Abstract
Non-Hodgkin lymphomas include a biologically and clinically heterogeneous group of cancers distinguished by genetics, histology, and treatment outcomes. New discoveries regarding the genomic alterations and epidemiological exposures associated with these lymphomas have enhanced our understanding of factors that contribute to lymphomagenesis for specific subtypes. We explore the impact of normal B-cell biology engineered for recognizing a wide variety of antigens on the development of specific lymphoma subtypes, review lymphoma genetics, and examine the epidemiology of B-cell NHLs including recent investigations of risk factors for particular lymphoma subtypes based on large pooled analyses. Burkitt lymphoma, an aggressive form of B-cell NHL involving translocation of the MYC gene and an immunoglobulin gene has been associated with a history of eczema, hepatitis C, and occupation as a cleaner. Increased risk of diffuse large B-cell lymphoma has been associated with increased young adult body mass index, history of B-cell-activating autoimmune diseases, hepatitis C, and several single nucleotide variants involving the human leukocyte antigen (HLA) region of chromosome 6 and non-HLA loci near EXOC2, PVT1, MYC, and NCOA1. Tumor sequencing studies suggest that multiple pathways are involved in the development of DLBCL. Additional studies of epidemiological exposures, genome wide associations, and tumor sequencing in follicular, lymphoplasmacytic, marginal zone, and mantle cell lymphoma demonstrate overlapping areas of increased risk factors and unique factors for specific subtypes. Integrating these findings is important for constructing comprehensive models of NHL pathogenesis, which could yield novel targets for therapy and strategies for lymphoma prevention in certain populations.
Introduction
B-cell non-Hodgkin lymphoma (NHL) comprises a heterogeneous group of hematologic malignancies distinguished by histology, genetic profile, and clinical behavior (Koff et al., 2015). In recent years, insights into normal B-cell biology, genetic and genomic alterations in lymphoma, and epidemiologic exposures associated with specific NHL subtypes have begun to inform our understanding of factors that may contribute to lymphomagenesis in general as well as development of certain NHL subtypes. Integrating these findings will be important to construct more comprehensive models of NHL pathogenesis, which could yield novel targets for therapy or even strategies for lymphoma prevention in certain high-risk populations.
Normal B Cell Biology: Primed for Lymphomagenesis
As part of the adaptive immune system, normal B-cells must be able to successfully recognize a wide variety of antigens to protect against infection and even malignancy. The transmembrane protein B-cell receptor (BCR), also known as antibody or immunoglobulin (Ig), is essential in this function: its variable regions bind to a specific antigen, while conserved portions of the protein engage in signal transduction that is key to B-cell survival and to the adaptive immune response. In order to generate a diverse antibody repertoire, maturing B-cells go through a rigorous selection process within lymphoid tissues. Cells that do not bind antigen strongly enough or bind self-antigen receive death signals, while those that bind antigen well receive survival signals. During this process, the Ig gene undergoes physiologic mutation and translocation to increase variety and strength of antigen binding as well as produce several antibody isotypes with distinct functions. Since such genetic alterations are intrinsic to normal B-cell development, perhaps it should come as no surprise that B-cells sometimes acquire mutations or translocations that predispose to malignant transformation to lymphoma. Indeed, several NHL subtypes are characterized by specific translocations involving the Ig gene, such as t(14,18) in follicular lymphoma (FL). In other lymphomas, mutations in genes encoding for key transcription factors such as NF-κB are known to play central roles in pathogenesis. However, these characteristic abnormalities have been shown to be insufficient to incite lymphoma development by themselves. Thus, much more work is needed to elucidate the complex interplay between host genetics and environmental factors that results in lymphomagenesis.
Epidemiology of Non-Hodgkin Lymphoma: Beyond Genetics
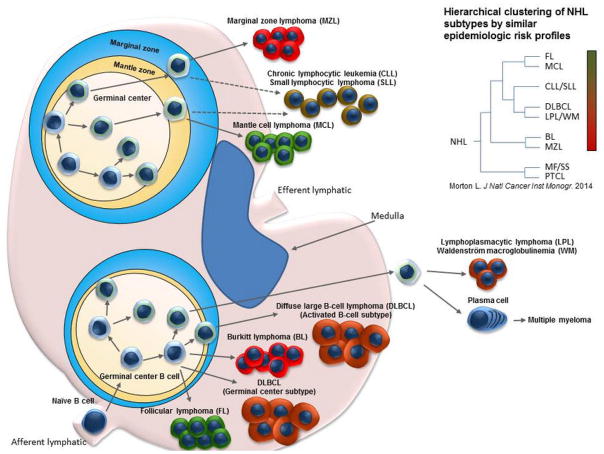
Historical studies have identified several strong risk factors for development of NHL, including severe immune suppression, autoimmune disorders, infections such as human immunodeficiency virus (HIV) and Epstein-Barr virus (EBV). However, until recently, investigation of risk factors for particular lymphoma subtypes remained difficult due to the relative rarity of each subtype, which limited statistical power. The International Lymphoma Epidemiology Consortium (InterLymph) overcame this limitation by pooling large case-control studies to increase statistical power in examination of risk factors for specific NHL subtypes (Bracci et al., 2014; Cerhan et al., 2014b; Linet et al., 2014; Mbulaiteye et al., 2014; Morton et al., 2014a; 2014b; Smedby et al., 2014; Vajdic et al., 2014). The InterLymph NHL Subtypes Project confirmed many known contributors to lymphomagenesis and also identified new factors associated with increased risk of certain NHL subtypes. Below, we describe these epidemiologic findings along with genetic risk factors identified for B-cell lymphoma subtypes. Table 1 organizes by B-cell NHL subtype the germline single nucleotide variants (SNVs) discovered in genome-wide association studies and epidemiologic risk factors found to confer increased NHL risk, as well as genetic abnormalities commonly found in these tumors. The illustration in Figure 1 graphically depicts the relationships between B-cell NHL subtypes and normal B-cell populations and the epidemiological relatedness and etiologic heterogeneity derived from the InterLymph analyses (Morton et al., 2014b).
Table 1.
Single Nucleotide Variants, Tumor Mutations, and Epidemiological Factors by NHL Subtype. Each NHL subtype harbors a distinct collection of genetic abnormalities and host/environmental risk factors.
| NHL Subtype |
Burkitt Lymphoma |
Diffuse Large B-cell Lymphoma |
Follicular Lymphoma |
Mantle Cell Lymphoma |
Marginal Zone Lymphoma |
Lymphoplasma- cytic Lymphoma/ Waldenström’s |
|---|---|---|---|---|---|---|
| Key Single Nucleotide Variants | NICL* | 543 total identified Newly identified: rs116446171 rs13255292 rs4733601 rs2523607 rs79480871 (Cerhan et al., 2014a; Morin et al., 2013) |
rs3117222 rs9275517 rs4938573 rs5947462 rs6444305 rs17749561 rs13254990 (Skibola et al., 2014; 2012) |
NICL* | rs9461741 rs2922994 (Vijai et at., 2015) |
|
| Key Tumor Mutations** | MYC (Love et al., 2012) | CECR1, MYD88 (Love et al., 2012) | CDKN2A/B (during tFL progression) (Pasqualucci et al., 2011) | ATM, CCND1, TP53 (Zhang et al., 2014; Bea et al., 2013) | BTNL2, NOTCH2 (SMZL) (Parry et al., 2013) | L265P (Ansell et al., 2014; Poulain et at., 2013) MYD88 (Ansell et al., 2014; Poulain et at., 2013) |
| Key Epidemiological Factors*** |
|
Increased young adult BMI
|
|
|
|
|
Note:
not identified in current literature;
most common;
increases risk
Figure 1.
Relationships between B-cell NHL subtypes and normal B-cell populations and the epidemiological relatedness and etiologic heterogeneity determined by hierachical clustering of medical history, lifestyle, family history, and occupational risk factors from the InterLymph analyses. Many NHL subtypes emerge from germinal center B-cells and each NHL subtype carries a collection of medical history, lifestyle, family history, and occupational risk factors.
Genetic and Epidemiological Risk Factors for B Cell Non-Hodgkin Lymphoma by Subtype
Burkitt Lymphoma (BL)
Burkitt lymphoma is an aggressive form of B-cell comprised of three variants: (1) endemic BL sporadic BL which is rarely associated with EBV NHL with endemic BL, which is associated with Epstein-Barr virus (EBV) in >95% of cases and occurs predominantly in the equatorial belt of Africa, (2) sporadic BL, which is rarely associated with EBV and amounts to <5% of NHL in the United States, and (3) HIV associated BL (Brady et al., 2007). All forms of BL involve a characteristic translocation of the MYC gene and an Ig gene. MYC is a regulatory gene that codes for a multifunctional transcription factor that activates expression of many genes involved in apoptosis, cell cycle progression, and cellular transformation. Interestingly, epidemiologic risk factors for BL varied according to age (Mbulaiteye et al., 2014). Among patients under 50 years of age, history of allergy (OR = 0.52; 95% CI = 0.30 to 0.90) or asthma (OR = 0.35; 95% CI = 0.13 to 0.95) decreased risk of BL, while history of eczema alone (OR = 2.82; 95% CI = 1.35 to 5.86) and occupation as a cleaner were associated with increased risk. Among older patients, history of infection with hepatitis C virus (HCV) resulted in increased risk of BL (OR = 4.10; 95% CI = 1.10 to 15.4), while history of blood transfusions was associated with a decreased risk. Regardless of age group, the tallest individuals (those in the 75th percentile of height or higher) were found to have a higher risk for BL, and those who consume alcohol had a decreased risk for BL.
A 2012 study characterized the genetic landscape in BL through genome and exome sequencing in an attempt to identify alterations that augment the ability of MYC translocation to incite development of BL (Love et al., 2012). As BL is characterized by dysregulation of the MYC oncogene (most often by translocation with the Ig gene), it is not surprising that sequencing of BL tumors revealed frequent MYC mutations. Mutations in ID3 that cause increased cell cycle progression and proliferation were also common in BL, but not in other subtypes such as diffuse large B-cell lymphoma (DLBCL). This finding could suggest that these altered genes cooperate to produce the BL phenotype, just as MYC translocation characterizes the BL subtype, but is not exclusive to it (Richter et al., 2012). In contrast, mutations in MYD88 and CD79A, while commonly concomitant in the activated B cell subtype (ABC) of DLBCL, were found to be mutually exclusive when present in BL. Interestingly, BL was also found to harbor mutations typically seen in the germinal center B cell (GCB) subtype of DLBCL, in genes such as GNA13, EZH2 and BCL2 (Love et al., 2012). Although these mutations are found in both BL and DLBCL, it is possible that the combinations in which they are found account for differences between the two subtypes.
Diffuse Large B-Cell Lymphoma (DLBCL)
Epidemiologic factors associated with an increased risk of DLBCL included increased young adult BMI, history of B cell-activating autoimmune diseases (OR = 2.36; 95% CI = 1.80 to 3.09), and HCV seropositivity (OR = 2.08; 95% CI = 1.23 to 3.49), among others. In contrast, atopic disorder and sunlight were associated with a decreased risk of DLBCL (Cerhan et al., 2014b). Interestingly, these factors did not appear to exert a summative effect on DLBCL risk, suggesting several distinct pathways of pathogenesis that may help to explain the genetic heterogeneity of DLBCL.
A genome wide association study (GWAS) examining 3,878 cases of DLBCL identified several single nucleotide variants (SNVs) associated with increased DLBCL risk. Importantly, almost all of these involved the human leukocyte antigen (HLA) region of chromosome 6, which encodes antigen-presenting proteins on the cell surface that are essential for immune function (Abdou et al., 2010). As noted above, HLA genes are polymorphic because they encode variations that promote specificity in the antigen binding epitopes and have been linked to susceptibility some B-cell NHL subtypes (Wang et al., 2010). Other studies described SNVs in three non-HLA loci near EXOC2, PVT1, MYC, and NCOA1 (Cerhan et al., 2014a). Beyond its role in BL, MYC appears to be an important gene across lymphoma subtypes. However, its role in DLBCL and the role of other genes are less clear, and this work is predominantly hypothesis-generating and requires confirmatory mechanistic studies.
Another study in which exome sequencing of 6 DLBCL patient samples was performed revealed novel inactivating mutations in MLL2, B2M, and CD58, thus providing evidence that dysregulated histone proteins and interaction with T cells may be important aspects of lymphomagenesis in this subtype (Pasqualucci et al., 2011). Mixed lineage leukemia protein 2 is a histone methyltransferase that controls access to gene transcription. Beta-2 microglobulin associates with the major histocompatibility complex (MHC) I in all nucleated cells. CD48 and CD58 are ligands of intercellular adhesion molecule (ICAM) 1 on T cells and NK cells. Another genomic study performing whole exome sequencing of DLBCL tumors found other commonly occurring mutations: BTG1, MEF2B, BCL2, MLL2, TNFRSF14, H1 protein, PCLO, P2RY8 and ACTB (Lohr et al., 2012). Additional mutations were found to have a functional role in DLBCL pathogenesis, but were less common (Lohr et al., 2012). MEF2B codes transcription factors that modulate histones, and H1 proteins are histone linkers that help DNA enter and exit the nucleosome. BTG1 regulates the cell cycle, while BCL2 is an oncogene that regulates apoptosis. TNFRSF14 is a TNF receptor in T cells and P2RY28 is a G-protein coupled receptor (GPCR) whose function is relatively unknown. Finally, ACTB is a cytoskeletal protein that activates B lymphocytes, and PCLO is a part of the cytoskeletal matrix. Another sequencing study identified 476 DLBCL cancer genes that were recurrently mutated in DLBCLs. Known oncogenes that were found to be somatically mutated included ARID1A, SETD2, CARD11, and PIK3R1 (Zhang et al., 2013). All of these findings suggest that multiple pathways are involved in the development of DLBCL. However, as many of these mutations involve immune function, they may likely explain the link between autoimmune disease and DLBCL risk discussed earlier.
Follicular Lymphoma
Another InterLymph subtype study indicated that family history of NHL increased FL risk by over two-fold (Linet et al., 2014). Individuals who were overweight as young adults also had an increased risk of FL. Those who had an increasing amount of recreational sun exposure were at a decreased risk of FL. Occupations that reduced risk of FL included baker, miller, and teachers in higher education, while working as a spray painter increased FL risk. Additional individual and lifestyle characteristics were identified to be sex-differential risk factors. Increased height was shown to increase risk of FL in males only, whereas a history of cigarette smoking increased FL risk in females only. The only medical condition associated with an increased risk of FL was history of Sjögren’s syndrome (OR = 13.2; 95% CI = 3.42 to 50.9). History of blood transfusion (OR = 0.78; 95% CI = 0.68 to 0.89) and allergies not including eczema (OR = 0.88; 95% CI = 0.79 to 0.98) were found to be associated with a decreased risk for FL (Linet et al., 2014). When stratified by gender, history of hay fever and food allergies reduced the risk for FL only among female patients.
A GWAS involving FL patients identified a SNV that was associated with changing levels of HLA-DPBL (a transcript involved in HLA class II expression) and decreased FL risk (Skibola et al., 2012). This association was confirmed in other GWAS, which also identified five new loci involved in HLA class II expression that were susceptible to genetic mutation in patients with FL (Skibola et al., 2014). Taken together, these findings may implicate DRβ1 (a protein of the beta subunit in HLA class II MHC) in FL pathogenesis.
The pathogenesis of FL is of particular clinical interest due to its ability to present as an indolent lymphoma with the potential to transform to DLBCL (known as transformed FL, or tFL). Whole exome sequencing has been used to identify mutations that promote transformation from FL to tFL (Pasqualucci et al., 2014). Genetic analysis suggests that both tFL and FL share a common precursor cell that goes on to acquire distinct mutations that define each subtype. While the two entities do share some mutations (e.g., in certain genes coding for histone modification enzymes), hundreds of mutations specific to tFL were found during exome sequencing, many more than were found in FL. One such example was the loss of CDKN2A/B, which is a regulator of G1 cell cycle progression; others include B2M and CD58 (involved in T cell recognition), PIM1, Pax5, and RhoH/TTF (involved in producing antibody diversity) (Pasqualucci et al., 2014). These findings indicate that the transformation from FL to tFL follows a divergent evolutionary path, and that tFL is genetically more complex than FL. Comparison of tFL’s genetic profile to that of GCB DLBCL showed that many mutations were shared between the two subtypes, but tFL retained several unique mutations (e.g., CDKN2A/B and FAS deletions). This genomic heterogeneity identifies an important roadblock in current clinical management of patients with either tFL or GCB DLBCL. Current initial treatment strategies are largely the same for these two patient populations, an approach that does not take into account the unique mutations that may influence pathogenesis and response to treatment (Pasqualucci et al., 2014).
Lymphoplasmacytic Lymphoma
Few factors have been associated with increased risk of lymphoplasmacytic lymphoma, including medical and familial histories of patients. Individuals with a history of Sjögren’s syndrome (OR = 3.2; 95% CI = 3.42 to 50.9), systemic lupus erthematosus (OR = 8.73; 95% CI = 2.91 to 26.2) or HCV infection, as well as family history of any hematologic malignancy carried an increased risk of LPL (Vajdic et al., 2014). Future data will need to be collected and analyzed to determine any further significant links between other identified risk factors and LPL diagnosis. Previously identified as a mutation found in DLBCL tumors, MYD88 mutations were observed to occur commonly in LPL patients and were also identified in whole exome sequencing of LPL (Poulain et al., 2013). MYD88 functions in several pathways, including JAK-STAT signaling, activation of NF-κB signaling, and IL-1 and IL-18 cytokine signaling. Most MYD88 mutations in LPL patients were gain-of-function (79%), while some patients carry copy number abnormalities. These findings eventually proved to be clinically valuable, as inhibition of MYD88 signaling resulted in cytotoxicity and cell growth inhibition in cell lines (Poulain et al., 2013). Further investigation of cellular mechanisms affected by MYD88 mutations has led to new targets for treatment of LPL. Immunoprecipitation experiments elucidated the mechanism by which MYD88 activates NF-κB signaling through TAK1 phosphorylation when TAK1 was found to be constitutively active in MYD88 mutant cell lines (Ansell et al., 2014). Importantly, TAK1 inhibitors decreased cell proliferation in these cell lines to a greater extent than NF-κB inhibitors.
Mantle Cell Lymphoma
Analysis of medical history, family history and occupational factors associated with MCL risk indicate that the etiology of MCL is likely multifactorial (Smedby et al., 2014). Decreased risk for MCL development was seen in individuals with history of any specific allergy (non-food related), hay fever, any atopic condition, asthma, eczema, or food allergy. Family history of hematological malignancy in first-degree relatives increased patient risk of MCL twofold. While working on a farm had no significant effect on MCL risk, individuals who lived on a farm had an increased risk of MCL (OR = 1.40; 95% CI = 1.30 to 1.90). The interaction between these factors remains unclear, as does the relative importance of each factor on MCL risk and their relationships to genetic risks and genomic abnormalities associated with lymphomagenesis. MCL is characterized by the t(11,14) translocation that results in constitutive expression of cyclin D1, which is the lesion that characterizes MCL. Whole exome sequencing has identified novel mutations present in MCL tumors, some of which are unique to MCL. These efforts also lend some insight into possible mechanisms of MCL pathogenesis. One such analysis associated presence of mutations in NOTCH2, a protein involved in cell differentiation, with dismal prognosis (Zhang et al., 2014). Patients with tumors harboring NOTCH2 mutation had a 3 year OS of 0% (versus 62% for patients without such a mutation). Other novel mutations recently identified include WHSC1, RB1, POT1 and SMARCA4 (Zhang et al., 2014). The mutations most commonly found were ataxia telangiectasia mutation (ATM), oncogene CCND11 and the tumor suppressor gene TP53. Furthermore, mirroring findings in other NHL subtype analyses, exome sequencing demonstrated heterogeneity across MCL patients (Bea et al., 2013). Furthermore, chromatin immunoprecipitation studies show that MCL may also be characterized by certain patterns of epigenetic gene modification (i.e., via DNA methylation and histone deacetylation) (Zhang et al., 2014). Therefore, in order to understand the pathogenesis of MCL, studies must take into account biology at the genetic and epigenetic levels.
Marginal Zone Lymphoma
Notably, when analyzing significant risk factors among each of the MZL subtypes (extranodal MZL, splenic MZL and nodal MZL,) some commonalities emerged, but other risk factors have been MZL subgroup-specific (Bracci et al., 2014). Regardless of MZL subtype, Sjögren’s syndrome (OR = 38.4; 95% CI = 17.04 to 86.48), systemic lupus erythematosus, peptic ulcers (OR = 6.57; 95% CI = 3.11 to 13.86), family history of a first degree relative with a history of a hematological malignancy, HCV positivity (OR = 3.04; 95% CI = 1.65 to 5.60), decreased years since quitting smoking, asthma without other atopic disorders, working as a carpenter OR = 2.43; 95% CI = 1.23 to 4.45), painter or metalworker and hair dye use (among women) were associated with increased risk of MZL. Factors associated with a decreased risk of MZL included increased recreational sun exposure, working as a teacher (OR = .50; 95% CI = 0.35 to 0.70), and alcohol consumption. History of B-cell-activating autoimmune disease increased the risk of all three subtypes.
Increased risk for EMZL was also associated with alcohol consumption. Asthma, without other atopic conditions, was associated only with SMZL risk (OR = 2.28; 95% CI = 1.23 to 4.23). Additionally, a family history of NHL was associated with increased risk for NMZL (OR = 2.82; 95% CI = 1.33 to 5.98), but not for other MZL subtypes. GWAS findings corroborate the epidemiologic link between autoimmune disease and MZL. One study found that the most significant genetic variant associated with MZL occurred proximally to HLA class II MHC and BTNL2 genes. Significantly, BTNL2 acts to regulate T-cell proliferation and cytokine production (Vijai et al., 2015). Exome sequencing has identified several novel mutations in SMZL, including NOTCH2, a potential target for therapy as discussed above in the MCL section (Rossi et al., 2012). Mutations in TNFAIP3, MAP3K14, MLL2 and SPEN also have been described in SMZL. The identification of MAP3K14 dysgregulation posits MAP kinase signaling as an important pathway not previously identified in MZL pathogenesis. Recurrent non-synonymous mutations specific to SMZL included those in CREBBP and CBFA2T3. CREBBP is associated with chromosome remodeling, while CBFA2T3 is involved in myeloid translocation, both of which contribute to cancer pathogenesis. Finally, whole exome sequencing of SMZL has also identified many non-recurrent mutations that affect several pathways involved in a variety of cellular processes: marginal zone development, chromatin remodeling, cytoskeleton and apoptosis (Martinez et al., 2014). These studies provide novel potential targets for therapy, but more work is needed to determine whether such targets will be relevant in SMZL treatment.
Conclusions and Future Directions
GWAS, whole exome sequencing and large cohort epidemiology studies of NHL subtypes have confirmed previously noted risk factors and identified novel exposures, genetic variants and tumor mutations that hint at the pathogenesis of these malignancies. While some factors are unique to a specific subtype, others are shared between two or more, and others still may characterize the transformation of one subtype to another. Such findings provide etiologic clues relating to both the commonality and the heterogeneity within NHL, which can be seen at the level of host genetics, tumor genomic profiling, and environmental factors. SNVs and mutations also help discern which molecular pathways may be integral to cancer development and maintenance in specific NHL subtypes. Identifying biological pathways through multidisciplinary studies thus provides a framework for developing new therapeutic drugs by highlighting the mechanisms by which particular lymphomas arise and survive. Investigation of risk factors common to several NHL subtypes may provide insight into pharmacologic agents that show activity in several different lymphomas. Alternatively, examining subtype-specific factors may shed light on possible therapeutic targets for a single subtype, such as ALK tyrosine kinase inhibitors in anaplastic large cell lymphoma. Thus, future efforts should integrate data obtained from GWAS, epidemiologic and sequencing studies to elucidate affected pathways and pursue subtype-specific therapies.
Acknowledgments
This work was supported in part by National Cancer Institute R21 CA158686-01A1 and U01CA195568, the Lymphoma Epidemiology of Outcomes (LEO) Cohort Study.
Footnotes
Disclosure
The authors report no conflicts of interest.
Contributor Information
Samantha Glass, University of Illinois at Chicago School of Medicine, Chicago, Illinois, 60612, United States. Specialty: Hematology, Oncology.
Anh Phan, Lymphoma Program, Department of Hematology/Oncology, Winship Cancer Institute, Emory University School of Medicine, Atlanta, Georgia, 30322, United States. Specialty: Hematology, Oncology.
Jessica N. Williams, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, 02115, United States. Specialty: Oncology
Christopher R. Flowers, Lymphoma Program, Department of Hematology/Oncology, Winship Cancer Institute, Emory University School of Medicine, Atlanta, Georgia, 30322, United States. Specialty: Hematology, Oncology
Jean L. Koff, Lymphoma Program, Department of Hematology/Oncology, Winship Cancer Institute, Emory University School of Medicine, Atlanta, Georgia, 30322, United States. Specialty: Hematology, Oncology
References
- Abdou AM, Gao X, Cozen W, Cerhan JR, Rothman N, Martin MP, Davis S, Schenk M, Chanock SJ, Hartge P, Carrington M, Wang SS. Human leukocyte antigen (HLA) A1-B8-DR3 (8.1) haplotype, tumor necrosis factor (TNF) G-308A, and risk of non-Hodgkin lymphoma. Leukemia. 2010;24(5):1055–1058. doi: 10.1038/leu.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell SM, Hodge LS, Secreto FJ, Manske M, Braggio E, Price-Troska T, Ziesmer S, Li Y, Johnson SH, Hart SN, Kocher JP, Vasmatzis G, Chanan-Kahn A, Gertz M, Fonseca R, Dogan A, Cerhan JR, Novak AJ. Activation of TAK1 by MYD88 L265P drives malignant B-cell Growth in non-Hodgkin lymphoma. Blood Cancer J. 2014;4:e183. doi: 10.1038/bcj.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bea S, Valdes-Mas R, Navarro A, Salaverria I, Martin-Garcia D, Jares P, Gine E, Pinyol M, Royo C, Nadeu F, Conde L, Juan M, Clot G, Vizan P, Di Croce L, Puente DA, Lopez-Guerra M, Moros A, Roue G, Aymerich M, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(45):18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracci PM, Benavente Y, Turner JJ, Paltiel O, Slager SL, Vajdic CM, Norman AD, Cerhan JR, Chiu BC, Becker N, Cocco P, Dogan A, Nieters A, Holly EA, Kane EV, Smedby KE, Maynadie M, Spinelli JJ, Roman E, Glimelius B, et al. Medical history, lifestyle, family history, and occupational risk factors for marginal zone lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014;2014(48):52–65. doi: 10.1093/jncimonographs/lgu011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G, Macarthur GJ, Farrell PJ. Epstein-Barr virus and Burkitt lymphoma. J Clin Pathol. 2007;60(12):1397–1402. doi: 10.1136/jcp.2007.047977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerhan JR, Berndt SI, Vijai J, Ghesquieres H, Mckay J, Wang SS, Wang Z, Yeager M, Conde L, De Bakker PI, Nieters A, Cox D, Burdett L, Monnereau A, Flowers CR, De Roos AJ, Brooks-Wilson AR, Lan Q, Severi G, Melbye M, et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet. 2014a;46(11):1233–1238. doi: 10.1038/ng.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerhan JR, Kricker A, Paltiel O, Flowers CR, Wang SS, Monnereau A, Blair A, Dal Maso L, Kane EV, Nieters A, Foran JM, Miligi L, Clavel J, Bernstein L, Rothman N, Slager SL, Sampson JN, Morton LM, Skibola CF. Medical history, lifestyle, family history, and occupational risk factors for diffuse large B-cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014b;2014(48):15–25. doi: 10.1093/jncimonographs/lgu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff JL, Chihara D, Phan A, Nastoupil LJ, Williams JN, Flowers CR. To Each Its Own: Linking the Biology and Epidemiology of NHL Subtypes. Curr Hematol Malig Rep. 2015;10(3):244–255. doi: 10.1007/s11899-015-0267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linet MS, Vajdic CM, Morton LM, De Roos AJ, Skibola CF, Boffetta P, Cerhan JR, Flowers CR, De Sanjose S, Monnereau A, Cocco P, Kelly JL, Smith AG, Weisenburger DD, Clarke CA, Blair A, Bernstein L, Zheng T, Miligi L, Clavel J, et al. Medical history, lifestyle, family history, and occupational risk factors for follicular lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014;2014(48):26–40. doi: 10.1093/jncimonographs/lgu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, Novak AJ, Dogan A, Ansell SM, Link BK, Zou L, Gould J, Saksena G, Stransky N, Rangel-Escareno C, Fernandez-Lopez JC, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, Lugar PL, Rizzieri DA, Lagoo AS, Bernal-Mizrachi L, Mann KP, Flowers CR, Naresh KN, Evens AM, Chadburn A, Gordon LI, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N, Almaraz C, Vaque JP, Varela I, Derdak S, Beltran S, Mollejo M, Campos-Martin Y, Agueda L, Rinaldi A, Kwee I, Gut M, Blanc J, Oscier D, Strefford JC, Martinez-Lopez J, Salar A, Sole F, Rodriguez-Peralto JL, Diez-Tascon C, et al. Whole-exome sequencing in splenic marginal zone lymphoma reveals mutations in genes involved in marginal zone differentiation. Leukemia. 2014;28(6):1334–1340. doi: 10.1038/leu.2013.365. [DOI] [PubMed] [Google Scholar]
- Mbulaiteye SM, Morton LM, Sampson JN, Chang ET, Costas L, De Sanjose S, Lightfoot T, Kelly J, Friedberg JW, Cozen W, Marcos-Gragera R, Slager SL, Birmann BM, Weisenburger DD. Medical history, lifestyle, family history, and occupational risk factors for sporadic Burkitt lymphoma/leukemia: the Interlymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014;2014(48):106–114. doi: 10.1093/jncimonographs/lgu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, Scott DW, Ding J, Roth A, Chiu R, Corbett RD, Chan FC, Mendez-Lago M, Trinh DL, Bolger-Munro M, Taylor G, Hadj Khodabakhshi A, Ben-Neriah S, Pon J, Meissner B, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton LM, Sampson JN, Cerhan JR, Turner JJ, Vajdic CM, Wang SS, Smedby KE, De Sanjose S, Monnereau A, Benavente Y, Bracci PM, Chiu BC, Skibola CF, Zhang Y, Mbulaiteye SM, Spriggs M, Robinson D, Norman AD, Kane EV, Spinelli JJ, et al. Rationale and Design of the International Lymphoma Epidemiology Consortium (InterLymph) Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014a;2014(48):1–14. doi: 10.1093/jncimonographs/lgu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton LM, Slager SL, Cerhan JR, Wang SS, Vajdic CM, Skibola CF, Bracci PM, De Sanjose S, Smedby KE, Chiu BC, Zhang Y, Mbulaiteye SM, Monnereau A, Turner JJ, Clavel J, Adami HO, Chang ET, Glimelius B, Hjalgrim H, Melbye M, et al. Etiologic heterogeneity among non-Hodgkin lymphoma subtypes: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014b;2014(48):130–144. doi: 10.1093/jncimonographs/lgu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry M, Rose-Zerilli MJ, Gibson J, Ennis S, Walewska R, Forster J, Parker H, Davis Z, Gardiner A, Collins A, Oscier DG, Strefford JC. Whole exome sequencing identifies novel recurrently mutated genes in patients with splenic marginal zone lymphoma. PloS One. 2013;8(12):e83244. doi: 10.1371/journal.pone.0083244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, Ouillette P, Trifonov V, Rossi D, Tabbo F, Ponzoni M, Chadburn A, Murty VV, Bhagat G, Gaidano G, Inghirami G, Malek SN, Rabadan R, Dalla-Favera R. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6(1):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, Chan J, Bhagat G, Chadburn A, Gaidano G, Mullighan CG, Rabadan R, Dalla-Favera R. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain S, Roumier C, Decambron A, Renneville A, Herbaux C, Bertrand E, Tricot S, Daudignon A, Galiegue-Zouitina S, Soenen V, Theisen O, Grardel N, Nibourel O, Roche-Lestienne C, Quesnel B, Duthilleul P, Preudhomme C, Leleu X. MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood. 2013;121(22):4504–4511. doi: 10.1182/blood-2012-06-436329. [DOI] [PubMed] [Google Scholar]
- Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, Rosolowski M, Ammerpohl O, Wagener R, Bernhart SH, Lenze D, Szczepanowski M, Paulsen M, Lipinski S, Russell RB, Adam-Klages S, Apic G, Claviez A, Hasenclever D, Hovestadt V, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, Monti S, Vaisitti T, Arruga F, Fama R, Ciardullo C, Greco M, Cresta S, Piranda D, Holmes A, Fabbri G, Messina M, Rinaldi A, Wang J, Agostinelli C, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola CF, Berndt SI, Vijai J, Conde L, Wang Z, Yeager M, De Bakker PI, Birmann BM, Vajdic CM, Foo JN, Bracci PM, Vermeulen RC, Slager SL, De Sanjose S, Wang SS, Linet MS, Salles G, Lan Q, Severi G, Hjalgrim H, et al. Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am J Hum Genet. 2014;95(4):462–471. doi: 10.1016/j.ajhg.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola CF, Conde L, Foo JN, Riby J, Humphreys K, Sille FC, Darabi H, Sanchez S, Hjalgrim H, Liu J, Bracci PM, Smedby KE. A meta-analysis of genome-wide association studies of follicular lymphoma. BMC Genomics. 2012;13:516. doi: 10.1186/1471-2164-13-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedby KE, Sampson JN, Turner JJ, Slager SL, Maynadie M, Roman E, Habermann TM, Flowers CR, Berndt SI, Bracci PM, Hjalgrim H, Weisenburger DD, Morton LM. Medical history, lifestyle, family history, and occupational risk factors for mantle cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014;2014(48):76–86. doi: 10.1093/jncimonographs/lgu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajdic CM, Landgren O, Mcmaster ML, Slager SL, Brooks-Wilson A, Smith A, Staines A, Dogan A, Ansell SM, Sampson JN, Morton LM, Linet MS. Medical history, lifestyle, family history, and occupational risk factors for lymphoplasmacytic lymphoma/Waldenstrom’s macroglobulinemia: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monographs. 2014;2014(48):87–97. doi: 10.1093/jncimonographs/lgu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijai J, Wang Z, Berndt SI, Skibola CF, Slager SL, De Sanjose S, Melbye M, Glimelius B, Bracci PM, Conde L, Birmann BM, Wang SS, Brooks-Wilson AR, Lan Q, De Bakker PI, Vermeulen RC, Portlock C, Ansell SM, Link BK, Riby J, et al. A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat Commun. 2015;6:5751. doi: 10.1038/ncomms6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Abdou AM, Morton LM, Thomas R, Cerhan JR, Gao X, Cozen W, Rothman N, Davis S, Severson RK, Bernstein L, Hartge P, Carrington M. Human leukocyte antigen class I and II alleles in non-Hodgkin lymphoma etiology. Blood. 2010;115(23):4820–4823. doi: 10.1182/blood-2010-01-266775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, Dunphy C, Choi W, Au WY, Srivastava G, Lugar PL, Rizzieri DA, Lagoo AS, Bernal-Mizrachi L, Mann KP, Flowers C, Naresh K, Evens A, Gordon LI, Czader M, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED, Fedoriw Y, Dunphy CH, Richards KL, Gill JI, Sun Z, Love C, Scotland P, Lock E, Levy S, Hsu DS, Dunson D, Dave SS. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123(19):2988–2996. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]