j are available in Table 6. All other data supporting the findings of this study are available from the corresponding author upon a reasonable request.