The human body is comprised of trillions of cells that are well organized into different functional compartments, tissues, and organs. To maintain homeostasis, coordinated intercellular communication is required. Some well established systems for communication between cells and their environment include the endocrine system and the complex circulation and neurological systems. Direct exchange of information through gap junctions and membrane nanotubes are also used by adjacent neighboring cells. In the last several years, investigators have identified cell-free nucleic acids in circulation within extracellular lipid vesicles, termed extracellular vesicles (EVs) (1). EVs represent small membrane phospholipid-based particles that are released by every cell in the human body. Harboring a variety of materials, including RNA, DNA, lipids, and proteins, EVs often reflect the cell of origin, and have emerged as important vehicles for the intercellular communication in human disease and maintenance of homeostasis (2). These initial observations have led to a large series of studies aimed at further defining the biogenesis, selective packaging, characterization of contents, and potential clinical applications of EVs in human health and disease. This is clearly evidenced by the exponential growth of the number of articles in the literature over the past few years. EVs are released by different cell types, and are largely defined by their physical characteristics instead of molecular properties. Therefore, it can be challenging to determine the precise function of EVs. For example, the initial observation of the release of transferrin receptor–containing lipid vesicles during the maturation of reticulocytes suggested that EVs may be essential for cells to remove unwanted or excessive molecules (3). We have now realized that EVs may have biological functions other than waste management. The mechanisms by which EVs can mediate intercellular signaling and changes in the micro- and macroenvironment are complex. When recipient cells absorb EVs from the extracellular environment, the contents can affect their biological activities. EVs may alter many physiopathological processes, including antigen presentation, cell proliferation, cell–cell communication, and others. For example, activated T cells within the lung could have their activities dampened through major histocompatibility complex (MHC) class II molecule–containing exosomes from dendritic cells (DCs) (4). Cancer cells use exosomes to promote angiogenesis and maintain or modify the microenvironment. CD147-positive EVs from epithelial ovarian cancer cells promoted angiogenesis in vitro (5). In addition, tumor-derived EVs converted fibroblasts or mesenchymal stem cells (MSCs) into myofibroblasts, a key player in the tumor microenvironment that facilitates tumor cell growth and invasion (6). The initiation and progression of lung diseases are often dependent on a well orchestrated interaction between environmental stressors, the immune system, genetic drivers, and communication between lung cells. EVs have emerged as potential vehicles for the transfer of molecular contents in the lung, thus providing improved understanding for the molecular underpinnings of lung disease and an opportunity for novel biomarkers and therapeutics.
EV Nomenclature and Biogenesis
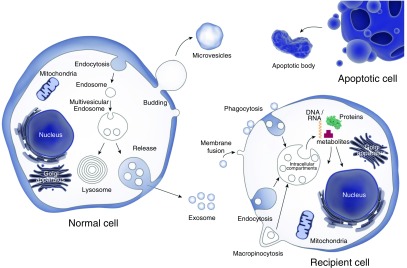
Although the nomenclature for EVs continues to evolve, they may be fundamentally divided into subgroups based on size and biogenesis (1). Thus, it is essential to be able to purify EVs so that their molecular content can be analyzed and biological functions assessed. Given the overlap that exists between subgroups, the term EVs is often used (Figure 1). EVs can be grouped into three main types: microvesicles (MVs), apoptotic bodies, and exosomes (Table 1). MVs, also known as ectosomes or shedding vesicles, are about 100–1,000 nm in diameter and are produced by shedding or outward budding of the plasma membrane from cells. Once MVs are generated, they are encapsulated as part of the cytosol. MVs, therefore, have a membrane structure similar to that of the cell of origin. Exosomes were first described more than 30 years ago by Pan and Johnstone (3) when they detected transferrin receptor–containing vesicles released by reticulocytes during the maturation of red blood cells. Exosomes are about 30–150 nm in diameter and produced within cells through inward budding of the endosomal membrane (Figure 1). The endosome system is an intracellular molecular transport system that drives both sorting and transportation of molecules between the plasma membrane, lysosomes, and trans-golgi network. Exosomes and their contents can be either processed through the lysosome or accumulate within multivesicular bodies and then be released to the extracellular environment once the multivesicular bodies fuse with cell membrane. The molecular contents of exosomes also often reflect those of its cell of origin. For example, exosomes released from DCs are enriched with CD1, CD80, CD86, and MHC class I and II molecules that are involved in the immune response (7). Exosome populations are enriched for tetraspanin membrane proteins, such as CD9, CD63, CD81, and vesicle-trafficking proteins, including tumor susceptibility (TSG) 101 and programmed cell death 6–interacting protein, and heat shock proteins (Hsps), like Hsp70 family A member 4 (HSPA4) (8). The CD63, programmed cell death 6 interacting protein and TSG101 are also enriched in the endosomal membrane. Apoptotic bodies (also called apoptotic blebs or apoptotic vesicles) are 50–5,000 nm in diameter and released from apoptotic cells (9). The density of apoptotic bodies ranges from 1.16 to 1.28 g/ml, overlapping with that of exosomes. In addition, the size of an apoptotic body is similar to that of a platelet. Apoptotic bodies contain DNA, and have been implicated as a vehicle for horizontal oncogene transfer. A number of proteins, including histones, thrombospondins, complement component C3, and annexin (ANXA) V, have been identified in apoptotic bodies (10).
Figure 1.
Extracellular vesicle biogenesis. Extracellular vesicles are divided into three main types: exosomes, microvesicles, and apoptotic bodies. Exosomes are formed in multivesicular endosomes and contain molecules including nucleic acids, proteins, lipids, and metabolites. The exosomal content can be transferred to other cells through different processes, including endocytosis, phagocytosis, macropinocytosis, or direct membrane fusion.
Table 1.
Extracellular Vesicle Classification
| Particle | Microvesicle | Exosome | Apoptotic body |
|---|---|---|---|
| Diameter, nm | 100–1,000 | 30–150 | 50–5,000 |
| Density, g/ml | Undetermined | 1.13–1.19 | 1.16–1.28 |
| Biogenesis | Shed from cell membrane | Inward budding of endosomal membrane | Released from blebs of apoptotic cells |
| Protein | VCAM1, ARF6, FLOT2, integrin, CD40, MMPs | CD9, CD63, CD81, TSG101, PDCD6IP, HSPA4 | Thrombospondins, C3, ANXA5, histones |
Definition of abbreviations: ARF6 = adenosine diphosphate–ribosylation factor 6; ANXA = annexin; C3 = complement component C3; FLOT = flotillin; HSPA4 = shock protein family A member 4; MMP = matrix metalloproteinase; PDCD6IP = programmed cell death 6–interacting protein; TSG101 = tumor susceptibility 101; VCAM = vascular cell adhesion protein.
Researchers continue to unravel the complexities of EV biogenesis. Changes in environmental stressors (e.g., hypoxia, pH, temperature) have all been identified as regulators of EV release (11–13). Proper EV release and function are dependent upon sorting, packaging, directed release, and uptake. The mechanisms that drive such processes are also slowly coming to light. For example, EV microRNA (miRNA) content does not always match that of the parent cell. Researchers have determined that an Exo motif of miRNA sequences can dictate miRNA sorting into EVs (14). This concept was further validated with the identification of a protein called synaptotagmin-binding, cytoplasmic RNA-interacting protein, the binding of which to select sequences within exosomal enriched miRNAs can guide packaging and release (15).
EV Isolation Characterization
EV Isolation
There are several different methods used for exosome purification, with most based on the physical characteristics of EVs (Table 2). One of the issues in studying EVs is overcoming the hurdle of obtaining pure populations of subgroups, as there is substantial overlap of characteristics between different lipid vesicles. For example, the density of exosomes overlaps with that of apoptotic bodies, and the range of particle size for exosomes overlaps with that of MVs.
Table 2.
Isolation Methods for Extracellular Vesicles
| Purification Method | Ultracentrifugation | Precipitation | Immunoaffinity | Size Exclusion Column |
|---|---|---|---|---|
| Time required | About 24–48 h | About 50 min | About 1 h | About 20 min |
| Purity | High | Low | High | Medium |
| Volume needed | About 1 ml | 100–200 μl | Variable | 100–200 μl |
| Other issues | • Centrifugation force may change the nature of lipid vesicle | • High protein contamination | • Obtain just a fraction of the exosome population | • Exosomes obtained, most maintain original structure |
| • Inconsistent results | • Polyethylene glycol may affect downstream analysis | • Suitable for functional studies | ||
| • May be contaminated with different types of vesicles |
Ultracentrifugation
Ultracentrifugation, combined with a density gradient method, results in a fairly pure exosome population. This method requires multiple rounds of sequential centrifugation to separate vesicles based on size (16). The drawbacks of this method include the long purification process, larger sample volume requirement, and low throughput. There is also a suggestion that the centrifugation forces are insufficient to pull down a significant fraction of exosomes in some samples, and may disrupt exosome structure and fuse lipid vesicles together.
Exosome precipitation kits
There are several precipitation-based commercial kits for exosome purification. These are most often based on water-clouding agents, such as polyethylene glycol to precipitate certain-sized particles. This method is quick, does not require ultracentrifugation, and results in very high throughput. It can also handle smaller sample volumes.
Immunoaffinity
Exosomes may be isolated by targeting common cell surface proteins, such as CD63. However, not all exosomes contain CD63, yielding only a fraction of the exosome population.
Size-exclusion column
Based on exosome particle size, size-exclusion columns yield relatively pure vesicles, with minimal protein contamination, within about 20 minutes. In addition, physical force is reduced, thus preserving shape. This method may still result in coelution with other subtypes, including MVs and apoptotic bodies (17).
EV Characterization
Through various high-throughput profiling platforms, we have gained insight into the spectrum of molecular contents of EVs. As most of these technologies require significant amount of materials, most profiling studies are based on in vitro cell line–secreted EVs. Based on ExoCarta (www.exocarta.org)—a database dedicated to cataloging exosome content—more than 5,000 different proteins, 2,000 RNAs, 1,300 miRNAs, and 800 lipid molecules have been identified (18).
Proteins
Protein content can be effectively analyzed to elucidate EV function. Mass spectrometry (MS)–based methods, such as one- or two-dimensional liquid chromatography–tandem MS and matrix-assisted laser desorption ionization–time of flight/time of flight MS are commonly used platforms to analyze the spectrum of proteins in EVs (19). Recently, using one-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis coupled with liquid chromatography–tandem MS, researchers studied the proteome of different types of vesicles and identified exosome-enriched proteins, including TSG101, syntenin-1, eps 15 homology domain–containing 4, ANXA11, and a disintegrin and metalloproteinase metallopeptidase domain 10 (20). To compare molecular contents from different samples, technologies, such as stable isotope labeling with amino acids in cell culture combined with MS can be used (21). Stable isotope labeling with amino acids is based on the metabolic incorporation of an isotopically “light” or “heavy” form of amino acids into proteins, the MS analysis of which results in not just identifying proteins in the EV, but also providing quantitative information on relative protein abundance. In addition to global profiling approaches, a targeted proteomics approach, called multiple (or selected) reaction monitoring, is often used to quantify specific proteins in EVs. The protein abundance determined by selected reaction monitoring is based on the spike-in of a known concentration of heavy labeled peptide standard (22). It is sensitive, specific, and does not require high-affinity antibodies to assess protein concentration. Other technologies, including antibody array, have also been used to profile protein content in EVs.
RNA
EVs contain a wide variety of RNA, including full-length protein coding mRNA, noncoding RNAs, such as miRNAs, long intergenic noncoding RNA, transfer RNAs, P-element induced Wimpy testis RNAs, ribosomal RNAs, vault RNAs, and cytoplasmic Y RNAs (23). Most of the RNAs in EVs are less than 700 nt, which suggests that some of the protein coding mRNAs and long intergenic noncoding RNAs are probably degraded RNA. Unlike cellular RNAs, RNAs from EVs lack significant amounts of ribosomal RNA, obviating the need for a ribosomal RNA depletion step (24). Commonly used technologies to assess the RNA, for both small and large RNA molecules in EVs, include complementary DNA microarrays, quantitative reverse transcription–polymerase chain reaction (RT-PCR), and next-generation sequencing (NGS). Quantitative RT-PCR is sensitive, and requires a small amount of RNA, but can only assess the concentration of specific RNA transcripts. Some of the RNAs in EVs are degraded transcripts, which may limit the assay. Microarray requires a higher concentration of RNA and, therefore, is not commonly used in EV RNA profiling. Recently, the NGS approach has gained significant interest, given low template requirements, ability to assess degraded RNA fragments, and lack of a requirement of prior knowledge on the identity of RNA in the sample. NGS requires specialized informatics tools to process the sequence reads, determine the sequence read identity, and quantify the level of specific transcripts. In addition, the library construction process, especially for small RNA, is long and may introduce significant variability.
Lipids
Due to its sensitivity and selectivity, MS is commonly used for lipid profiling (25). Unlike other biomolecules, lipids do not have a general guideline on ionization efficiency and rules of fragmentation in MS; therefore, it is difficult to develop standard methods for global lipid analysis. Electrospray ionization and matrix-assisted laser desorption ionization are the commonly used ionization technologies, because they do not induce significant fragmentation.
One limitation to current technologies for EV characterization of content lies in the heterogeneity of EV populations that exist in body fluids. Thus, detection of “disease- or cell-specific” EVs and signal may be masked by larger numbers of EVs originating from a multitude of cell types. To overcome these challenges, novel platforms for targeted EV capture and characterization, including lipid biospheres and lipoplex-based nanochips, are under development (26, 27)
EVs and Intercellular Communication in the Lung
The exchange of genetic information mediated through EV transfer between cells of the lung has become an increasingly recognized mechanism for initiating the immune response, development of disease, and maintenance of homeostasis. The environmental cues that drive the release of EVs from the lung and shuttling of contents include hypoxia, LPS, pollutants, cigarette smoke, and microbial pathogens. However, although our knowledge of the underlying mechanisms for cell-specific interactions and downstream signaling is minimal, it appears that EV release is cell, stimulus, and disease specific. Multiple cell types, including epithelial cells, macrophages, mast cells, and endothelial cells, have been identified as mediators of EV intercellular communication.
Immune Response
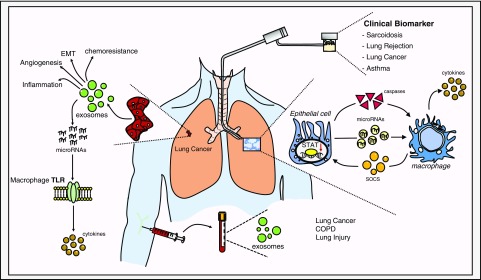
Early studies revealed that exosomes derived from human tracheobronchial epithelium harbor typical EV characteristics and epithelial-specific markers, including mucins (28). The release of epithelial-derived EVs in the setting of environmental exposures may be a critical step in the triggering the immune response, and serve as a link to the development of diseases of the airway, including chronic obstructive pulmonary disease (COPD) and asthma. For example, epithelial cell–derived EVs promote leukotriene conversion of leukotriene C4 (LTC4) to LTD4 in monocytic cells, potentially contributing to tissue remodeling and bronchoconstriction (29). Stimulated monocytes induce apoptosis through EV transfer of caspase-1 (30). In the case of environmental allergens, dust-derived EVs drive a neutrophilic response in the lung, characterized by T-helper cell type 1 (Th1) and Th17 accumulation in the lung (31). In addition, preclinical models of inhalation of LPS led to an increase in EVs within bronchoalveolar lavage fluid (BALF) that, in turn, drives a Th1 and Th17 response (32). The molecular contents that facilitate an inflammatory response are varied. For example, select miRNAs, including miR-146 and miR-155 within immune cells, can regulate the inflammatory response (33). Reciprocal inflammatory signaling may occur between cell types of the lung, with epithelial and alveolar macrophages (AMs) representing one of the best studied examples (34) (Figure 2). AMs release EV-based suppressor of cytokine signaling (SOCS) proteins (SOCS1 and SOCS2) to abrogate activation of cytokine-induced signal transducers and activators of transcription in alveolar epithelial cells (35). The mechanisms underlying AM EV-selective packaging of SOCS proteins remain largely unknown. There is a suggestion that exposure to infectious agents may induce SOCS3 release from AM-derived EVs (36). In vivo hyperoxia treatment resulted in a release of primarily epithelial-derived EVs, both in circulation and in BALF. Exposure of AMs to these same epithelial cell–derived EVs led to a potent induction in proinflammatory cytokines in vitro and neutrophil and macrophage accumulation in vivo (37). These observed effects were partially mediated through a caspase-3–Rho-associated protein kinase pathway (37).
Figure 2.
Applications for extracellular vesicles in lung disease. COPD = chronic obstructive pulmonary disease; EMT = epithelial–mesenchymal transition; SOCS = suppressor of cytokine protein signaling; STAT = signal transducers and activators of transcription; TLR = Toll-like receptor.
The relative contribution of subfamily members of EVs to intercellular transfer of contents and downstream signaling remains largely unknown, but there are likely to be both overlapping and divergent functions. In the setting of oxidative stress after hypoxia exposure in mice, the majority of EVs in both BALF and in vitro were MVs and epithelial in origin (38). Furthermore, these miRNA-enriched MVs induced a potent inflammatory (tumor necrosis factor α) and migratory response in recipient macrophages.
Lung Cancer
EVs have perhaps been best studied in the hallmarks of cancer biology and cancer initiation and progression. In lung cancer, EVs and their contents can drive angiogenesis, proliferation, epithelial–mesenchymal transition, and chemosensitivity (39–43) (Figure 2). Novel studies demonstrate that tumor-derived exosomes carry cell surface integrins that direct organ-specific metastatic deposition (44). Exosomes expressing integrin (ITG) α6β and ITGα6αβ1 were more likely to target lung cells, whereas ITGαvβ5 targets cells in the liver (44). One particularly intriguing role for EVs has been as a biological link between benign and malignant disease. For example, exosomes derived from plasma of mice exposed to intermittent hypoxia and plasma from patients with obstructive sleep apnea enhanced malignant properties of recipient cancer cells (45).
Toll-Like Receptor Signaling
The mechanisms by which EV contents facilitate downstream signaling are not fully understood. However, a recent and exciting development in the field of EV biology involves the recognition that EV content may function as ligands for Toll-like receptors (TLRs). Investigators demonstrated that miRNAs released by cancer cell–derived EVs can induce macrophage-mediated production of proinflammatory cytokines through binding of TLR8 (46) (Figure 2). This novel link may also serve as a mechanism for the development of chronic lung disease. LPS-mediated TLR4 activation was shown to increase the release of exosome-packaged proteases, including prolyl endopeptidase from airway epithelial cells (47).
Noninvasive Biomarkers of Lung Disease
A number of clinical diagnostic applications have been developed based on the concentration changes of specific molecules in circulation. The observed release of EVs in blood and other body fluids after environmental stressors, or in the setting of disease states, suggests that they provide a less-invasive means for disease monitoring, making them particularly compelling candidates for clinical biomarkers. The presence of these cell-free molecules in body fluids are likely the result of normal or pathological conditions associated with cell death; therefore, disease states may affect this spectrum of circulating molecules. The reported number of exosomes ranges from 0.84 to 4.32 × 108 particles/ml of plasma and contains about 40–60 ng of RNA (48). The exosome has a density that ranges between 1.10 and 1.14 g/ml, and a molecular weight of approximately 3.3 × 107 g/mole. Furthermore, an independent study by Chevillet and colleagues (49) suggested that the copies of miRNAs in body fluid exosomes, when isolated by differential ultracentrifugation, are minimal, raising questions regarding their biological significance.
Lung Cancer
Several studies have suggested the utility of blood-based EV signatures as potential biomarkers in lung cancer. A high-throughput analysis of circulating exosomes resulted in the identification of a panel of miRNAs that could distinguish between malignant and benign nodules of the lung (96.0% sensitivity; 60.0% specificity; area under the curve, 76.0%) (50). An independent study determined that a six-miRNA panel in plasma could distinguish between patients with adenocarcinoma of the lung and healthy control subjects (51). Finally, patterns of exosome-bound proteins in circulation have been correlated with lung cancer survival (52). Although encouraging, such studies are limited by small cohort sizes, and thus require larger independent validation.
Acute Lung Injury
Few studies to date have investigated the impact of lung injury on circulating EV populations. In vivo murine exposure to 4 hours of high–tidal volume mechanical ventilation resulted in physiological characteristics of acute lung injury and a marked increase in the release of endothelial (platelet endothelial cell adhesion molecule/ANXA5+)–derived microparticles (300–1,000 nm) in circulation (53).
COPD
Cigarette smoke exposure leads to a potent induction of endothelial-derived microparticles, which may signify endothelial injury. In COPD, elevated subpopulations of microparticles in circulation correlated with presence of COPD and exacerbations (54). Environmentally derived EVs may also elicit a response in the periphery linked to disease activity or risk. In one small study, investigators demonstrated that elevated serum IgG against dust-derived EVs was a risk factor for the development of lung cancer, COPD, and asthma (55).
EVs are detectable in BALF; however, the cell origin of BALF EVs remains unclear. One of the first reports of EVs in BALF by Admyre and colleagues (56) described BALF EVs that express MHC class I and II, CD53, and CD63, suggesting possible antigen-presenting cell origin. The molecular contents of BALF EVs may be applied as diagnostic biomarkers of clinical disease.
Sarcoidosis
Using mass spectroscopy, Martinez-Bravo and colleagues (57) conducted an analysis of protein expression in BALF-derived EVs from patients with sarcoidosis versus control subjects. They identified 690 proteins, including several up-regulated inflammatory proteins. An independent study demonstrated elevated numbers of BALF-derived EVs from patients with sarcoidosis inducing a proinflammatory response in recipient cells (58).
Lung Transplant
A recent study among patients undergoing lung transplantation demonstrated that BALF EVs in those with acute lung rejection carried distinct RNA transcripts linked to both the innate and adaptive immune responses (59). Comparing patients with and without chronic rejection, investigators identified higher levels of red blood cell– and epithelial cell–derived EVs among those with chronic rejection and an association with poor survival (60).
Asthma
Several studies have demonstrated that BALF-derived EVs in asthma harbor distinct properties that drive inflammation. Epithelial-derived exosomes were detectable in BALF of subjects with asthma and stimulated both CXCL-8 and LTC4 release in recipient epithelial cells (61). In addition, BALF EVs derived from subjects with asthma induced cytokine and leukotriene production in recipient epithelial cells (62). Preclinical murine models for asthma also exhibited a marked increase in epithelial-derived EVs that can induce IL-13–mediated monocyte proliferation (63).
EVs as Targeted Therapeutics
The clinical use of EVs for therapy is in the near future. One such example involves the use of EVs as modifiers of response to immunotherapy for lung cancer. Investigators have explored the use of DC-derived exosomes (Dex) as a means for augmenting natural killer and T cell function in the setting of immunotherapy (64). Treatment of patients with advanced-stage non–small cell lung cancer with Dex resulted in an increased natural killer cell function in a subgroup of patients, suggesting potential for Dex as an adjuvant therapeutic strategy for immunotherapy (64). Aptamer-based targeting represents another potential avenue for augmenting the immune response. Targeting cancer-derived Hsp70-enriched EVs attenuated myeloid-derived suppressor cell activation (65)
EVs have also been applied as therapies in pulmonary hypertension (PH). Aliotta and colleagues (66) demonstrated that the exosome fraction of circulating EVs from mice treated with monocrotaline, when delivered by tail vein injection, induced PH-caused anatomical changes consistent with PH in healthy mice. Conversely, intravenous exosomes derived from MSCs both prevented and reversed monocrotaline-induced PH.
The use of MSCs represents an exciting and novel direction for therapeutics in lung disease. For example, MSCs have been shown to reduce inflammation, permeability, and bacterial growth in in vivo models of pneumonia, and reduce the Th2/Th17–mediated airway hyperresponsiveness in models of allergic airway inflammation. In vivo administration of MSC-conditioned media and EVs reduced Th2/Th17 inflammation and hyperresponsiveness of the airway after Aspergillus hyphal extract exposure (67). Immunization with Staphylococcus aureus–exposed EVs conferred significant protective effects on the immune system in the setting of both lethal and sublethal exposure to S. aureus infection (68). Translation of MSCs as therapies in human disease is most evident in acute lung injury (69, 70). However, the challenges inherent to isolation and purification of MSCs and potential toxicity may limit the potential for large-scale human studies. The effects of MSC-derived conditioned media on lung cellular repair and inflammation mirrors that of primary MSCs, suggesting the presence of therapeutic components within the media (71, 72). MSC-derived EVs are protective in abrogating inflammation and improving alveolar fluid clearance in preclinical models of lung injury (73). Although the underlying mechanisms have yet to be full elucidated, an independent study showed that MSC-derived EV miRNAs could mediate both autophagy and mitophagy in macrophages (74).
Conclusions: The Future of EV Biology in Lung Disease
Since their initial discovery over 30 years ago, EVs have assumed an increasingly prominent role as mediators of intercellular communication in human disease. To date, investigators have demonstrated that EV-mediated transfer of RNAs and proteins contribute to the lung immune response to environmental insults and biology fundamental to cancer initiation and progression. We have observed early studies targeting EVs in augmenting the immune response to cancers. Such exciting potential should be tempered by the fact that our current understanding of their role in the lung is minimal, and there remains a great deal to be learned. Standardized protocols for the isolation, quantification, and characterization of EV family members will be essential in parsing out the relative contributions of each subtype to disease. The majority of studies to date have focused on smaller exosomes, with minimal attention to the potential contribution from larger MVs and apoptotic bodies. Second, the observed stimulus, cell- and disease-specific release of EVs, highlights the complex “networks” underlying EV-mediated biology. Third, we are just beginning to understand that the mechanisms by which select EV subpopulations and their contents are packaged and released. Finally, the identification and relative stability of EVs in body fluids provides us with an opportunity to develop novel, clinically informative biomarkers, but will also depend on improved understanding of EV member contributions to disease pathogenesis and characterization of EV contents. However, further stoichiometric studies are required to better quantify body fluid–based EV contents and their potential contribution to downstream pathophysiology. Although it is still early, one should be optimistic about the future for EV biology in lung disease and the potential for additional intense investigation to eventually impact lung health.
Footnotes
Supported by National Institutes of Health grants 1R01CA190740-01 (C.M.C. and S.P.N.-S.) and 1R01DA040395-01 (S.P.N.-S.).
Originally Published in Press as DOI: 10.1164/rccm.201612-2457PP on July 5, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Araldi E, Krämer-Albers EM, Hoen EN, Peinado H, Psonka-Antonczyk KM, Rao P, van Niel G, Yáñez-Mó M, Nazarenko I. International Society for Extracellular Vesicles: first annual meeting, April 17–21, 2012: ISEV-2012. J Extracell Vesicles. 2012;1:19995. doi: 10.3402/jev.v1i0.19995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aucamp J, Bronkhorst AJ, Badenhorst CP, Pretorius PJ. A historical and evolutionary perspective on the biological significance of circulating DNA and extracellular vesicles. Cell Mol Life Sci. 2016;73:4355–4381. doi: 10.1007/s00018-016-2370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 4.Buschow SI, Nolte-’t Hoen EN, van Niel G, Pols MS, ten Broeke T, Lauwen M, Ossendorp F, Melief CJ, Raposo G, Wubbolts R, et al. MHC II in dendritic cells is targeted to lysosomes or T cell–induced exosomes via distinct multivesicular body pathways. Traffic. 2009;10:1528–1542. doi: 10.1111/j.1600-0854.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- 5.Millimaggi D, Mari M, D’Ascenzo S, Carosa E, Jannini EA, Zucker S, Carta G, Pavan A, Dolo V. Tumor vesicle–associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia. 2007;9:349–357. doi: 10.1593/neo.07133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webber JP, Spary LK, Sanders AJ, Chowdhury R, Jiang WG, Steadman R, Wymant J, Jones AT, Kynaston H, Mason MD, et al. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2015;34:290–302. doi: 10.1038/onc.2013.560. [DOI] [PubMed] [Google Scholar]
- 7.Théry C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes: selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jerez S, Araya H, Thaler R, Charlesworth MC, López-Solís R, Kalergis AM, Céspedes PF, Dudakovic A, Stein GS, van Wijnen AJ, et al. Proteomic analysis of exosomes and exosome-free conditioned media from human osteosarcoma cell lines reveals secretion of proteins related to tumor progression. J Cell Biochem. 2017;118:351–360. doi: 10.1002/jcb.25642. [DOI] [PubMed] [Google Scholar]
- 9.Orozco AF, Lewis DE. Flow cytometric analysis of circulating microparticles in plasma. Cytometry A. 2010;77:502–514. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lleo A, Zhang W, McDonald WH, Seeley EH, Leung PS, Coppel RL, Ansari AA, Adams DH, Afford S, Invernizzi P, et al. Shotgun proteomics: identification of unique protein profiles of apoptotic bodies from biliary epithelial cells. Hepatology. 2014;60:1314–1323. doi: 10.1002/hep.27230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray WD, French KM, Ghosh-Choudhary S, Maxwell JT, Brown ME, Platt MO, Searles CD, Davis ME. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ Res. 2015;116:255–263. doi: 10.1161/CIRCRESAHA.116.304360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakase I, Kobayashi NB, Takatani-Nakase T, Yoshida T. Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci Rep. 2015;5:10300. doi: 10.1038/srep10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, Guo J, Yang M, Zhu X, Cao X. Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft–dependent pathway and act as efficient tumor vaccine. J Immunol. 2011;186:2219–2228. doi: 10.4049/jimmunol.1002991. [DOI] [PubMed] [Google Scholar]
- 14.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M. The RNA-binding protein syncrip is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Reports. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 17.de Menezes-Neto A, Sáez MJ, Lozano-Ramos I, Segui-Barber J, Martin-Jaular L, Ullate JM, Fernandez-Becerra C, Borrás FE, Del Portillo HA. Size-exclusion chromatography as a stand-alone methodology identifies novel markers in mass spectrometry analyses of plasma-derived vesicles from healthy individuals. J Extracell Vesicles. 2015;4:27378. doi: 10.3402/jev.v4.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, et al. ExoCarta: a Web-based compendium of exosomal cargo. J Mol Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S, Mishra R, Bigham GE, Wehman B, Khan MM, Xu H, Saha P, Goo YA, Datla SR, Chen L, et al. A deep proteome analysis identifies the complete secretome as the functional unit of human cardiac progenitor cells. Circ Res. 2017;120:816–834. doi: 10.1161/CIRCRESAHA.116.309782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi DS, Choi DY, Hong BS, Jang SC, Kim DK, Lee J, Kim YK, Kim KP, Gho YS. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles. 2012;1(1) doi: 10.3402/jev.v1i0.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, ’t Hoen PA. Deep sequencing of RNA from immune cell–derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lässer C, Shelke GV, Yeri A, Kim DK, Crescitelli R, Raimondo S, Sjöstrand M, Gho YS, Van Keuren Jensen K, Lötvall J. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017;14:58–72. doi: 10.1080/15476286.2016.1249092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallejo MC, Nakayasu ES, Longo LV, Ganiko L, Lopes FG, Matsuo AL, Almeida IC, Puccia R. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS One. 2012;7:e39463. doi: 10.1371/journal.pone.0039463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McVey MJ, Spring CM, Semple JW, Maishan M, Kuebler WM. Microparticles as biomarkers of lung disease: enumeration in biological fluids using lipid bilayer microspheres. Am J Physiol Lung Cell Mol Physiol. 2016;310:L802–L814. doi: 10.1152/ajplung.00369.2015. [DOI] [PubMed] [Google Scholar]
- 27.Lee LJ, Yang Z, Rahman M, Ma J, Kwak KJ, McElroy J, Shilo K, Goparaju C, Yu L, Rom W, et al. Extracellular mRNA detected by tethered lipoplex nanoparticle biochip for lung adenocarcinoma detection. Am J Respir Crit Care Med. 2016;193:1431–1433. doi: 10.1164/rccm.201511-2129LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kesimer M, Scull M, Brighton B, DeMaria G, Burns K, O’Neal W, Pickles RJ, Sheehan JK. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23:1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukic A, Ji J, Idborg H, Samuelsson B, Palmberg L, Gabrielsson S, Rådmark O. Pulmonary epithelial cancer cells and their exosomes metabolize myeloid cell–derived leukotriene C4 to leukotriene D4. J Lipid Res. 2016;57:1659–1669. doi: 10.1194/jlr.M066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra S, Wewers MD, Sarkar A. Mononuclear phagocyte–derived microparticulate caspase-1 induces pulmonary vascular endothelial cell injury. PLoS One. 2015;10:e0145607. doi: 10.1371/journal.pone.0145607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YS, Choi EJ, Lee WH, Choi SJ, Roh TY, Park J, Jee YK, Zhu Z, Koh YY, Gho YS, et al. Extracellular vesicles, especially derived from Gram-negative bacteria, in indoor dust induce neutrophilic pulmonary inflammation associated with both Th1 and Th17 cell responses. Clin Exp Allergy. 2013;43:443–454. doi: 10.1111/cea.12085. [DOI] [PubMed] [Google Scholar]
- 32.Shin TS, Kim JH, Kim YS, Jeon SG, Zhu Z, Gho YS, Kim YK. Extracellular vesicles are key intercellular mediators in the development of immune dysfunction to allergens in the airways. Allergy. 2010;65:1256–1265. doi: 10.1111/j.1398-9995.2010.02359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander M, Hu R, Runtsch MC, Kagele DA, Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM, O’Connell RM. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider DJ, Speth JM, Peters-Golden M. Signed, sealed, delivered: microenvironmental modulation of extracellular vesicle–dependent immunoregulation in the lung. Front Cell Dev Biol. 2016;4:94. doi: 10.3389/fcell.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourdonnay E, Zasłona Z, Penke LR, Speth JM, Schneider DJ, Przybranowski S, Swanson JA, Mancuso P, Freeman CM, Curtis JL, et al. Transcellular delivery of vesicular SOCS proteins from macrophages to epithelial cells blunts inflammatory signaling. J Exp Med. 2015;212:729–742. doi: 10.1084/jem.20141675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speth JM, Bourdonnay E, Penke LR, Mancuso P, Moore BB, Weinberg JB, Peters-Golden M. Alveolar epithelial cell–derived prostaglandin E2 serves as a request signal for macrophage secretion of suppressor of cytokine signaling 3 during innate inflammation. J Immunol. 2016;196:5112–5120. doi: 10.4049/jimmunol.1502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon HG, Cao Y, Yang J, Lee JH, Choi HS, Jin Y. Lung epithelial cell–derived extracellular vesicles activate macrophage-mediated inflammatory responses via ROCK1 pathway. Cell Death Dis. 2015;6:e2016. doi: 10.1038/cddis.2015.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H, Zhang D, Zhu Z, Dela Cruz CS, Jin Y. Epithelial cell–derived microvesicles activate macrophages and promote inflammation via microvesicle-containing microRNAs. Sci Rep. 2016;6:35250. doi: 10.1038/srep35250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, Ratajczak MZ. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 40.Xiao X, Yu S, Li S, Wu J, Ma R, Cao H, Zhu Y, Feng J. Exosomes: decreased sensitivity of lung cancer A549 cells to cisplatin. PLoS One. 2014;9:e89534. doi: 10.1371/journal.pone.0089534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XQ, Liu JT, Fan LL, Liu Y, Cheng L, Wang F, Yu HQ, Gao J, Wei W, Wang H, et al. Exosomes derived from gefitinib-treated EGFR-mutant lung cancer cells alter cisplatin sensitivity via up-regulating autophagy. Oncotarget. 2016;7:24585–24595. doi: 10.18632/oncotarget.8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng Z, Rong Y, Teng Y, Zhuang X, Samykutty A, Mu J, Zhang L, Cao P, Yan J, Miller D, et al. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene. 2017;36:639–651. doi: 10.1038/onc.2016.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman MA, Barger JF, Lovat F, Gao M, Otterson GA, Nana-Sinkam P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016;7:54852–54866. doi: 10.18632/oncotarget.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almendros I, Khalyfa A, Trzepizur W, Gileles-Hillel A, Huang L, Akbarpour M, Andrade J, Farré R, Gozal D. Tumor cell malignant properties are enhanced by circulating exosomes in sleep apnea. Chest. 2016;150:1030–1041. doi: 10.1016/j.chest.2016.08.1438. [DOI] [PubMed] [Google Scholar]
- 46.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szul T, Bratcher PE, Fraser KB, Kong M, Tirouvanziam R, Ingersoll S, Sztul E, Rangarajan S, Blalock JE, Xu X, et al. Toll-like receptor 4 engagement mediates prolyl endopeptidase release from airway epithelia via exosomes. Am J Respir Cell Mol Biol. 2016;54:359–369. doi: 10.1165/rcmb.2015-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, et al. Characterization of human plasma–derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, Pass HI. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8:1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, Cheng W, Wang F, Qi LW, Chen Y, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandfeld-Paulsen B, Aggerholm-Pedersen N, Bæk R, Jakobsen KR, Meldgaard P, Folkersen BH, Rasmussen TR, Varming K, Jørgensen MM, Sorensen BS. Exosomal proteins as prognostic biomarkers in non–small cell lung cancer. Mol Oncol. 2016;10:1595–1602. doi: 10.1016/j.molonc.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabrera-Benítez NE, Valladares F, García-Hernández S, Ramos-Nuez Á, Martín-Barrasa JL, Martínez-Saavedra MT, Rodríguez-Gallego C, Muros M, Flores C, Liu M, et al. Altered profile of circulating endothelial-derived microparticles in ventilator-induced lung injury. Crit Care Med. 2015;43:e551–e559. doi: 10.1097/CCM.0000000000001280. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi T, Kobayashi S, Fujino N, Suzuki T, Ota C, He M, Yamada M, Suzuki S, Yanai M, Kurosawa S, et al. Increased circulating endothelial microparticles in COPD patients: a potential biomarker for COPD exacerbation susceptibility. Thorax. 2012;67:1067–1074. doi: 10.1136/thoraxjnl-2011-201395. [DOI] [PubMed] [Google Scholar]
- 55.Kim YS, Choi JP, Kim MH, Park HK, Yang S, Kim YS, Kim TB, Cho YS, Oh YM, Jee YK, et al. IgG sensitization to extracellular vesicles in indoor dust is closely associated with the prevalence of non-eosinophilic asthma, COPD, and lung cancer. Allergy Asthma Immunol Res. 2016;8:198–205. doi: 10.4168/aair.2016.8.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Admyre C, Grunewald J, Thyberg J, Gripenbäck S, Tornling G, Eklund A, Scheynius A, Gabrielsson S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur Respir J. 2003;22:578–583. doi: 10.1183/09031936.03.00041703. [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Bravo MJ, Wahlund CJ, Qazi KR, Moulder R, Lukic A, Rådmark O, Lahesmaa R, Grunewald J, Eklund A, Gabrielsson S. Pulmonary sarcoidosis is associated with exosomal vitamin D–binding protein and inflammatory molecules. J Allergy Clin Immunol. 2017;139:1186–1194. doi: 10.1016/j.jaci.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 58.Qazi KR, Torregrosa Paredes P, Dahlberg B, Grunewald J, Eklund A, Gabrielsson S. Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis. Thorax. 2010;65:1016–1024. doi: 10.1136/thx.2009.132027. [DOI] [PubMed] [Google Scholar]
- 59.Gregson AL, Hoji A, Injean P, Poynter ST, Briones C, Palchevskiy V, Weigt SS, Shino MY, Derhovanessian A, Sayah D, et al. Altered exosomal RNA profiles in bronchoalveolar lavage from lung transplants with acute rejection. Am J Respir Crit Care Med. 2015;192:1490–1503. doi: 10.1164/rccm.201503-0558OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harms A, Fuehner T, Warnecke G, Haverich A, Gottlieb J, Trummer A. Epithelial and erythrocyte microvesicles from bronchoalveolar lavage fluid are elevated and associated with outcome in chronic lung allograft dysfunction. Transplantation. 2015;99:2394–2400. doi: 10.1097/TP.0000000000000881. [DOI] [PubMed] [Google Scholar]
- 61.Levänen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, Pollack JL, Sköld CM, Svartengren M, Grunewald J, Gabrielsson S, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. 2013;131:894–903. doi: 10.1016/j.jaci.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torregrosa Paredes P, Esser J, Admyre C, Nord M, Rahman QK, Lukic A, Rådmark O, Grönneberg R, Grunewald J, Eklund A, et al. Bronchoalveolar lavage fluid exosomes contribute to cytokine and leukotriene production in allergic asthma. Allergy. 2012;67:911–919. doi: 10.1111/j.1398-9995.2012.02835.x. [DOI] [PubMed] [Google Scholar]
- 63.Kulshreshtha A, Ahmad T, Agrawal A, Ghosh B. Proinflammatory role of epithelial cell–derived exosomes in allergic airway inflammation. J Allergy Clin Immunol. 2013;131:1194–1203. doi: 10.1016/j.jaci.2012.12.1565. 1203.e1–14. [DOI] [PubMed] [Google Scholar]
- 64.Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, Le Chevalier T, Livartoski A, Barlesi F, Laplanche A, et al. Dendritic cell–derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. OncoImmunology. 2015;5:e1071008. doi: 10.1080/2162402X.2015.1071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gobbo J, Marcion G, Cordonnier M, Dias AMM, Pernet N, Hammann A, Richaud S, Mjahed H, Isambert N, Clausse V, et al. Restoring anticancer immune response by targeting tumor-derived exosomes with a HSP70 peptide aptamer. J Natl Cancer Inst. 2015;108 doi: 10.1093/jnci/djv330. djv330. [DOI] [PubMed] [Google Scholar]
- 66.Aliotta JM, Pereira M, Wen S, Dooner MS, Del Tatto M, Papa E, Goldberg LR, Baird GL, Ventetuolo CE, Quesenberry PJ, et al. Exosomes induce and reverse monocrotaline-induced pulmonary hypertension in mice. Cardiovasc Res. 2016;110:319–330. doi: 10.1093/cvr/cvw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cruz FF, Borg ZD, Goodwin M, Coffey AL, Wagner DE, Rocco PR, Weiss DJ. CD11b+ and Sca-1+ cells exert the main beneficial effects of systemically administered bone marrow–derived mononuclear cells in a murine model of mixed Th2/Th17 allergic airway inflammation. Stem Cells Transl Med. 2016;5:488–499. doi: 10.5966/sctm.2015-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi SJ, Kim MH, Jeon J, Kim OY, Choi Y, Seo J, Hong SW, Lee WH, Jeon SG, Gho YS, et al. Active immunization with extracellular vesicles derived from Staphylococcus aureus effectively protects against staphylococcal lung infections, mainly via Th1 cell–mediated immunity. PLoS One. 2015;10:e0136021. doi: 10.1371/journal.pone.0136021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson JG, Liu KD, Zhuo H, Caballero L, McMillan M, Fang X, Cosgrove K, Vojnik R, Calfee CS, Lee JW, et al. Mesenchymal stem (stromal) cells for treatment of ARDS: a phase 1 clinical trial. Lancet Respir Med. 2015;3:24–32. doi: 10.1016/S2213-2600(14)70291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsai HL, Chang JW, Yang HW, Chen CW, Yang CC, Yang AH, Liu CS, Chin TW, Wei CF, Lee OK. Amelioration of paraquat-induced pulmonary injury by mesenchymal stem cells. Cell Transplant. 2013;22:1667–1681. doi: 10.3727/096368912X657765. [DOI] [PubMed] [Google Scholar]
- 71.Lee JW, Fang X, Gupta N, Serikov V, Matthay MA. Allogeneic human mesenchymal stem cells for treatment of E. coli endotoxin–induced acute lung injury in the ex vivo perfused human lung. Proc Natl Acad Sci U S A. 2009;106:16357–16362. doi: 10.1073/pnas.0907996106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu YG, Feng XM, Abbott J, Fang XH, Hao Q, Monsel A, Qu JM, Matthay MA, Lee JW. Human mesenchymal stem cell microvesicles for treatment of Escherichia coli endotoxin–induced acute lung injury in mice. Stem Cells. 2014;32:116–125. doi: 10.1002/stem.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gennai S, Monsel A, Hao Q, Park J, Matthay MA, Lee JW. Microvesicles derived from human mesenchymal stem cells restore alveolar fluid clearance in human lungs rejected for transplantation. Am J Transplant. 2015;15:2404–2412. doi: 10.1111/ajt.13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phinney DG, Di Giuseppe M, Njah J, Sala E, Shiva S, St Croix CM, Stolz DB, Watkins SC, Di YP, Leikauf GD, et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat Commun. 2015;6:8472. doi: 10.1038/ncomms9472. [DOI] [PMC free article] [PubMed] [Google Scholar]