Abstract
The immune checkpoint ligand PD-L1 and the transmembrane mucin MUC1 are upregulated in triple-negative breast cancer (TNBC) where they contribute to its aggressive pathogenesis. Here we report that genetic or pharmacological targeting of the oncogenic MUC1 subunit MUC1-C is sufficient to suppress PD-L1 expression in TNBC cells. Mechanistic investigations showed that MUC1-C acted to elevate PD-L1 transcription by recruitment of MYC and NF-κB p65 to the PD-L1 promoter. In an immunocompetent model of TNBC in which Eo771/MUC1-C cells were engrafted into MUC1 transgenic mice, we showed that targeting MUC1-C associated with PD-L1 suppression, increases in tumor infiltrating CD8+ T-cells and tumor cell killing. MUC1 expression in TNBCs also correlated inversely with CD8, CD69 and GZMB, and downregulation of these markers associated with decreased survival. Taken together, our findings show how MUC1 contributes to immune escape in TNBC, and they offer a rationale to target MUC1-C as a novel immunotherapeutic approach for TNBC treatment.
Keywords: TNBC, MUC1-C, PD-L1, immune evasion, IFN-γ
Introduction
Triple-negative breast cancer (TNBC) typically has an aggressive clinical course (1). Additionally, treatment of TNBC has been limited by availability of actionable targets (1). Most patients with operable disease therefore receive adjuvant chemotherapy (1). Moreover, patients with newly diagnosed stage II or III disease are often treated with neoadjuvant chemotherapy (1). Interestingly, the presence of tumor-infiltrating lymphocytes (TILs) is associated with improved survival in TNBC patients treated with adjuvant or neoadjuvant chemotherapy (1,2). In addition, the programmed death ligand 1 (PD-L1) is overexpressed in TNBC (3–5) and is associated with high tumor grades and poor clinical outcomes (6–9), further indicating that evasion of immune destruction contributes to TNBC pathogenesis. Indeed, several Phase I trials performed with antibodies targeting the programmed death 1 (PD-1)/PD-L1 axis have demonstrated objective and durable responses in patients with heavily pretreated, metastatic TNBC (10,11). These findings have thus supported the importance of PD-L1 as a target for the immunotherapy of TNBC; however, little is known about the regulation of PD-L1 expression in TNBC cells.
Mucin 1 (MUC1) is a heterodimeric protein that is overexpressed in approximately 90% of TNBCs in association with amplification of the MUC1 gene and the activation of auto-inductive transcriptional circuits (12). The MUC1 C-terminal (MUC1-C) transmembrane subunit functions as an oncoprotein by interacting with diverse effectors that have been linked to hallmarks of the cancer cell (12). MUC1-C includes an intrinsically disordered cytoplasmic domain, which acts as a node for integrating multiple signaling pathways at the cell membrane and in the nucleus (12,13). In this way, MUC1-C activates PI3K→AKT and MEK→ERK signaling in TNBC and other breast cancer cells (14,15). MUC1-C also directly activates the β-catenin→TCF4→MYC and NF-κ B p65 pathways and thereby the induction of their target genes (16,17). In concert with these findings, MUC1-C has been linked to induction of the epithelial-mesenchymal transition (EMT), epigenetic reprogramming, stemness and self-renewal of basal B TNBC cells (18–22). Other studies have shown that overexpression of MUC1, and specifically the oncogenic MUC1-C subunit, by cancer cells is associated with protection from killing by (i) TRAIL, (ii) Fas ligand, (iii) T-cell perforin/granzyme B-mediated lysis (23,24), supporting the notion that MUC1-C contributes to immune evasion.
The present work demonstrates that MUC1-C activates the CD274/PD-L1 gene in basal B TNBC cells. The results show that (i) MUC1-C drives PD-L1 transcription by MYC- and NF-κB p65-mediated mechanisms, and (ii) targeting MUC1-C with genetic and pharmacologic approaches results in the suppression of PD-L1. Targeting MUC1-C in MUC1.Tg mice harboring mouse Eo771/MUC1-C tumors further showed suppression of PD-L1 expression by tumor cells and activation of the tumor immune microenvironment. These results and those from analysis of TNBC datasets provide evidence for involvement of MUC1-C in immune evasion of these cancers. Our findings further support a model in which MUC1-C integrates the induction of PD-L1 expression with the EMT program, CSC state and epigenetic programming in basal B TNBC cells.
Materials and Methods
Cell culture
Human BT-549, SUM-159 and mouse Eo771 TNBC cells were propagated in RPMI1640 medium (ATCC, Manassas, VA, USA). Human MDA-MB-468 and MDA-MB-231 TNBC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Manassas, VA, USA). Human BT-20 TNBC cells were cultured in Eagle’s Minimum Essential Medium (EMEM) (ATCC). Media were supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 µg/ml streptomycin. Authentication of the cells upon thawing of stocks was performed by short tandem repeat (STR) analysis. Cells were monitored every 3 months for mycoplasma contamination by the MycoAlert® Mycoplasma Detection Kit (Lonza, Rockland, MA, USA). Cells were passaged for 3 months and then replaced with fresh stocks. BT-549 and MDA-MB-231 cells were transfected with lentiviral vectors to stably express a scrambled control shRNA (CshRNA; Sigma, St. Louis, MO, USA) and a NF-κB p65 shRNA (Sigma). Human BT-20 and mouse Eo771 cells were transfected to express an empty vector or one encoding MUC1-C and selected for growth in the presence of puromycin. Cells were treated with the I B inhibitor BAY-11–7085 (Sigma), the BET bromodomain inhibitor JQ-1 (Delmore JE, Cell, 2011) or DMSO as the vehicle control. Cells were also treated with empty nanoparticles (NPs) or GO-203/NPs (25).
Tetracycline-inducible MUC1 and MYC silencing
MUC1shRNAs (shRNA TRCN0000122938 and shRNA#2 TRCN0000122937; MISSION shRNA; Sigma), MYCshRNA (TRCN0000039642; MISSION shRNA, Sigma) or a control scrambled CshRNA (Sigma) were cloned into the pLKO-tetpuro vector (Addgene, Cambridge, MA, USA; Plasmid #21915). The viral vectors were co-transfected with the lentivirus packaging plasmids into 293T cells and the supernatant was collected at 48 h after transfection. BT-549 or MDA-MB-231 cells were incubated with the supernatant for 12 h in the presence of 8 µg/ml polybrene. Tet-inducible cells were selected for growth in 1–2 µg/ml puromycin and treated with doxycycline (DOX; Sigma).
Quantitative real-time, reverse transcriptase PCR (qRT-PCR)
Whole cell RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The High Capacity cDNA Reverse Transcritpion kit (Life Technologies, Carlsbad, CA, USA) was used to synthesize cDNAs from 2 µg RNA. cDNA samples were then amplified using the Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and ABI Prism Sequence Detector (Applied Biosystems). Primers used for qRT-PCR are listed in Supplementary Table S1.
Immunoblot analysis
Whole cell extracts were obtained using NP-40 buffer composed of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, protease inhibitor cocktail and DTT. Immunoblotting was performed with anti-MUC1-C (ThermoFischer Scientific, Waltham, MA, USA), anti-PD-L1, anti-MYC, anti-phospho-p65(Ser-536)(Cell Signaling Technology, Danvers, MA, USA), anti-NF-κB p65 (Santa Cruz Biotechnology, Dallas, TX), mouse PD-L1 (Bio-Techne, Minneapolis, MN, USA) and anti-β-actin (Sigma). Immunoreactive complexes were detected using horseradish peroxidase-conjugated secondary antibodies (GE Healthcare Life Sciences, Marlborough, MA, USA) and an enhanced chemiluminescence (ECL) detection reagents (Perkin Elmer Health Sciences, Waltham, MA, USA).
Promoter-reporter assays
Cells were transfected with 1.5 µg of PD-L1 promoter-luciferase reporter (pPD-L1-Luc) or control vector (Active Motif, Carlsbad, CA, USA) in the presence of Superfect (Qiagen, Germantown, MD, USA). After 48 h, the cells were lysed in passive lysis buffer. Lysates were analyzed using the Lightswitch Luciferase Assay Kit (Active Motif).
Chromatin immunoprecipitation (ChIP) assays
Soluble chromatin was prepared from 3×106 cells and precipitated with anti-MYC, anti-NF-κB p65 (Santa Cruz Biotechnology), anti-MUC1-C or a control nonimmune IgG. Power SYBR Green PCR Master Mix (Applied Biosystems) and ABI Prism Sequence Detector (Applied Biosystems) were used for amplification of ChIP qPCRs. Primers used for qPCR of the PD-L1 promoter and GAPDH control region are listed in Supplementary Table S2. Relative fold enrichment was calculated as described (26).
Mouse model studies
Eo771/MUC1-C cells (0.5×106 cells) were subcutaneously injected into the flanks of six-week old human MUC1.Tg mice. After reaching a tumor size of ~150 mm3, mice were pair-matched into two groups and treated with empty NPs or 15 mg/kg GO-203/NPs once a week for 2 weeks. At the end of the treatment, mice were sacrificed for harvesting of the tumors. In an additional experiment, mice bearing Eo771/MUC1-C tumors were treated with vehicle control (PBS) or 10 mg/kg anti-PD-L1 (BioXCell, West Lebanon, NH, USA) on days 1 and 5 as described (27). Animal studies were performed with approval from the Dana-Farber Cancer Institute Animal Care and Use Committee.
FACS analysis
Eo771/MUC1-C tumors were harvested, cut into small pieces and incubated in dissociation medium containing 100 units/ml Collagenase IV (ThermoFisher Scientific, Grand Island, NY, USA) and 50 µg/ml DNase I (Roche, Indianapolis, IN, USA) for 30 min at 37°C. Tumor cell suspensions were passed through 70 µm strainers (ThermoFisher Scientific). After lysis of red blood cells with ACK buffer (ThermoFisher Scientific), tumor cells were counted, and an aliquot of each sample was analyzed by FACS staining for CD69 and granzyme B (BioLegend, San Diego, CA, USA) expression on CD8+ T-cells (BD LSR II Flow Cytometer, BD Pharmingen, San Diego, CA, USA). Spleen cells were used for adjusting compensation during the analysis. After Ficoll separation, 3×106 cells were incubated with Leucocyte Activation Cocktail (BD Pharmingen) and Alexa 488 labeled anti-mouse CD107α antibody (BioLegend) for 6 h at 37°C. Cells were processed for FACS analysis of IFN-γ (ThermoFisher Scientific), granzyme B and CD107α.
CTL assays
The day before mice sacrifice, Eo771/MUC1-C cells (6×103 per well) were plated in 96-well plates and incubated overnight. Lymph nodes were digested with ACK lysis buffer (GIBCO, Waltham, MA, USA) and rinsed with PBS. Cells (effector cells) were incubated with Eo771/MUC1-C cells (target cells) in 96 well-plates for 6 h. The percentage cytotoxicity was assayed measuring LDH release following the manufacturer’s recommendations (CytoTox 96® Non-Radioactive Cytotoxicity Assay; Promega, Madison, WI, USA) and calculated using the formula: (Experimental-Effector spontaneous-Target spontaneous)/(Target maximum-Target spontaneous)x100.
Bioinformatic analyses
Datasets of TNBC patients were downloaded from the Gene Expression Omnibus (GEO) under the accession number GSE25066 (28). Raw signal intensities were RMA normalized across patients (29). Multiple probe sets corresponding to the same gene were averaged. Expression values of MUC1, CD8, CD69 and GZMB in TNBC samples were assessed for correlations using the Spearman coefficient. The prognostic value of CD8, CD69 and GZMB expression in TNBC datasets was determined as described (30). Expression values were averaged and TNBC patients were segregated by median expression. The Kaplan-Meier survival probability plot with the hazard ratio (95% confidence interval) and log-rank p-value were calculated and plotted in R.
Statistical analysis
Analyses were performed using GraphPad Prism version 7.0 (GraphPad Software Inc, San Diego, CA, USA) and p values <0.05 were considered statistically significant differences.
Results
MUC1 drives PD-L1 expression in TNBC cells
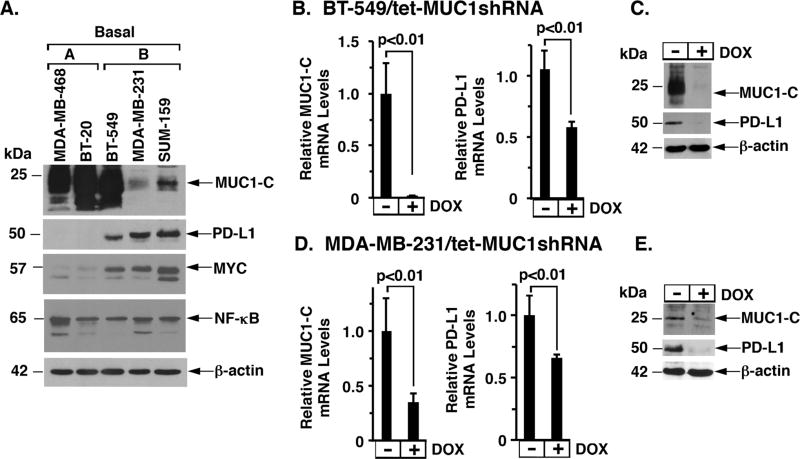
MUC1-C induces the EMT state, CSC characteristics and epigenetic reprogramming in basal B TNBC cells (14,18–20,26,31). To investigate the potential relationships between MUC1-C and PD-L1, we first performed immunoblot analysis of TNBC cell lines and found readily detectable PD-L1 levels in the mesenchymal basal B BT-549, MDA-MB-231 and SUM159 cells, as compared to that in basal A MDA-MB-468 and BT-20 cells (Fig. 1A). The results further showed that, in contrast to NF-κB p65, MYC is upregulated in basal B, but not basal A, TNBC cells (Fig. 1A). We therefore established BT-549, MDA-MB-231 and SUM159 cells with stable expression a tetracycline-inducible control shRNA (tet-CshRNA) or MUC1 shRNA (tet-MUC1shRNA) to determine whether MUC1-C contributes to the regulation of PD-L1 expression. As a control, doxycycline (DOX) treatment of BT-549/tet-CshRNA cells had no effect on MUC1 or PD-L1 expression (Supplementary Fig. S1A). By contrast, treatment of BT-549/tet-MUC1shRNA cells with DOX was associated with slowing of proliferation (Supplementary Fig. S1B) and downregulation of MUC1-C and PD-L1 mRNA (Fig. 1B) and protein (Fig. 1C). In addition, DOX treatment of BT-549/tet-MUC1shRNA#2 cells expressing a different MUC1 shRNA resulted in downregulation of PD-L1 expression (Supplementary Fig. S1C and S1D). Similar results were obtained with DOX-treated (i) MDA-MB-231/tet-CshRNA (Supplementary Fig. S1E) and MDA-MB-231/tet-MUC1shRNA (Fig. 1D and 1E) cells, and (ii) SUM-159/tet-MUC1shRNA (Supplementary Fig. S1F and S1G) cells, further supporting the premise that MUC1-C promotes the induction of PD-L1 expression.
Figure 1. MUC1-C induces PD-L1 expression.
A. Lysates from the designated basal A and basal B TNBC cells were immunoblotted with the indicated antibodies. B–C. BT-549 cells were transduced to stably express a tetracycline-inducible MUC1 shRNA (tet-MUC1shRNA). Cells treated with or without 500 ng/ml DOX for 4 d were analyzed for MUC1 (left) and PD-L1 mRNA levels (right) by qRT-PCR. The results (mean±SD of 3 determinations) are expressed as relative mRNA levels compared with that obtained for control DOX-untreated cells (assigned a value of 1) (B). Lysates from cells treated with or without 500 ng/ml DOX for 7 d were immunoblotted with the indicated antibodies (C). D–E. MDA-MB-231/tet-MUC1shRNA cells treated with or without 200 ng/ml DOX for 4 d were analyzed for MUC1 (left) and PD-L1 mRNA levels (right) by qRT-PCR (mean±SD of 3 determinations) (D). Lysates from cells treated with or without 200 ng/ml DOX for 7 d were immunoblotted with the antibodies (E).
Targeting the MUC1-C cytoplasmic domain downregulates PD-L1 expression
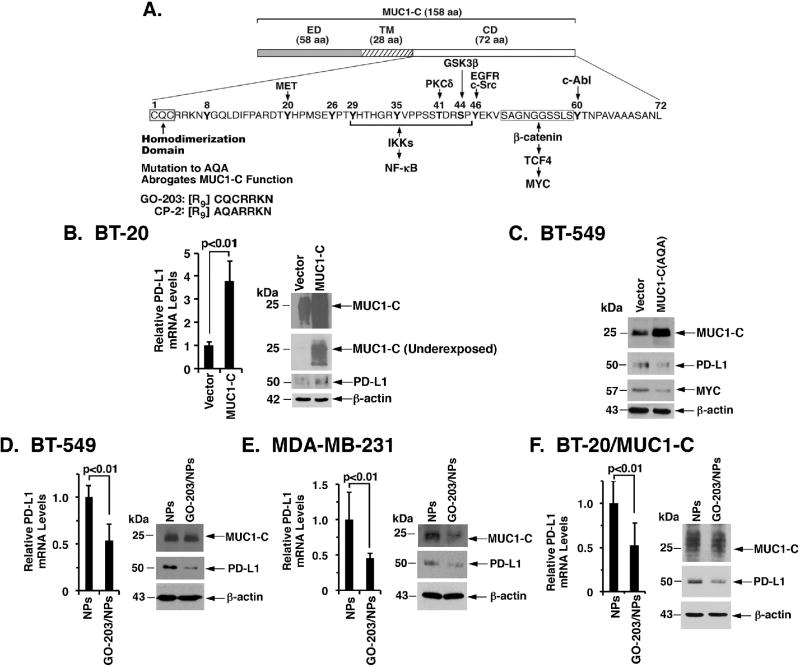
The MUC1-C subunit consists of a 58-amino acid (aa) ectodomain, a 28-aa transmembrane domain, and a 72-aa intrinsically disordered cytoplasmic domain (CD) (Fig. 2A). Enforced expression of MUC1-C has been linked to the induction of EMT (26). In concert with these and the above findings, we found that BT-20 cells transduced with an empty or MUC1-C vector and selected for growth in the presence of puromycin exhibit (i) modest upregulation of constitutive PD-L1 expression in BT-20/vector cells associated with the selective pressure, and (ii) more pronounced increases in PD-L1 mRNA and protein levels in BT-20/MUC1-C cells (Fig. 2B, left and right; an underexposed blot is shown to document MUC1-C upregulation). These results demonstrate that MUC1-C, and not the shed MUC1-N subunit, is sufficient for the induction of PD-L1 expression. Of note, the MUC1-C cytoplasmic domain includes a CQC motif (Fig. 2A), which is essential for the formation of MUC1-C homodimers and their import into the nucleus (12,32). In this regard, mutation of the CQC motif to AQA abrogates MUC1-C function (33) and, in the present studies, expression of the MUC1-C(AQA) mutant in BT-549 cells (20) resulted in downregulation of PD-L1 expression (Fig. 2C). The findings that the CQC motif is of importance to MUC1-C signaling provided the basis for developing the cell-penetrating GO-203 peptide to target this site (Fig. 2A)(33,34). In addition, GO-203 has been encapsulated in polymeric nanoparticles (GO-203/NPs) for sustained delivery in vitro and in animal models (25). Treatment of BT-549 (Fig. 2D, left and right) and MDA-MB-231 (Fig. 2E, left and right) cells with GO-203/NPs, but not empty NPs, was associated with downregulation of PD-L1 expression. Moreover, studies in BT-20 cells with MUC1-C overexpression (BT-20/MUC1-C) further demonstrated that targeting MUC1-C with GO-203 results in suppression of PD-L1 mRNA and protein (Fig. 2F, left and right). These findings thus demonstrated that MUC1-C is sufficient for the induction of PD-L1 expression and that this pathway is inhibited by targeting the MUC1-C CQC motif.
Figure 2. Targeting MUC1-C suppresses PD-L1 expression.
A. Schema of MUC1-C with the 58 amino acid (aa) extracellular domain (ED), the 28 aa transmembrane domain (TM), and the 72 aa cytoplasmic domain (CD). The CQC motif of the CD domain is indispensable for MUC1-C homodimerization, and is targeted by the cell-penetrating GO-203 peptide. Highlighted are interactions of the MUC1-C cytoplasmic domain with the NF-κB p65 and MYC pathways. B. BT-20 cells stably transduced to express a control or MUC1-C vector were analyzed for PD-L1 mRNA levels by qRT-PCR. The results (mean±SD of 3 determinations) are expressed as relative PD-L1 mRNA levels compared to that obtained for vector cells (assigned a value of 1) (left). Lysates were immunoblotted with the indicated antibodies (right). C. BT-549 cells were transfected to stably express an empty vector or MUC1-C(AQA) mutant. Lysates were immunoblotted with the indicated antibodies. D–F. BT-549 (D), MDA-MB-231 (E), and BT-20/MUC1-C (F) cells treated with empty NPs or 2.5 µM GO-203/NPs for 5 d were analyzed for PD-L1 mRNA levels by qRT-PCR. The results (mean±SD of 3 determinations) are expressed as relative PD-L1 mRNA levels compared to that obtained for empty NPs (assigned a value of 1)(left). Lysates from cells treated with empty NPs or 2.5 µM GO-203/NPs for 7 d were immunoblotted with the indicated antibodies (right).
MUC1-C drives PD-L1 by a MYC-dependent mechanism
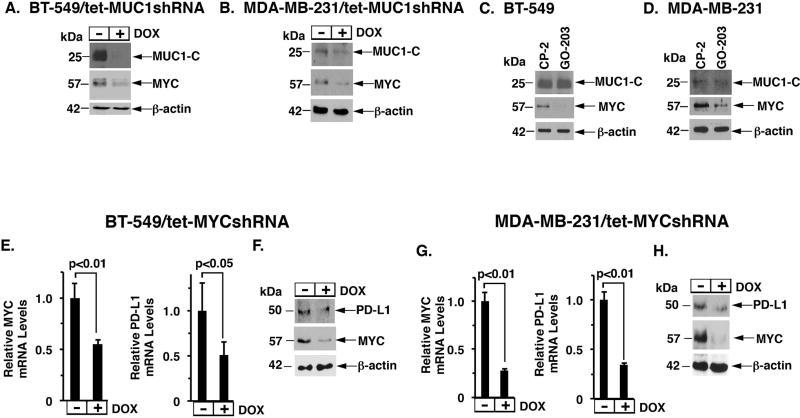
MUC1-C is associated with the upregulation of MYC (29,35) and drives MYC-mediated epigenetic reprogramming (20); however, there is no known relationship between MUC1-C→MYC signaling and PD-L1. In searching for evidence, we found that DOX treatment of BT-549/tet-MUC1shRNA (Fig. 3A) and MDA-MB-231/tet-MUC1shRNA (Fig. 3B) cells results in the downregulation of MYC expression. Treatment of BT-549 (Fig. 3C) and MDA-MB-231 (Fig. 3D) cells with GO-203, but not the control CP-2 peptide, was also associated with the suppression of MYC, supporting the premise that MUC1-C induces MYC expression in TNBC cells. To determine if MYC drives PD-L1 in TNBC cells, we established BT-549 and MDA-MB-231 cells with stable expression of a tet-MYCshRNA. DOX treatment of BT-549/tet-MYCshRNA was associated with suppression of MYC and PD-L1 mRNA (Fig. 3E, left and right) and protein (Fig. 3F). Similar results were obtained in DOX-treated MDA-MB-231/tet-MYCshRNA cells (Fig. 3G, left and right; and Fig. 3H). Treatment of BT-549 with JQ1, a BET bromodomain inhibitor, was also associated with downregulation of PD-L1 expression in BT-549 (Supplementary Fig. S2A) and BT-20/MUC1-C (Supplementary Fig. S2B) cells, providing further support for a MUC1-C→MYC→PD-L1 pathway in basal B TNBC cells.
Figure 3. MUC1-C→MYC signaling induces PD-L1 expression.
A-B. Lysates from BT-549/tet-MUCshRNA (A) and MDA-MB-231/tet-MUCshRNA (B) cells treated with or without DOX for 7 d were immunoblotted with the indicated antibodies. C–D. Lysates from BT-549 (C) and MDA-MB-231 (D) cells treated with 5 µM CP-2 or 5 µM GO-203 for 3 d were immunoblotted with the indicated antibodies. E–F. BT-549/tet-MYCshRNA cells treated with or without 200 ng/ml DOX for 1 d were analyzed for MYC and PD-L1 levels by qRT-PCR. The results (mean±SD of 3 determinations) are expressed as relative mRNA levels compared with that obtained for control DOX-untreated cells (assigned a value of 1)(E). Lysates from cells treated with or without 200 ng/ml DOX for 3 d were immunoblotted with the indicated antibodies (F). G–H. MDA-MB-231/tet-MYCshRNA cells treated with or without 200 ng/ml DOX for 1 d were analyzed for MYC and PD-L1 levels by qRT-PCR (mean±SD of 3 determinations) (G). Lysates from cells treated with or without 200 ng/ml DOX for 3 d were immunoblotted with the indicated antibodies (H).
MUC1-C induces PD-L1 expression by the NF-κB p65 pathway
MUC1-C activates the proinflammatory TAK1→IKK→NF-κB p65 pathway in cancer cells (Fig. 2A) (17,36,37). MUC1-C also binds directly to NF-κB p65 and thereby drives its downstream target genes, including (i) MUC1 itself in an autoinductive loop (17), and (ii) ZEB1 with activation of the EMT program in basal B TNBC cells (26). To investigate whether MUC1-C activates PD-L1 by an NF-κB p65-mediated pathway, we first showed that downregulation of MUC1-C in DOX-treated BT-549/tet-MUC1shRNA (Supplementary Fig. S3A) and MDA-MB-231/tet-MUC1shRNA (Supplementary Fig. S3B) cells results in the suppression of phospho-p65, but not p65, levels. To extend this analysis, we established BT-549 and MDA-MB-231 cells expressing a p65shRNA. Targeting NF-κB p65 in BT-549/p65shRNA (Supplementary Fig. S3C, left and right) and MDA-MB-231/p65shRNA (Supplementary Fig. S3D, left and right) cells was associated with decreases in PD-L1 mRNA and protein, indicating that MUC1-C drives PD-L1 expression by the NF-κB p65 pathway. In support of this contention, treatment of BT-549 (Supplementary Fig. S3E, left and right) and BT-20/MUC1-C (Supplementary Fig. S3F, left and right) cells with BAY-11–7085, an irreversible inhibitor of IκB phosphorylation, resulted in suppression of PD-L1 expression.
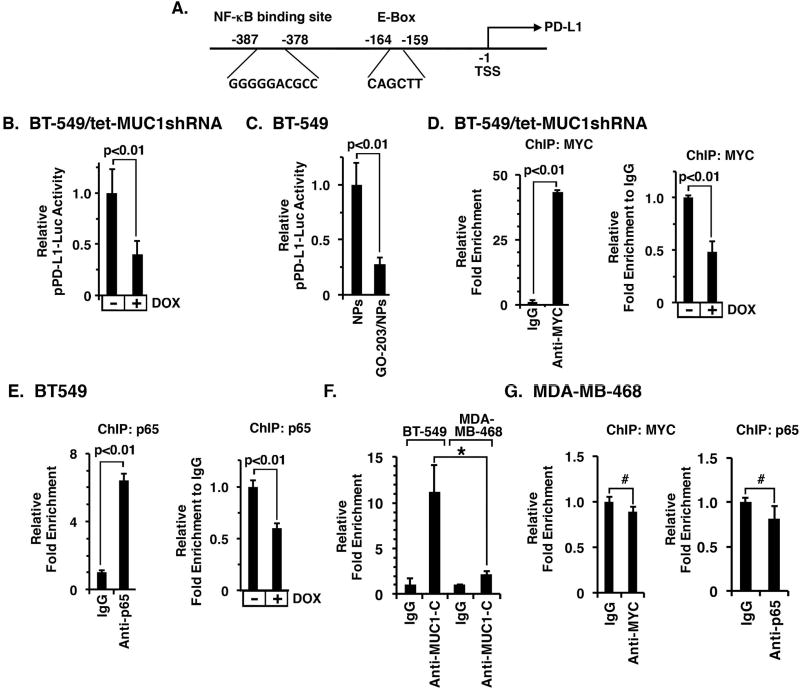
MUC1-C enhances MYC and NF-κB p65 occupancy on the PD-L1 promoter
The PD-L1 promoter contains (i) an E-box sequence (CAGCTT) for MYC binding at positions −164 to −159, and (ii) an NF-κB p65 binding site (GGGGGACGCC) at positions −387 to −378 upstream to the transcription start site (Fig. 4A) (35). To determine whether MUC1-C activates the PD-L1 promoter, we transfected DOX-treated BT-549/tet-MUC1shRNA cells with a PD-L1 promoter-luciferase reporter (pPD-L1-Luc). The results demonstrated that silencing MUC1-C suppresses pPD-L1-Luc activity (Fig. 4B). Targeting MUC1-C in BT-549 cells with GO-203/NP treatment also decreased activity of the pPD-L1-Luc reporter (Fig. 4C), indicating that MUC1-C activates the PD-L1 promoter. To our knowledge, it is not known if MYC or NF-κB p65 occupies the PD-L1 promoter in TNBC cells. Accordingly, we performed ChIP studies of chromatin from BT-549/tet-MUC1shRNA cells which demonstrated that MYC is detectable on the PD-L1 promoter (Fig. 4D, left) and that silencing MUC1-C decreases MYC occupancy (Fig. 4D, right). In a similar way, we found that NF-κB p65 also occupies the PD-L1 promoter by a MUC1-C-dependent mechanism (Fig. 4E, left and right). Notably, MUC1-C occupancy on the PD-L1 promoter was substantially greater in basal B BT-549 cells as compared to that found in basal A MDA-MB-468 cells, which have low to undetectable levels of PD-L1 expression (Fig. 4F). In addition, there was no significant detection of MYC or NF-κB p65 occupancy on the PD-L1 promoter in MDA-MB-468 cells (Fig. 4G, left and right), providing mechanistic evidence for the findings that MUC1-C drives PD-L1 in basal B, and not basal A, TNBC cells.
Figure 4. MUC1-C enhances MYC and NF-κB p65 occupancy on the PD-L1 promoter.
A. Schema of the pPD-L1 promoter with highlighting of the E-box at −159 to −164 and NF-κB binding site at −378 to −387 upstream to the transcription start site (TSS). B. BT-549/tet-MUC1shRNA cells cultured with or without DOX for 5 d were transfected with the pPD-L1-Luc reporter for 48 h and then assayed for luciferase activity. The results (mean±SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained for control DOX-untreated cells (assigned a value of 1). C. BT-549 cells treated with NPs or GO-203/NPs for 4 d were transfected with pPD-L1-Luc reporter for 48 h and then assayed for luciferase activity. The results (mean±SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained with empty NP-treated cells (assigned a value of 1). D. Soluble chromatin from BT-549/tet-MUC1shRNA cells was precipitated with anti-MYC or a control IgG (left). The final DNA samples were amplified by qPCR with primers for the PD-L1 promoter MYC binding region or GAPDH as a control. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). Soluble chromatin from 549/tet-MUC1shRNA cells cultured with or without DOX for 5 d was precipitated with anti-MYC or a control IgG. The final DNA samples were amplified by qPCR. The results (mean±SEM of three determinations) are expressed as the relative fold enrichment compared to that obtained for control DOX-untreated cells (assigned a value of 1) (right). E. Soluble chromatin from BT-549/tet-MUC1shRNA cells was precipitated with anti-NF-κB p65 or a control IgG (left). The final DNA samples were amplified by qPCR with primers for the PD-L1 promoter NF-κB binding region or GAPDH as a control. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). Soluble chromatin from BT-549/tet-MUC1shRNA cells cultured with or without DOX for 5 d was precipitated with anti-NF-κB p65 or a control IgG (right). The final DNA samples were amplified by qPCR. The results (mean±SEM of three determinations) are expressed as the relative fold enrichment compared to that obtained for control DOX-untreated cell chromatin (assigned a value of 1). F. Soluble chromatin from BT-549 and MDA-MB-468 cells was precipitated with anti-MUC1-C or a control IgG. The final DNA samples were amplified by qPCR with primers for the PD-L1 promoter or GAPDH as a control. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG controls (assigned a value of 1). *p<0.05. G. Soluble chromatin from MDA-MB-468 cells was precipitated with anti-MYC (left), anti-NF-κB p65 (right) or a control IgG (left). The final DNA samples were amplified by qPCR with primers for the PD-L1 promoter or GAPDH as a control. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG controls (assigned a value of 1). #p>0.05.
MUC1-C drives PD-L1 expression in mouse Eo771 TNBC cells
To extend this line of investigation, we studied mouse Eo771 TNBC cells stably expressing human MUC1-C (Eo771/MUC1-C). Notably, Eo771/MUC1-C cells exhibited increased levels of PD-L1 mRNA (Supplementary Fig. S4A) and protein (Supplementary Fig. S4B) relative to that in control cells expressing an empty vector (Eo771/vector). In concert with the above studies in human TNBC cells, we also found that the MUC1-C→PD-L1 response is inhibited by treatment with JQ1 (Supplementary Fig. S4C, left and right) and BAY-11 (Supplementary Fig. S4D, left and right). In addition, treatment of the Eo771/MUC1-C cells with GO-203/NPs was associated with downregulation of PD-L1 mRNA and protein (Supplementary Fig. S4E, left and right), confirming that MUC1-C drives PD-L1 expression in mouse Eo771 cells by MYC- and NF-κB p65-mediated mechanisms.
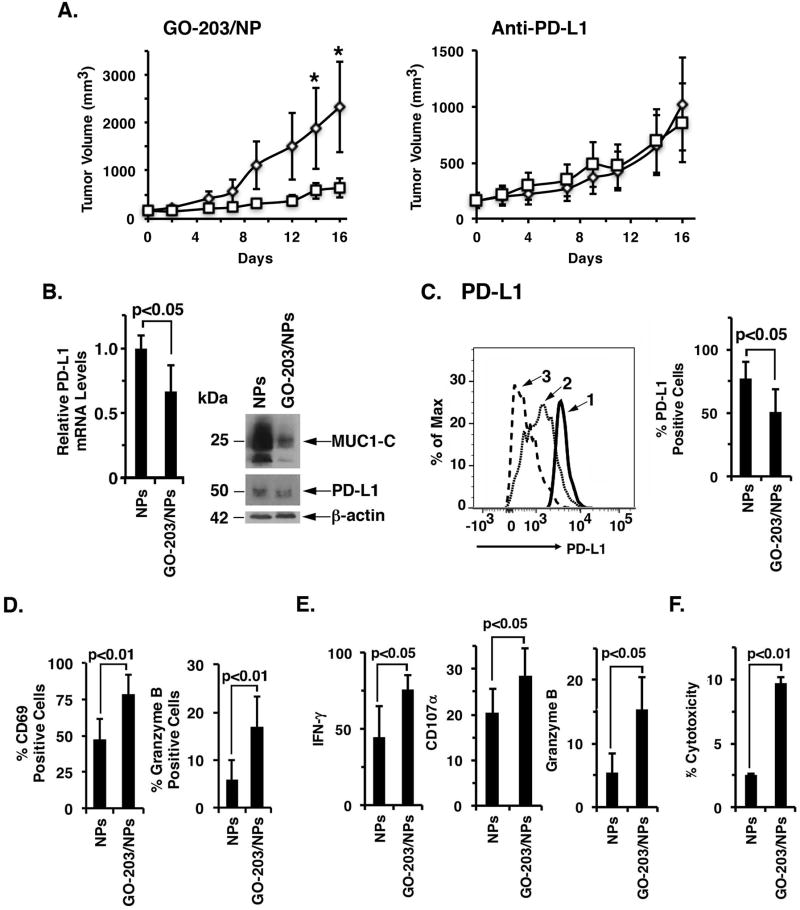
Targeting MUC1-C in Eo771/MUC1-C tumors suppresses PD-L1 expression and activates the tumor immune microenvironment
We next performed studies in the human MUC1 transgenic (MUC1.Tg) mouse model. The immune competent MUC1.Tg mice express the MUC1 transgene in normal tissues in a pattern and at levels consistent with that in human s (38). In addition, MUC1.Tg mice are tolerant to MUC1, thereby providing an experimental setting for engraftment of Eo771/MUC1-C cells. MUC1.Tg mice with established Eo771/MUC1-C tumors were treated with GO-203/NPs to assess the effects of targeting MUC1-C on the tumor microenvironment. GO-203/NP, but not anti-PD-L1, treatment was associated with inhibition of Eo771/MUC1-C tumor growth as compared to that obtained with respective controls (Fig. 5A, left and right). Analysis of the GO-203/NP-treated tumors on day 16 showed that targeting MUC1-C results in the downregulation of PD-L1 mRNA and protein (Fig. 5B, left and right). In addition, GO-203/NP treatment decreased PD-L1 expression on the Eo771/MUC1-C cell surface (Fig. 5C, left and right). Analysis of the TIL population also revealed that expression of the CD69 activation marker and granzyme B is upregulated in CD8+ T-cells after GO-203/NP treatment (Fig. 5D, left and right; Supplemental Figs. S5A and S5B). Moreover, and consistent with these results, in vitro stimulation assays demonstrated that tumor-infiltrating CD8+ T-cells from GO-203/NP-treated mice exhibit increases in expression of IFN-γ (Fig. 5E, left; Supplemental Fig. S6A), the degranulation marker CD107α (Fig. 5E, middle; Supplemental Fig. S6B) and granzyme B (Fig. 5E, right; Supplemental Fig. S6C). In support of these findings indicative of enhanced function, TILs from the GO-203/NP-treated mice were more effective in killing Eo771/MUC1-C tumor cells (Fig. 5F).
Figure 5. Targeting MUC1-C in Eo771/MUC1-C tumors activates the immune microenvironment.
A. Eo771/MUC1-C cells were injected subcutaneously into the flanks of MUC1.Tg mice. Left panel. Mice with established tumors of approximately 150 mm3 were pair-matched and then treated with empty NPs (diamonds) or 15 mg/kg GO-203/NPs (squares)(left). The results are expressed as tumor volume (mean±SEM; 5 mice per group). One of the tumors in the GO-203/NP-treated group was undetectable at the time of harvest. *p<0.05. Tumors were harvested on day 16 when the controls showed signs of necrosis. Right panel. In a subsequent experiment, mice were treated with PBS or 10 mg/kg anti-PD-L1 on days 1 and 6. Tumors in the control group showed signs of necrosis on day 17 when the study was terminated according to the animal protocol. B. Tumor cells were analyzed for PD-L1 mRNA levels by qRT-PCR. The results (mean±SD of 4 determinations) are expressed as relative mRNA levels compared with that obtained for empty NP-treated tumors (assigned a value of 1)(left). Lysates were immunoblotted with the indicated antibodies (right). C–E. Single cell suspensions were prepared for FACS analysis. C. In a representative histogram, tumor cells from NP-treated (profile #1) and GO-203/NP-treated (profile #2) mice were analyzed for PD-L1 expression (left). An isotype identical antibody was used as a control (profile #3)(left). The percentage of PD-L1-positive tumor cells is expressed as the mean±SD (4 tumors per group)(right). D. Tumor-infiltrating CD8+ T-cells were analyzed for CD69 and granzyme B expression. The results are expressed as the percentage (mean±SD; n=4) of CD69 (left) and granzyme B (right) positive cells. E. Tumor-infiltrating immune cells were isolated by Ficoll separation and stimulated with the Leucocyte Activation Cocktail. CD8+ T-cells were analyzed for expression of the CD107α degranulation marker (left), IFN-γ (middle) and granzyme B (right). The results are expressed as the percentage (mean±SD; n=4) of positive cells. F. Lymph nodes obtained from NP- and GO-203/NP-treated mice were disrupted into cell single suspensions. Effectors were plated in 96-well plates with Eo771/MUC1-C target cells at a 3:1 ratio. After 6 h, T-cell mediated cytotoxicity was assayed measuring LDH release. The results are expressed as percentage cytotoxicity (mean±SD; n=4).
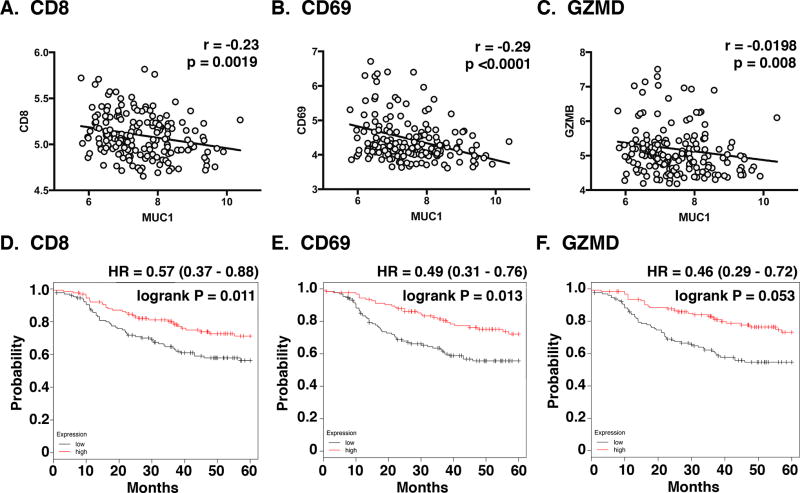
Correlation of MUC1 with T-cell activation in TNBC
To further understand the relationship between MUC1-C and T-cell activation in TNBCs, we performed bioinformatics analyses on the microarray dataset from the Gene Expression Omnibus (GSE25066). The results demonstrated that MUC1 expression correlates inversely with that obtained for CD8 (Fig. 6A), CD69 (Fig. 6B) and GZMB (Fig. 6C). Additionally, we found that higher levels of CD8 (Fig. 6D), CD69 (Fig. 6E) and GZMB (Fig. 6F) expression are associated with significant increases in disease-free survival of TNBC patients.
Figure 6. Correlation between MUC1 and T-cell activation in TNBCs.
A–C. Gene expression data of TNBCs was obtained from GSE25066 datasets. Correlation between MUC1 and CD8A/B (A), CD69 (B) and GZMB (C) expression (C) was assessed using the Spearman’s correlation coefficient, where p<0.05 was considered as statistically significant. D–F. Kaplan-Meier plots comparing the Relapse-Free Survival (RFS) of TNBC patients. Patients were stratified with high (red) or low (black) expression of CD8 (D), CD69 (E) and GZMB (F) against the median. The survival curves were compared using the log-rank test. HR, hazard ratio.
Discussion
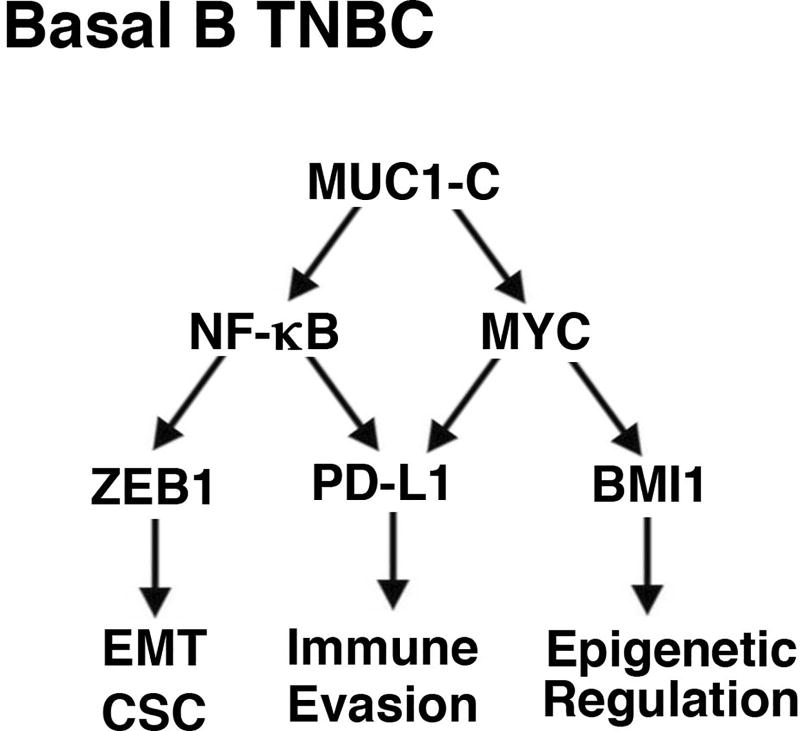
Evidence is accumulating that immunotherapy may represent an option for the treatment of TNBC. Inhibition of the PD-1/PD-L1 axis has been associated with responses in patients with advanced TNBC, indicating that immune evasion contributes to the pathogenesis of this refractory disease (10,11). In addition, the overexpression of PD-L1 in TNBCs, albeit by unclear mechanisms, has been linked to more aggressive disease and poor clinical outcomes (6–8). The present studies demonstrate that MUC1-C drives (i) constitutive PD-L1 expression in basal B BT-549, MDA-MB-231 and SUM-159 TNBC cells, which display mesenchymal and CSC characteristics (39–41)(Fig. 7), and (ii) inducible PD-L1 expression in basal A BT-20 TNBC cells. The results support a model in which MUC1-C activates the PD-L1 promoter in part by a MYC-dependent pathway. MUC1-C has been shown to activate MYC-mediated BMI1 expression and epigenetic alterations in basal B TNBC cells (20)(Fig. 7). Here, we show that targeting MUC1-C results in the downregulation of MYC, decreased occupancy of MYC on the PD-L1 promoter and suppression of pPD-L1-Luc reporter activation, all in support of a transcriptional mechanism. A MUC1-C→MYC→PD-L1 pathway was further supported by the findings that targeting MYC with inducible silencing or the JQ1 inhibitor suppresses PD-L1 expression. In concert with studies in NSCLC cells (30), the present results also demonstrate that MUC1-C induces PD-L1 by an NF-κB p65-mediated mechanism. Along these lines, MUC1-C activates the inflammatory NF-κB p65 pathway in basal B TNBC cells (17,26,37). MUC1-C binds directly to NF-κB p65 and promotes NF-κB p65 occupancy on its target gene, ZEB1, which in turn drives the ZEB1→miR-200c loop and the induction of EMT (17,26)(Fig. 7). By extension, we show here that targeting MUC1-C decreases NF-κB p65 occupancy on the PD-L1 promoter and suppresses activation of the pPD-L1-Luc reporter. Targeting NF-κB p65 also resulted in downregulation of PD-L1 expression, supporting activation of a MUC1-C→NF-κB p65→PD-L1 pathway. Of note, the MUC1-C→MYC and MUC1-C→NF-κB p65 pathways both have significant roles in driving PD-L1 expression in the basal B TNBC cells (Fig. 7), supporting potential cross-talk of these two transcription factors in activating the PD-L1 promoter. Further studies will thus be needed to determine whether MUC1-C occupancy the PD-L1 promoter is conferred by MYC- and/or NF-κB p65-dependent mechanisms.
Figure 7. Proposed model for MUC1-C-induced integration of PD-L1 expression with EMT, CSC state and epigenetic programming in basal B TNBC cells.
The present results demonstrate that MUC1-C activates the PD-L1 gene by NF-κB p65- and MYC-mediated mechanisms. MUC1-C→NF-κB p65 signaling also activates the ZEB1 gene and thereby represses miR-200c with induction of the EMT program and CSC state (26,31). Additionally, the MUC1-C→NF-κB p65 pathway promotes epigenetic reprogramming by induction of genes encoding DNMT1/3b and components of the PRC2 complex, including EZH2 (18,21). Moreover, MUC1-C-induced activation of the MYC pathway induces BMI1 expression and PRC1-mediated epigenetic alterations (20). In this way, MUC1-C integrates PD-L1 expression with the EMT program, CSC state and epigenetic reprogramming in basal B TNBC cells.
As an extension of the studies in human TNBC cells, we established mouse Eo771 TNBC cells that stably express human MUC1-C and confirmed that MUC1-C induces PD-L1 expression in this model. The results further indicate that, as observed in human TNBC cells, MUC1-C-induced increases in PD-L1 in Eo771/MUC1-C cells are mediated by MYC and NF-κB. The Eo771/MUC1-C cells also provided an opportunity to assess the effects of targeting MUC1-C in immune competent MUC1.Tg mice bearing established Eo771/MUC1-C tumors. Notably, and in contrast to GO-203/NPs, anti-PD-L1 treatment had little if any effect on growth of the Eo771/MUC1-C tumors. Importantly, GO-203/NP treatment of Eo771/MUC1-C cells growing in vitro and as tumors in MUC1.Tg mice was associated with downregulation of PD-L1 expression. We also found that targeting MUC1-C and thereby suppression of PD-L1 in Eo771/MUC1-C tumors is associated with activation of the CD8+ T-cell population. In support of that contention and in concert with studies in NSCLC cells (42), we found that GO-203/NP treatment results in upregulation of the CD69 activation marker and granzyme B in the CD8+ T-cell population. The CD8+ T-cells obtained from GO-203/NP-treated mice were also more effective in killing Eo771/MUC1-C cells. In patients with TNBCs treated with adjuvant or neoadjuvant chemotherapy, the presence of TILs is associated with improved clinical outcomes (2,43,44). Noteworthy, however, is the lack of information regarding whether the TILs were activated or not in these trials. For these reasons, we analyzed datasets obtained from basal A and B TNBC patients and, interestingly, found that MUC1 expression predicts for decreases in mRNA levels of intratumoral (i) CD8, and (ii) the CD69 and granzyme B markers of T-cell activation. In addition, our analysis of the databases showed that decreases in CD8, CD69 and GZMB expression each correlated with more aggressive disease. A more detailed analysis will be of interest as larger datasets that distinguish between basal A and B TNBCs become available. Nonetheless, these findings and those in our in vitro and mouse model studies collectively support a previously unreported role for MUC1-C in suppressing immune recognition and destruction.
Upregulation of PD-L1 has been linked to EMT and poor clinical outcomes in breast, lung and other types of cancers (45–47), suggesting that immune evasion contributes to the invasive and metastatic phenotype. The present studies provide new insights into the integration of increased PD-L1 expression with the EMT process. In this way, MUC1-C drives EMT in basal B TNBC cells by activation of the inflammatory NF-κB p65 pathway and thereby induction of the EMT transcription factor ZEB1 (26)(Fig. 7). In turn, ZEB1 (i) promotes loss of polarity by suppression of polarity factors, such as CRB3, and (ii) activates the HIPPO/YAP pathway with induction of MYC in TNBC cells (19). ZEB1 also decreases expression of the miR-200c tumor suppressor, which is a negative regulator of PD-L1 (26). In this regard, recent work has demonstrated that MUC1-C increases PD-L1 expression in NSCLC cells by an NF-κB-mediated mechanism (30,42) and in AML cells by suppression of miR-200c (48), supporting the premise that MUC1-C regulates PD-L1 by transcriptional and posttranscriptional mechanisms which are dependent on cell context. The MUC1-C→NF-κB p65 and MUC1-C→MYC pathways also have the capacity to induce epigenetic modifications needed for the associated changes in gene expression for the EMT program and CSC state (18,20–22)(Fig. 7). Of potential interest is why PD-L1 and EMT would be integrated in basal B TNBC cells. One explanation is that invasive and metastatic cancer cells require a defense against immune recognition. Another possibility is that the overexpression of MUC1-C with induction of PD-L1 and EMT represents an appropriation and exploitation by cancer cells of an epithelial stress response that evolved to repair damaged epithelia (22).
Regarding translational relevance of targeting MUC1-C, a Phase I trial of GO-203 in patients with advanced solid tumors has demonstrated an acceptable safety profile for this agent. Based on these and the present findings, the formulation of GO-203 in NPs is being advanced for more prolonged and less frequent dosing of patients with TNBC in Phase I-II studies as a potential approach for suppressing PD-L1 in the tumor cells themselves. These trials of GO-203/NPs will be integrated with the administration of immune checkpoint inhibitors and other immunotherapeutic approaches.
Supplementary Material
Acknowledgments
Financial Support: This publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers R01 CA097098 (D.W. Kufe), R01 CA166480 (D.W. Kufe) and R21 CA216553 (D.W. Kufe).
Abbreviations
- TNBC
triple-negative breast cancer
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- PD-L1
programmed death ligand 1
- PD-1
programmed death 1
- EMT
epithelial-mesenchymal transition
- NSCLC
non-small cell lung cancer
- MUC1.Tg mice
human MUC1 transgenic mice
- TILs
tumor-infiltrating lymphocytes
- IFN-γ
interferon-γ
- CD
cytoplasmic domain
- DOX
doxycycline
- NP
polymeric nanoparticle
- NSCLC
non-small cell lung cancer
Footnotes
Potential Conflict of Interest: The authors declare competing financial interests, D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
References
- 1.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Teijido P, Cabal ML, Fernandez IP, Perez YF. Tumor-infiltrating lymphocytes in triple negative breast cancer: The future of immune targeting. Clin Med Insights Oncol. 2016;10:31–9. doi: 10.4137/CMO.S34540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groschel S, Bommer M, Hutter B, Budczies J, Bonekamp D, Heining C, et al. Integration of genomics and histology revises diagnosis and enables effective therapy of refractory cancer of unknown primary with PDL1 amplification. Cold Spring Harb Mol Case Stud. 2016;2:a001180. doi: 10.1101/mcs.a001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soliman H, Khalil F, Antonia S. PD-L1 expression is increased in a subset of basal type breast cancer cells. PLoS One. 2014;9:e88557. doi: 10.1371/journal.pone.0088557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckers RK, Selinger CI, Vilain R, Madore J, Wilmott JS, Harvey K, et al. Programmed death ligand 1 expression in triple-negative breast cancer is associated with tumour-infiltrating lymphocytes and improved outcome. Histopathology. 2016;69:25–34. doi: 10.1111/his.12904. [DOI] [PubMed] [Google Scholar]
- 6.Guo L, Li W, Zhu X, Ling Y, Qiu T, Dong L, et al. PD-L1 expression and CD274 gene alteration in triple-negative breast cancer: implication for prognostic biomarker. Springerplus. 2016;5:805. doi: 10.1186/s40064-016-2513-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muenst S, Schaerli AR, Gao F, Daster S, Trella E, Droeser RA, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2014;146:15–24. doi: 10.1007/s10549-014-2988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabatier R, Finetti P, Mamessier E, Adelaide J, Chaffanet M, Ali HR, et al. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget. 2015;6:5449–64. doi: 10.18632/oncotarget.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali HR, Glont SE, Blows FM, Provenzano E, Dawson SJ, Liu B, et al. PD-L1 protein expression in breast cancer is rare, enriched in basal-like tumours and associated with infiltrating lymphocytes. Ann Oncol. 2015;26:1488–93. doi: 10.1093/annonc/mdv192. [DOI] [PubMed] [Google Scholar]
- 10.Pusztai L, Karn T, Safonov A, Abu-Khalaf MM, Bianchini G. New strategies in breast cancer: Immunotherapy. Clin Cancer Res. 2016;22:2105–10. doi: 10.1158/1078-0432.CCR-15-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: Phase Ib KEYNOTE-012 study. J Clin Oncology. 2016 May 2 doi: 10.1200/JCO.2015.64.8931. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–81. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raina D, Agarwal P, Lee J, Bharti A, McKnight C, Sharma P, et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10:e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK→C/EBPβ-mediated induction of aldehyde dehydrogenase activity in breast cancer cells. J Biol Chem. 2013;288:30829–903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raina D, Uchida Y, Kharbanda A, Rajabi H, Panchamoorthy G, Jin C, et al. Targeting the MUC1-C oncoprotein downregulates HER2 activation and abrogates trastuzumab resistance in breast cancer cells. Oncogene. 2014;33:3422–31. doi: 10.1038/onc.2013.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajabi H, Ahmad R, Jin C, Kosugi M, Alam M, Joshi M, et al. MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem. 2012;287:10703–13. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res. 2009;69:7013–21. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajabi H, Tagde A, Alam M, Bouillez A, Pitroda S, Suzuki Y, et al. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016;35:6439–45. doi: 10.1038/onc.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alam M, Bouillez A, Tagde A, Ahmad R, Rajabi H, Maeda T, et al. MUC1-C represses the Crumbs complex polarity factor CRB3 and downregulates the Hippo pathway. Mol Cancer Res. 2016;14:1266–76. doi: 10.1158/1541-7786.MCR-16-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiraki M, Maeda T, Bouillez A, Alam M, Tagde A, Hinohara K, et al. MUC1-C activates BMI1 in human cancer cells. Oncogene. 2016 Nov 28; doi: 10.1038/onc.2016.439. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajabi H, Hiraki M, Tagde A, Alam M, Bouillez A, Christensen CL, et al. MUC1-C activates EZH2 expression and function in human cancer cells. Sci Rep. 2017 Aug 7;7 doi: 10.1038/s41598-017-07850-0. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajabi H, Kufe D. MUC1-C oncoprotein integrates a program of EMT, epigenetic reprogramming and immune evasion in human carcinomas. BBA Reviews on Cancer. 2017;1868:117–22. doi: 10.1016/j.bbcan.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agata N, Kawano T, Ahmad R, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68:6136–44. doi: 10.1158/0008-5472.CAN-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David JM, Hamilton DH, Palena C. MUC1 upregulation promotes immune resistance in tumor cells undergoing brachyury-mediated epithelial-mesenchymal transition. Oncoimmunology. 2016;5:e1117738. doi: 10.1080/2162402X.2015.1117738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, et al. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res. 2015;21:2338–47. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33:1680–9. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Lee LF, Fisher TS, Jessen B, Elliott M, Evering W, et al. Combination of 4-1BB agonist and PD-1 antagonist promotes antitumor effector/memory CD8 T cells in a poorly immunogenic tumor model. Cancer immunology research. 2015;3:149–60. doi: 10.1158/2326-6066.CIR-14-0118. [DOI] [PubMed] [Google Scholar]
- 28.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–81. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, et al. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127:2587–97. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouillez A, Rajabi H, Jin C, Samur M, Tagde A, Alam M, et al. MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small cell lung cancer. Oncogene. 2017 Mar 13; doi: 10.1038/onc.2017.47. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget. 2014;5:2622–34. doi: 10.18632/oncotarget.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–30. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 33.Kufe D. Functional targeting of the MUC1 oncogene in human cancers. Canc Bio Ther. 2009;8:1201–7. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–9. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouillez A, Rajabi H, Pitroda S, Jin C, Alam M, Kharbanda A, et al. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76:1538–48. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34:5187–97. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, et al. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–27. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowse GJ, Tempero RM, VanLith ML, Hollingsworth MA, Gendler SJ. Tolerance and immunity to MUC1 in a human MUC1 transgenic murine model. Cancer Res. 1998;58:315–21. [PubMed] [Google Scholar]
- 39.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32:35–48. doi: 10.3233/BD-2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouillez A, Adeegbe D, Jin C, Hu X, Tagde A, Alam M, et al. MUC1-C promotes the suppressive immune microenvironment in non-small cell lung cancer. Oncoimmunology. 2017;6:e1338998. doi: 10.1080/2162402X.2017.1338998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959–66. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol. 2015;33:983–891. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 45.Alsuliman A, Colak D, Al-Harazi O, Fitwi H, Tulbah A, Al-Tweigeri T, et al. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol Cancer. 2015;14:149. doi: 10.1186/s12943-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou Y, Diao L, Parra Cuentas ER, Denning WL, Chen L, Fan YH, et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016;22:3630–42. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noman MZ, Janji B, Abdou A, Hasmim M, Terry S, Tan TZ, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology. 2017;6:e1263412. doi: 10.1080/2162402X.2016.1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pyzer AR, Stroopinsky D, Rajabi H, Washington A, Tagde A, Coll M, et al. MUC1 mediated induction of myeloid-derived suppressor cells in patients with acute myeloid leukemia. Blood. 2017;129:1791–801. doi: 10.1182/blood-2016-07-730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.