Abstract
Studies in mammals, including humans, have reported age-related changes in microbiota dynamics. A major challenge, however, is to dissect the cause and effect relationships involved. Invertebrate model organisms such as the fruit fly Drosophila and the nematode Caenorhabditis elegans have been invaluable in studies of the biological mechanisms of aging. Indeed, studies in flies and worms have resulted in the identification of a number of interventions that can slow aging and prolong life span. In this review, we discuss recent work using invertebrate models to provide insight into the interplay between microbiota dynamics, intestinal homeostasis during aging and life span determination. An emerging theme from these studies is that the microbiota contributes to cellular and physiological changes in the aging intestine and, in some cases, age-related shifts in microbiota dynamics can drive health decline in aged animals.
Keywords: Intestinal barrier, Microbiome, Dysbiosis, Longevity, Mortality
Introduction
Providing insight into the biological mechanisms of aging is a pressing challenge with enormous biomedical significance. For many years, studies of the biology of aging were largely descriptive and correlative in nature. However, in the last two decades, pioneering work primarily using invertebrate model organisms has shed light on the molecular and cellular mechanisms of aging [1]. The nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster have been at the forefront of these efforts. Studies in both flies and worms have identified a number of genetic and pharmacological interventions that ameliorate aging and prolong life span [2, 3]. The ultimate goal of this work is to identify novel therapeutic approaches to promote healthy aging, i.e. to delay the onset of pathology or disease, in humans [3–5].
Although invertebrate models have been successfully exploited to identify prolongevity interventions, the pathophysiological underpinnings of most of these interventions are not clearly defined.
In recent years, a number of studies have shown that the intestine represents a critical target organ for genetic manipulations that prolong life span [6–15]. One interpretation of these findings is that maintaining intestinal homeostasis during aging is an important determinant of health and viability at the organismal level [11, 15]. A logical extension of this idea is that alterations in the intestinal microbiota could influence organismal health during aging. In this review, we discuss recent work using invertebrate models to better understand the interplay between age-related changes in the gut microbiota, intestinal homeostasis and longevity.
Modeling the human microbiota: the role of invertebrate model systems
The human gastrointestinal tract houses a complex microbial population, the taxonomic membership of which is greatly impacted by environmental, lifestyle and dietary factors. Healthy adult individuals show significant inter-individual variability in microbiota composition. However, within a healthy individual this population appears to remain relatively stable over time [16, 17]. In contrast, the microbiota of individuals with ill health differs significantly from that of healthy individuals. Microbial populations associated with ill health have been described as imbalanced relative to those of healthy controls, and this state of imbalance is referred to as dysbiosis. The inherent variability of healthy microbiota populations makes dysbiosis difficult to define and no consistent marker has yet been identified. However, a reduced overall diversity of taxa characterized by preferential loss of beneficial organisms and increased abundance of pathogens or pathobiont species appears to be an accepted signature of dysbiosis in much of the current literature [18].
Dysbiosis has now been associated with a large number of disease states, many of which are age related, and has also been linked with frailty in the elderly (reviewed in [19–21]). This has led to speculation regarding the role of the intestinal microbiota in maintaining host health throughout the life course. Given that the correlations between dysbiosis and ill health have primarily been identified in cross-sectional studies, the cause and effect relationship between microbial dynamics and health status remains uncertain. Longitudinal studies of the onset of type 2 diabetes and inflammatory bowel disease are currently being carried out by the Integrative Human Microbiome Project [22] and these studies should shed light on the temporal relationship between microbial dynamics and disease onset/development. However, experimental models are required to establish causality and to identify the molecular mechanisms that define the microbiota–host health axis.
The mouse is currently the most commonly used model organism in microbiota-based studies, largely because, as a mammal, its gastrointestinal tract shares significant similarity with that of humans and because of the availability of a large number of relevant disease models. However, high maintenance costs limit its use in aging studies and particularly in life span analysis. The application of the invertebrate model organisms, the fruit fly D. melanogaster and the nematode worm C. elegans, to the study of host–microbe interactions during aging is a natural extension of their productive history of use in understanding the molecular mechanisms that modulate longevity. Both the fly and the worm have short life spans, lasting on average 2 months and 2–3 weeks, respectively, in addition to fast reproduction, an extensive repertoire of molecular and genetic tools and low maintenance costs. This makes them ideal for life span analyses that require large population sizes.
The fly and the worm provide model systems of differing complexity. The intestinal tract of the worm consists of a simple tube of enterocyte cells [23] typically associated with a single bacterial species, Escherichia coli, in laboratory populations. In contrast, the Drosophila intestine is more complex and contains multiple cell types and regional compartmentalization [24, 25] associated with a more diverse microbial population. Extensive research into the aging process of both animals has yielded a number of biomarkers of intestinal and organismal aging, several of which can be measured without killing the animal [15, 26, 27]. This ensures that the aging process, and health status, of individual animals can be followed in longitudinal studies that encompass the entire life span. Critically, in both organisms the molecular pathways regulating metabolic and innate immune function are evolutionarily conserved. Therefore, these model organisms are ideally placed to drive the discovery of molecular mechanisms that underlie host–microbe associations, and to study their relevance to life span and the pathophysiology of aging.
Age-related changes in microbiota dynamics
The human intestinal microbiota is dominated by the bacterial phyla Bacteroidetes, Firmicutes, Actinobacteria and Proteobacteria [28]. Other microorganisms, such as viruses and fungi, are also present but have not been well studied. While there are some species that are believed to be present in the majority of healthy individuals, the high inter-individual variability in microbiota composition has led to debate regarding the existence of common community structures. Despite efforts to group microbiota profiles into enterotypes, the current literature continues to support the view that each microbiota profile may be uniquely situated along a continuous gradient of taxa abundances [16, 17, 29, 30]. In addition to the resident intestinal microbiota, ingestion of food-borne microbes contributes a transient microbial population that has the potential to impact host health (reviewed in [31]).
While in healthy adults the intestinal microbial community appears relatively stable over a period of months, aging results in significant changes to community structure. Reported changes in taxonomic composition with age vary somewhat between studies, but appear to include shifts in the Bacteriodetes:Firmicutes ratio and Clostridium groups, and increased carriage of Proteobacteria members, particularly the Gammaproteobacteria [32–34]. Notably, the most significant changes in microbiota composition are correlated with health status rather than with chronological age. Age-related changes have been shown to correlate with diet, residence status and health [35, 36], levels of inflammation [34] and measures of frailty [37, 38]. While the aged intestinal microbiota often shows the features of dysbiosis detailed above, i.e., reduced diversity and a loss of beneficial taxa, this is complicated by the significant differences in lifestyle, diet and health among the aged population. Importantly, reduced diversity is associated with frailty and reduced cognitive performance, rather than with chronological age [19]. In contrast, the risk of malnutrition in the elderly is associated with increased microbiota diversity, and particularly the Clostridiales subpopulation. Counterintuitively, this population is also associated with frailty and with long stay residential care [19]. Overall, inter-individual variability is increased when compared with young, healthy controls [31] and this variability challenges attempts to define age-associated signatures of dysbiosis. In the mouse, aging also influences microbiota composition. However, whether these changes correlate with frailty is unclear due to a low sample size [39].
Both C. elegans and Drosophila are associated with complex bacterial populations in their native habitats. These communities are dominated by species from the phyla Proteobacteria and Firmicutes, with members of Bacteroidetes and Actinobacteria also fairly abundant, albeit at lower levels than found in the human intestinal microbiota [40–45]. Therefore, at this high taxonomic level there is significant overlap in the bacterial taxa associated with the worm, fly, mouse and humans. There is little evidence of a core microbiota in either the fly or the worm, and inter-individual variability is high [41, 42]. Both invertebrate models feed on microbes present in their environment, so their intestinal microbiota is closely associated with their food substrate. The food source is primarily bacteria for C. elegans and yeasts for Drosophila. However, in both cases intestinal microbiota composition is not simply determined by the bacterial proportions present in the food substrate, but is driven by the host and impacted by developmental stage and genotype [41, 46]. Remarkably, there is also strong evidence that the impact of these intestinal microbes is not exclusively nutritional [40, 47, 48].
In laboratory populations, C. elegans are typically maintained on a diet of E. coli, historically due to the availability of E. coli in research laboratories. This C. elegans–E. coli association is therefore the simplest host–microbe model system available. Several groups have reported that aged worms show a significant accumulation of E. coli in their intestines [49–51]. Laboratory Drosophila strains host an intestinal microbiota with a composition that ranges from 3 to 30 bacterial species and is highly variable depending on the laboratory, genotype and diet [52–54]. In addition to bacterial species, fungi and other microbial eukaryotes, Archaea and viral taxa add to the complexity of this microbial population [54]. However, only the bacterial component has been extensively studied to date. A number of studies have demonstrated that microbial load in the Drosophila intestine increases with age [9, 54–57]. Our recent work has shown that increased microbial loads are also associated with a shift in bacterial composition toward a greater proportion of Proteobacteria species, in particular members of the Gammaproteobacteria (including the genera Escherichia, Providencia, Haemophilus, Actinobacteria, Pseudomonas and Vibrio), and reduced Firmicutes levels [54]. Critically, we have demonstrated that, as in humans, the largest changes in both microbial load and species composition in the Drosophila intestinal microbiota are more closely associated with the health status of the animal than they are with chronological age [54]. This suggests that the fundamental principles that underlie intestinal functional decline, and organismal decline, may be highly evolutionarily conserved. A key challenge for the field is the identification of these principles, and how the intestinal microbiota may contribute to this process.
Microbial dynamics and the pathophysiology of aging
Recent studies have demonstrated the importance of maintaining intestinal homeostasis to whole organism health and longevity [11, 15]. To understand the role of the intestinal microbiota in age-related decline, we, therefore, need to identify both the mechanisms underlying microbial contributions to intestinal decline, and the impact of intestinal decline on distal tissues. The aging intestines of both worms and flies show morphological decline. In C. elegans, this includes a loss of nuclei, changes to the shape and size of the intestine, and a loss of microvilli [58]. Increased bacterial loads correlate with age-related decline in the worm intestine [58]. In Drosophila, a breakdown of regional identities [14, 24] and epithelial dysplasia [59–61] are driven by changes in intestinal stem cell proliferation and differentiation (the regulation of intestinal stem cells and dysplasia has been recently reviewed in [62]). Moreover, there is now clear evidence that preventing microbial exposure delays the onset of intestinal dysplasia [9, 55, 56, 63]. These studies suggest that changes in the microbial population during aging drive immune activation in the intestinal epithelium, which in turn drives epithelial dysplasia [9, 63]. However, the cause and effect relationships between dysbiosis, dysplasia and age-related immune activation during aging are not clear-cut. While interventions that reduce microbial exposure also reduce age-related immune activation [9, 56, 57], in Drosophila reducing age-related immune activation is itself sufficient to prevent microbial dysbiosis [9]. In addition, interventions that prevent intestinal stem cell proliferation and maintain gut compartmentalization are sufficient to reduce both age-related immune activation and microbial dysbiosis [14, 64]. Taken together, these studies demonstrate that the regulation of epithelial regeneration, control of the microbiota, and immune function is tightly networked. Whether age-related decline in the intestine occurs in a stereotyped manner and is always initiated by particular changes in one of these factors or whether it can be driven by stochastic changes in any of them remains an open question.
One way to address the timing of events that drive intestinal decline is to utilize markers of health status that do not require animal killing and to allow individual animals to be followed during aging. One such marker of age-related intestinal decline is intestinal barrier dysfunction. In the fly, the loss of intestinal barrier function can be assayed by the feeding of a non-absorbable dye, which in healthy animals is retained within the intestine but which in animals with intestinal barrier dysfunction will spread throughout the body cavity, visible through the cuticle [12, 65]. We have shown that the loss of barrier function in the fly is a clear indicator of age-related health decline and is a better predictor of organismal death than chronological age [65]. More recent work has demonstrated that intestinal barrier loss is an evolutionarily conserved biomarker of aging [66–69]. Using this dye-based assay to track flies prior to, and after, intestinal barrier loss we have demonstrated that while the most dramatic changes in microbial load occur following barrier dysfunction, changes in microbial composition are apparent several days prior to the loss of barrier function [54]. Indeed, using fecal sampling we discovered that alterations in the microbiota precede and predict the onset of intestinal barrier dysfunction in aged flies [54]. Flies that are raised axenically, without microbial exposure, also show a significant delay in the onset of intestinal barrier dysfunction [54]. Taken together, these data suggest that changes in the microbial population in the Drosophila intestine are an early event, and potentially a driving factor, in intestinal decline. In fact, a recent study by Thevaranjan, Bowdish and colleagues has demonstrated that the composition of the aged microbiota contributes to increased intestinal permeability in old mice [69], suggesting that the relationship between dysbiosis and intestinal permeability is conserved over large evolutionary distances.
While the studies discussed above have identified a number of biomarkers of age-related intestinal decline, work to characterize the impact of these changes on key intestinal functions such as nutrient uptake and water absorption is in its infancy. Changes in the expression of key metabolic regulators, such as the transcription factor Foxo, which is activated in response to reduced insulin signaling, and digestive enzymes suggest significant changes in nutrient uptake and processing may occur in the aging fly intestine [24, 70]. Similarly, we have shown that changes in the pH and water content of the fly’s fecal output occur with age, suggestive of declines in epithelial transport and absorption [54]. While we have shown that antibiotic treatment that prevents age-related dysbiosis can ameliorate some of these functional changes [54], more work is needed to establish the contribution of the microbiota to the digestive and absorptive functions of the intestinal epithelium.
In addition to a dramatic expansion in the intestinal microbial population, flies that have lost intestinal barrier function show reduced fat and glycogen stores, reduced mobility and systemic immune activation [65]. Changes in nutrient uptake and energy metabolism in the intestine and the loss of intestinal barrier function itself may be the causative factors driving whole organism health decline. This suggestion is supported by the ability of interventions that maintain intestinal homeostasis during aging to prolong life span (reviewed in [11]). Our recent work has shown that, in the fly, intestinal barrier dysfunction alone does not drive organismal decline. Rather, it is the combination of intestinal barrier dysfunction with expansion of the intestinal microbial population that drives systemic immune activation and mortality [54]. Indeed, antibiotic treatment of flies that have lost intestinal barrier function restores their remaining life span to that of age-matched healthy controls [54]. In an interesting parallel, it has been reported that the extent of bacterial accumulation in the intestine is inversely correlated with worm life span [50], again supporting a key role for host–microbe interactions in determining health and longevity.
Influence of gut microbiota on host life span
The most direct approach to examine the influence of the gut microbiota on host longevity is to examine the impact of axenic culture. There is a twofold increase in C. elegans life span under axenic culture [71]. However, because bacteria are also a food source it is not possible to exclude the possibility that dietary restriction contributes to extended life span under axenic conditions. Several studies have examined the impact of axenic culture on Drosophila life span. However, the results have not always been consistent. In a pioneering work, Brummel, Benzer and colleagues reported that the presence of bacteria in the first week of adult life was required for a normal life span [72]. In other words, axenic culture was reported to shorten life span in male flies. Interestingly, in the same study, it was reported that, late in life, the presence of bacteria can shorten life span. More specifically, treating flies with antibiotics from midlife onwards extended life span. However, a comprehensive follow-up study from Ren, Tower and colleagues reported that reducing bacterial load by axenic culture or antibiotics had no effect on male fly life span [57]. Differing nutrient conditions between experiments/laboratories may represent one potential explanation for these inconsistent reports of the impact of bacterial exposure on fly life span. Indeed, the presence of microbes can promote fly longevity under low nutrient conditions [73]. More recently, we have reported that axenic culture prolongs female fly life span [54]. Our findings are consistent with a report showing that antibiotic treatment can also extend fly life span [64]. In addition, our observation that axenic female flies are long-lived is consistent with reports from different laboratories showing that axenic flies show a delay in intestinal aging at a cellular level [9, 55, 56]. We also observed that preventing bacterial growth, via antibiotic feeding, significantly extended the life span of aged flies showing intestinal barrier failure [54]. To further assess the impact of the microbiota on life span, we fed homogenate from aged flies to young flies. As a result, there was an increase in bacterial load and shift in microbiota composition. Importantly, feeding homogenate from aged flies to young flies significantly decreased life span [54]. Taken together, these findings support the notion that the microbiota can contribute to age-onset mortality in Drosophila.
Beyond the impact of axenic culture, there have been numerous studies examining the impact of bacteria on worm life span. As discussed above, worms fed the standard laboratory E. coli strain OP50 have been reported to show an age-related accumulation of bacteria in the gut [49–51]. Moreover, feeding worms dead bacteria or treating with kanamycin can increase C. elegans life span perhaps by preventing or delaying E. coli accumulation in the intestinal lumen [49, 74]. Interestingly, worms fed a diet of E. coli deficient in coenzyme Q or with defects in ATP synthase live longer than worms fed OP50 [75, 76]. Follow-up studies revealed that respiratory-deficient Q-less E. coli do not accumulate in the aging intestinal tract to the same degree as OP50 [51]. Thus, there is a growing body of evidence to suggest that the age-related accumulation of bacteria in the worm intestine contributes to mortality. However, this hypothesis has been challenged by a recent study examining individual worms maintained on E. coli OP50 expressing GFP [77]. In this study, it was reported that there is considerable heterogeneity in the accumulation of bacteria in individual worms during aging, with many worms not appearing to accumulate bacteria at all [77]. This finding suggests that bacterial accumulation is not a universal contributor to age-onset mortality in worms. It is, therefore, possible that individual worms die from distinct pathologies. Interestingly, it has been reported that variable pathogenicity from E. coli is a major source of life span variability in individual worms [78]. In summary, while there may be individual variability involved, there is an emerging understanding that bacterial pathogenicity can contribute to mortality in aged worms.
Identifying mechanisms of microbial impact on host longevity
Beyond studies of age-related bacterial accumulation, work in the C. elegans–E. coli system has begun to characterize the molecular mechanisms by which the intestinal microbiota may influence host longevity. The simplicity of this model system has allowed the screening of E. coli mutants for their impact on host longevity. Recent studies have identified a number of mutants that increase host life span and demonstrated the specific genetic control of microbial regulation of host longevity [77, 79]. Inhibition of folate synthesis by E. coli, through mutation or pharmacologically, extends worm life span in a growth-independent manner [77, 80]. Interestingly, however, supplementation or restriction of C. elegans folate levels has no impact on longevity [77]. Altogether, these data demonstrate that the process of folate synthesis in E. coli, and not folate levels themselves, limits host life span. Whether microbial folate synthesis contributes to age-related decline in mammals remains to be investigated. Nevertheless, this work demonstrates the utility of simple model organisms in the rapid identification and characterization of the molecular mechanisms underlying microbial contributions to functional decline in the aging host. A number of additional studies have demonstrated that diffusible molecules originating in bacteria can impact worm life span [81]. C. elegans lack the enzyme nitric oxide (NO) synthase and, hence, rely upon bacterially derived NO. Remarkably, it has been demonstrated that bacterially derived NO enhances C. elegans life span [82]. In addition, small non-coding RNAs (ncRNAs) expressed endogenously by E. coli can modulate C. elegans longevity [83]. More specifically, feeding worms E. coli deficient in the ncRNA DsrA significantly increases life span. Therefore, bacterial-derived molecules can both shorten and prolong worm life span.
Future studies/outstanding questions
Genetic studies using flies and worms have resulted in the identification of genes and pathways that modulate aging and life span in mammals, including the TOR, AMPK and autophagy pathways [2, 5, 13, 84–86]. Hence, there is growing optimism regarding the ability to develop safe interventions to slow aging and increase healthy life span in humans [3, 4]. However, even in invertebrate models, the relevance of the microbiota in anti-aging interventions is only beginning to be explored. While bacterial-specific effects associated with worm longevity interventions have been reported [87–89], the pathophysiological mechanisms involved remain unclear. Moving forward, it will be important to determine whether the prolongevity effects of manipulating pathways such as AMPK/autophagy are mediated through improved commensal homeostasis. A better understanding of whether anti-aging interventions are influenced by or dependent upon microbiota composition may be critical in assessing whether these interventions are likely to prove effective in promoting healthy aging in a broad spectrum of aged individuals.
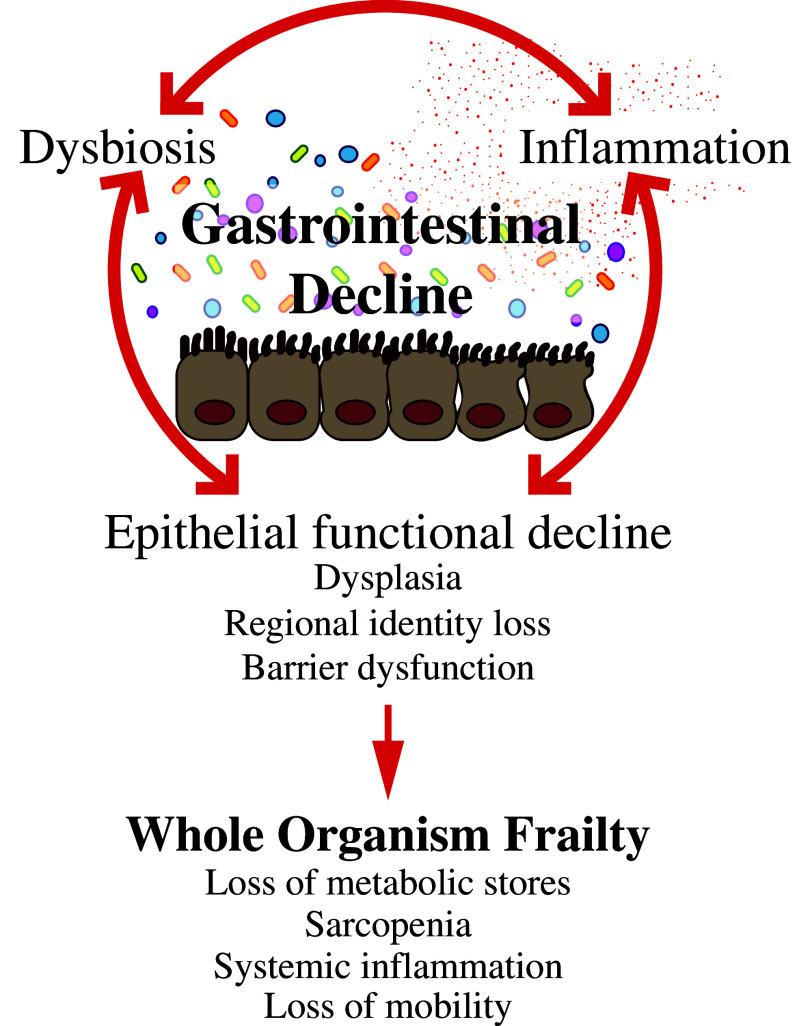
While this work is in its infancy, the apparent similarities in the nature of age-onset dysbiosis and its association with ill health between the invertebrate models and mammals are evident. The contribution of dysbiosis to the development of inflammation and the role of inflammation itself have been recognized in the context of both disease and age-related ill health. However, the importance of the reciprocal nature of the relationship between inflammation and dysbiosis has only recently become apparent. In line with data from mammalian models of inflammatory bowel disorders [90], studies in Drosophila have demonstrated that chronic immune activation can drive dysbiosis [91]. In addition, in Drosophila, prevention of age-related immune activation is sufficient to prevent dysbiosis [9], and in the mouse anti-TNF treatment can reduce age-related dysbiosis [69]. These findings are consistent with the reduction of dysbiosis following reduction of intestinal inflammation in pediatric Crohn’s disease patients [92]. Together, these data suggest a model whereby the development of dysbiosis and increased inflammation feed into each other resulting in a downward spiral that drives intestinal, and ultimately organismal, decline (Fig. 1). Therefore, it is critical to identify the factors that initiate this process and to characterize the molecular mechanisms that underpin microbiota–immune cross talk during aging.
Fig. 1.
Age-related gastrointestinal decline drives whole organism frailty. The cause and effect relationships between age-related dysbiosis, immune activation and functional decline of the intestinal epithelium are unclear. The experimental systems available in the worm and fly are currently being used to identify the molecular mechanisms that underlie these complex relationships
Despite the distinct differences between the invertebrate gut, and its associated microbiota, and that of mammalian models and humans, striking similarities in the relationship between dysbiosis, immune function and intestinal decline, in particular intestinal permeability, are apparent. These similarities are relevant in the context of both disease models and aging. Consequently, while we must be cautious in drawing direct parallels between any model system and humans, these findings demonstrate the value of the invertebrate models as discovery tools. The strength of invertebrate studies lies in the rapid identification of potential associations, markers, candidate genes or pathways, or other molecular processes for targeted testing in more complex and costly model systems. For example, a key finding from invertebrate studies has been an age-related increase in bacterial load [9, 50, 54, 57]. To our knowledge, the occurrence of changes in bacterial load in the mammalian intestine has not been fully investigated. Moving forward, a consideration of bacterial load, in addition to species composition, may be critical in understanding the impact of the microbiota on the aging intestine.
The host–microbiota association brings together a complex microbial ecosystem with host metabolic, immune and repair systems to form an intricate network of interacting parts. Identifying specific cause and effect relationships between microbial dynamics and host health outcomes, and the mechanistic basis of these relationships, therefore represents a significant challenge. In addressing this complexity, the experimental systems provided by the invertebrate models are a critical part of our toolkit.
Acknowledgements
We apologize to our colleagues whose work we were unable to discuss due to space limitations. D.W.W is supported by the National Institute on Aging (R01AG037514, R01AG049157, and R01AG040288). This review was written while D.W.W was a Julie Martin Mid‐Career Awardee in Aging Research supported by the Ellison Medical Foundation and AFAR.
Contributor Information
Rebecca I. Clark, Email: rebecca.clark2@durham.ac.uk
David W. Walker, Email: davidwalker@ucla.edu
References
- 1.Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol. 2013;75:621–644. doi: 10.1146/annurev-physiol-030212-183712. [DOI] [PubMed] [Google Scholar]
- 2.Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 3.Longo VD, Antebi A, Bartke A, Barzilai N, Brown-Borg HM, Caruso C, Curiel TJ, de Cabo R, Franceschi C, Gems D, Ingram DK, Johnson TE, Kennedy BK, Kenyon C, Klein S, Kopchick JJ, Lepperdinger G, Madeo F, Mirisola MG, Mitchell JR, Passarino G, Rudolph KL, Sedivy JM, Shadel GS, Sinclair DA, Spindler SR, Suh Y, Vijg J, Vinciguerra M, Fontana L. Interventions to slow aging in humans: are we ready? Aging Cell. 2015;14(4):497–510. doi: 10.1111/acel.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaeberlein M, Rabinovitch PS, Martin GM. Healthy aging: the ultimate preventative medicine. Science. 2015;350(6265):1191–1193. doi: 10.1126/science.aad3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115(4):489–502. doi: 10.1016/S0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 7.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6(10):e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156(1–2):109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hur JH, Bahadorani S, Graniel J, Koehler CL, Ulgherait M, Rera M, Jones DL, Walker DW. Increased longevity mediated by yeast NDI1 expression in Drosophila intestinal stem and progenitor cells. Aging (Albany NY) 2013;5(9):662–681. doi: 10.18632/aging.100595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rera M, Azizi MJ, Walker DW. Organ-specific mediation of lifespan extension: more than a gut feeling? Ageing Res Rev. 2013;12(1):436–444. doi: 10.1016/j.arr.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14(5):623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulgherait M, Rana A, Rera M, Graniel J, Walker DW. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 2014;8(6):1767–1780. doi: 10.1016/j.celrep.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Qi Y, Jasper H. Preventing age-related decline of gut compartmentalization limits microbiota dysbiosis and extends lifespan. Cell Host Microbe. 2016;19(2):240–253. doi: 10.1016/j.chom.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jasper H. Exploring the physiology and pathology of aging in the intestine of Drosophila melanogaster . Invertebr Reprod Dev. 2015;59(sup1):51–58. doi: 10.1080/07924259.2014.963713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoetendal EG, Akkermans ADL, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64(10):3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajilić-Stojanović M, Heilig HGHJ, Molenaar D, Kajander K, Surakka A, Smidt H, de Vos WM. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11(7):1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis M. An introduction to microbial dysbiosis. In: Henderson B, Nibali L, editors. The human microbiota and chronic disease. Dysbiosis as a cause of human pathology. Hoboken, New Jersey, United States: Wiley, Blackwell Publishing (Holdings) Ltd; 2016. [Google Scholar]
- 19.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350(6265):1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 21.Belizário JE, Napolitano M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front Microbiol. 2015 doi: 10.3389/fmicb.2015.01050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium TIHiRN The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 2014;16(3):276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGhee JD (2007) The C. elegans intestine. In: Community TCer (ed) WormBook. doi:10.1895/wormbook.1.133.1 [DOI] [PMC free article] [PubMed]
- 24.Buchon N, Osman D, David FP, Fang HY, Boquete JP, Deplancke B, Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3(5):1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster . Annu Rev Genet. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- 26.Tissenbaum HA. Using C. elegans for aging research. Invertebr Reprod Dev. 2015;59(sup1):59–63. doi: 10.1080/07924259.2014.940470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melov S. Geroscience approaches to increase healthspan and slow aging. F1000Res. 2016 doi: 10.12688/f1000research.7583.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium THMP Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R. Rethinking “Enterotypes”. Cell Host Microbe. 2014;16(4):433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derrien M, Vlieg JETvH. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23(6):354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, Weerd Hd, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, Dv Sinderen, O’Connor M, Harnedy N, O’Connor K, Henry C, O’Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O’Toole PW. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci. 2011;108(Supplement 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariat D, Firmesse O, Levenez F, Guimarăes VD, Sokol H, Doré J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, Brigidi P, De Vos W. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010 doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 36.Jeffery IB, Lynch DB, O’Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10(1):170–182. doi: 10.1038/ismej.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Tongeren SP, Slaets JPJ, Harmsen HJM, Welling GW. Fecal microbiota composition and frailty. Appl Environ Microbiol. 2005;71(10):6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, O’Toole PW, Spector TD, Steves CJ. Signatures of early frailty in the gut microbiota. Genome Med. 2016 doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langille MGI, Meehan CJ, Koenig JE, Dhanani AS, Rose RA, Howlett SE, Beiko RG. Microbial shifts in the aging mouse gut. Microbiome. 2014 doi: 10.1186/s40168-014-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samuel BS, Rowedder H, Braendle C, Félix M-A, Ruvkun G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc Natl Acad Sci. 2016;113(27):E3941–E3949. doi: 10.1073/pnas.1607183113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, Félix M-A, Schulenburg H. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 2016 doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong ACN, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7(10):1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandler JA, Morgan Lang J, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 2011 doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, Promislow DEL. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster . Appl Environ Microbiol. 2007;73(11):3470–3479. doi: 10.1128/AEM.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 2007;75(4):1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong ACN, Luo Y, Jing X, Franzenburg S, Bost A, Douglas AE. The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster . Appl Environ Microbiol. 2015;81(18):6232–6240. doi: 10.1128/AEM.01442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14(3):403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Shin SC, Kim S-H, You H, Kim B, Kim AC, Lee K-A, Yoon J-H, Ryu J-H, Lee W-J. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334(6056):670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 49.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161(3):1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Portal-Celhay C, Bradley ER, Blaser MJ. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans . BMC Microbiol. 2012;12:49. doi: 10.1186/1471-2180-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomez F, Monsalve GC, Tse V, Saiki R, Weng E, Lee L, Srinivasan C, Frand AR, Clarke CF. Delayed accumulation of intestinal coliform bacteria enhances life span and stress resistance in Caenorhabditis elegans fed respiratory deficient E. coli . BMC Microbiol. 2012;12:300. doi: 10.1186/1471-2180-12-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erkosar B, Storelli G, Defaye A, Leulier F. Host-intestinal microbiota mutualism: “learning on the fly”. Cell Host Microbe. 2013;13(1):8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster . Gut Microbes. 2012;3(4):307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, Rana A, Rera M, Pellegrini M, Ja WW, Walker DW. Distinct shifts in microbiota composition during drosophila aging impair intestinal function and drive mortality. Cell Rep. 2015;12(10):1656–1667. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio. 2014;5(3):e01117–e01214. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23(19):2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6(2):144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 58.McGee MD, Weber D, Day N, Vitelli C, Crippen D, Herndon LA, Hall DH, Melov S. Loss of intestinal nuclei and intestinal integrity in aging C. elegans . Aging Cell. 2011;10(4):699–710. doi: 10.1111/j.1474-9726.2011.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3(4):442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi N-H, Kim J-G, Yang D-J, Kim Y-S, Yoo M-A. Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging Cell. 2008;7(3):318–334. doi: 10.1111/j.1474-9726.2008.00380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park J-S, Kim Y-S, Yoo M-A. The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging. 2009;1(7):637–651. doi: 10.18632/aging.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Jasper H. Gastrointestinal stem cells in health and disease: from flies to humans. Dis Models Mech. 2016;9(5):487–499. doi: 10.1242/dmm.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen H, Zheng X, Zheng Y. Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia. Cell. 2014;159(4):829–843. doi: 10.1016/j.cell.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petkau K, Parsons BD, Duggal A, Foley E. A deregulated intestinal cell cycle program disrupts tissue homeostasis without affecting longevity in Drosophila. J Biol Chem. 2014;289(41):28719–28729. doi: 10.1074/jbc.M114.578708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci USA. 2012;109(52):21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dambroise E, Monnier L, Ruisheng L, Aguilaniu H, Joly JS, Tricoire H, Rera M. Two phases of aging separated by the Smurf transition as a public path to death. Sci Rep. 2016 doi: 10.1038/srep23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gelino S, Chang JT, Kumsta C, She X, Davis A, Nguyen C, Panowski S, Hansen M. Intestinal autophagy improves healthspan and longevity in C. elegans during dietary restriction. PLoS Genet. 2016 doi: 10.1371/journal.pgen.1006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kavanagh K, Brown RN, Davis AT, Uberseder B, Floyd E, Pfisterer B, Shively CA. Microbial translocation and skeletal muscle in young and old vervet monkeys. Age (Dordrecht, Netherlands) 2016;38(3):58. doi: 10.1007/s11357-016-9924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, Schertzer JD, Larché MJ, Davidson DJ, Verdú EF, Surette MG, Bowdish DME. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21(4):455.e454–466.e454. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karpac J, Biteau B, Jasper H. Misregulation of an adaptive metabolic response contributes to the age-related disruption of lipid homeostasis in Drosophila. Cell reports. 2013 doi: 10.1016/j.celrep.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans . Exp Gerontol. 2002;37(12):1371–1378. doi: 10.1016/S0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- 72.Brummel T, Ching A, Seroude L, Simon AF, Benzer S. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci USA. 2004;101(35):12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamada R, Deshpande SA, Bruce KD, Mak EM, Ja WW. Microbes promote amino acid harvest to rescue undernutrition in Drosophila. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans . Genetics. 2000;154(4):1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Larsen PL, Clarke CF. Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science. 2002;295(5552):120–123. doi: 10.1126/science.1064653. [DOI] [PubMed] [Google Scholar]
- 76.Saiki R, Lunceford AL, Bixler T, Dang P, Lee W, Furukawa S, Larsen PL, Clarke CF. Altered bacterial metabolism, not coenzyme Q content, is responsible for the lifespan extension in Caenorhabditis elegans fed an Escherichia coli diet lacking coenzyme Q. Aging Cell. 2008;7(3):291–304. doi: 10.1111/j.1474-9726.2008.00378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Virk B, Jia J, Maynard CA, Raimundo A, Lefebvre J, Richards SA, Chetina N, Liang Y, Helliwell N, Cipinska M, Weinkove D. Folate acts in E. coli to accelerate C. elegans aging independently of bacterial biosynthesis. Cell Rep. 2016;14(7):1611–1620. doi: 10.1016/j.celrep.2016.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez-Blanco A, Kim SK. Variable pathogenicity determines individual lifespan in Caenorhabditis elegans . PLoS Genet. 2011;7(4):e1002047. doi: 10.1371/journal.pgen.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khanna A, Kumar J, Vargas MA, Barrett L, Katewa S, Li P, McCloskey T, Sharma A, Naude N, Nelson C, Brem R, Killilea DW, Mooney SD, Gill M, Kapahi P. A genome-wide screen of bacterial mutants that enhance dauer formation in C. elegans . Sci Rep. 2016;6:38764. doi: 10.1038/srep38764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, Briggs Z, Hodge E, Edwards R, Ward J, Gems D, Weinkove D. Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biol. 2012;10:67. doi: 10.1186/1741-7007-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell. 2014;156(3):408–411. doi: 10.1016/j.cell.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Bacterial nitric oxide extends the lifespan of C. elegans . Cell. 2013;152(4):818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 83.Liu H, Wang X, Wang HD, Wu J, Ren J, Meng L, Wu Q, Dong H, Wu J, Kao TY, Ge Q, Wu ZX, Yuh CH, Shan G. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans . Nat Commun. 2012;3:1073. doi: 10.1038/ncomms2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burkewitz K, Zhang Y, Mair WB. AMPK at the nexus of energetics and aging. Cell Metab. 2014;20(1):10–25. doi: 10.1016/j.cmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gelino S, Hansen M (2012) Autophagy—an emerging anti-aging mechanism. J Clin Exp Pathol (Suppl 4):006 [DOI] [PMC free article] [PubMed]
- 86.Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11(6):453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans . Genes Dev. 2009;23(4):496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pang S, Curran SP. Adaptive capacity to bacterial diet modulates aging in C. elegans . Cell Metab. 2014;19(2):221–231. doi: 10.1016/j.cmet.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153(1):228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Craven M, Egan CF, Dowd SE, McDonough SP, Dogan B, Denkers EY, Bowman D, Scherl EJ, Simpson KW. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn’s disease. PLoS One. 2012;7(7):e41594. doi: 10.1371/journal.pone.0041594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dantoft W, Lundin D, Esfahani S, Engstrom Y. The POU/Oct transcription factor Pdm1/nub Is necessary for a beneficial gut microbiota and normal lifespan of Drosophila. J Innate Immun. 2016;8(4):412–426. doi: 10.1159/000446368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lewis James D, Chen Eric Z, Baldassano Robert N, Otley Anthony R, Griffiths Anne M, Lee D, Bittinger K, Bailey A, Friedman Elliot S, Hoffmann C, Albenberg L, Sinha R, Compher C, Gilroy E, Nessel L, Grant A, Chehoud C, Li H, Wu Gary D, Bushman Frederic D. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18(4):489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]