Abstract
Rationale: Idiopathic pulmonary fibrosis (IPF) involves the accumulation of α-smooth muscle actin–expressing myofibroblasts arising from interactions with soluble mediators such as transforming growth factor-β1 (TGF-β1) and mechanical influences such as local tissue stiffness. Whereas IPF fibroblasts are enriched for aerobic glycolysis and innate immune receptor activation, innate immune ligands related to mitochondrial injury, such as extracellular mitochondrial DNA (mtDNA), have not been identified in IPF.
Objectives: We aimed to define an association between mtDNA and fibroblast responses in IPF.
Methods: We evaluated the response of normal human lung fibroblasts (NHLFs) to stimulation with mtDNA and determined whether the glycolytic reprogramming that occurs in response to TGF-β1 stimulation and direct contact with stiff substrates, and spontaneously in IPF fibroblasts, is associated with excessive levels of mtDNA. We measured mtDNA concentrations in bronchoalveolar lavage (BAL) from subjects with and without IPF, as well as in plasma samples from two longitudinal IPF cohorts and demographically matched control subjects.
Measurements and Main Results: Exposure to mtDNA augments α-smooth muscle actin expression in NHLFs. The metabolic changes in NHLFs that are induced by interactions with TGF-β1 or stiff hydrogels are accompanied by the accumulation of extracellular mtDNA. These findings replicate the spontaneous phenotype of IPF fibroblasts. mtDNA concentrations are increased in IPF BAL and plasma, and in the latter compartment, they display robust associations with disease progression and reduced event-free survival.
Conclusions: These findings demonstrate a previously unrecognized and highly novel connection between metabolic reprogramming, mtDNA, fibroblast activation, and clinical outcomes that provides new insight into IPF.
Keywords: mitochondria, interstitial lung disease, mechanotransduction, biomarkers
At a Glance Commentary
Scientific Knowledge on the Subject
Idiopathic pulmonary fibrosis (IPF) is a fatal disease defined by the presence of highly activated, glycolytically programmed myofibroblasts. The relationship of these cells to mitochondrial DNA (mtDNA), a danger-associated molecular pattern that participates in inflammation and tissue remodeling, has not been defined.
What This Study Adds to the Field
We show that glycolytic reprogramming in normal human lung fibroblasts induced by either transforming growth factor-β1 or local substrate stiffness, and spontaneously exhibited by IPF fibroblasts, is accompanied by excessive local concentrations of extracellular mtDNA. Exposure to mtDNA augments α-smooth muscle actin expression in otherwise normal lung fibroblasts. In a clinical correlate of our findings, mtDNA concentrations are increased in the lungs and blood of patients with IPF, where, in the latter compartment, they display an impressive association with disease progression and reduced event-free survival. These results demonstrate a previously unrecognized and highly novel connection between metabolic remodeling, mtDNA, fibroblast activation, and clinical outcomes that should be investigated as a new therapeutic area in IPF.
Idiopathic pulmonary fibrosis (IPF) is a fatal interstitial lung disease characterized by radiographic or histologic findings of usual interstitial pneumonia in the absence of identifiable causes (1). With a median survival of 2.5–3.5 years (2), it predominately affects individuals aged 60–75 years old (3) and exhibits a highly variable disease course (4) that would benefit from prognostic models incorporating clinical parameters with circulating biomarkers (5, 6). A characteristic feature of IPF is the accumulation of α-smooth muscle actin (α-SMA)–expressing myofibroblasts (1) that may arise from normal lung fibroblasts in response to biochemical influences such as transforming growth factor-β1 (TGF-β1) (1) and biophysical responses to the stiffened microenvironment (7). TGF-β1 also critically regulates the alterations in mitochondrial function and bioenergetics that are required to fuel these activated cells (8, 9). However, because TGF-β1’s pleiotropic functions make therapeutic targeting difficult (10), interruption of its downstream effects may be both safer and more efficacious.
The contribution of innate immune activation to IPF is an emerging field of study (11–14). Innate immune cells such as macrophages are believed to amplify fibrosis via a variety of mechanisms (11); mutations in genes related to innate immunity are associated with IPF (12), and expression of innate immune receptors such as Toll-like receptor 9 (TLR9) (13) demonstrate outcome-predictive properties in IPF (14). Although these data suggest that innate immune agonists might be enriched in IPF, their identification remains elusive. Innate immune agonists are stratified into pathogen-associated molecular patterns, which are derived from microbes, and danger-associated molecular patterns (DAMPS), which are released by injured or activated cells (15). Because IPF is not believed to result from infectious stimuli, DAMPS may be more relevant in this clinical context. One such ligand is the hypomethylated, CpG-rich mitochondrial DNA (mtDNA) that is released by both necrotic cells (16) and viable cells in response to various stressors (17). Exposure to mtDNA (18) or synthetic analogs (14) is sufficient to activate macrophages and fibroblasts in several experimental settings, including experimentally induced lung fibrosis (18), and mtDNA participates in the formation of neutrophil extracellular traps that contribute to inflammation and tissue remodeling in a variety of human conditions, including pulmonary fibrosis (19). Interestingly, despite the distinct alterations in fibroblast metabolism caused by TGF-β1 (8, 9), as well as the demonstration of mitochondrial abnormalities as a driver of myofibroblast activation and experimentally induced lung fibrosis (20), an association of IPF with the extracellular accumulation of mtDNA has not been explored. Furthermore, although elevations in circulating mtDNA predict mortality in several clinical settings (21, 22), a clinical relationship with IPF outcomes has yet to be defined.
We aimed to define an association between fibroblast activation, extracellular mtDNA, and clinical outcomes in IPF. Our experimental platform combined TGF-β1 stimulation of normal human lung fibroblasts (NHLFs) with a biomechanical model of the IPF microenvironment to demonstrate that glycolytic reprogramming induced by fibrotic stimuli is paralleled by enhanced concentrations of mtDNA in the extracellular milieu and replicates the spontaneous phenotype of IPF fibroblasts. Exogenous mtDNA mediates α-SMA expression in normal lung fibroblasts and is enriched in the bronchoalveolar lavage (BAL) fluid and plasma of patients with IPF, where, in the latter compartment, it displays a robust association with reduced event-free survival. Some of the results of these studies were previously reported in the form of an abstract (23).
Methods
Detailed methods are provided in the online supplement.
Human Subjects
All human studies were performed with informed consent according to protocols approved by the institutional review boards at the University of Pittsburgh and Yale School of Medicine. IPF diagnosis was based on current consensus guidelines (24, 25) rendered through expert multidisciplinary discussion at each institution. Demographic and clinical data were obtained at the time of the blood draw. Patients were followed for 43 months for time-to-event analysis, where death resulting from any cause was considered an event.
Culture and TGF-β1 Stimulation of Human Lung Fibroblasts
NHLFs and IPF fibroblasts obtained from Lonza (Allendale, NJ) and Asterand Bioscience (Detroit, MI) were used between passages 3 and 7 and stimulated with and without 5 ng/ml of recombinant human TGF-β1 as previously described (14).
Seahorse Metabolism Assay
Cellular metabolism was measured with the Seahorse XFe96 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA) for real-time analysis of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR).
Fluorescence-activated Cell Sorting
Fluorescence-activated cell sorting–based determination of mitochondrial membrane potential using MitoTracker Red and MitoTracker Green probes (Life Technologies, Carlsbad, CA) was performed as described in the online supplement.
Immunostaining and Microscopy
Immunostaining and evaluation of cells and tissues via conventional or fluorescence microscopy or structured illumination microscopy were performed as described in the online supplement.
Transmission Electron Microscopy
TGF-β1–stimulated cells were evaluated via transmission electron microscopy as described in the online supplement.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Assay
Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described in the online supplement.
DNA Isolation and Quantification
Isolation and quantification of the human MT-ATP6 gene and/or ACTB from fibroblasts, Escherichia coli, cell-free culture supernatants, BAL fluid, and plasma were performed as previously described (17, 22).
mtDNA Isolation and Stimulation of NHLFs
Purification of DNA from mitochondria isolated from NHLFs and stimulation of NHLFs with this DNA were performed as described in the online supplement.
RNA Isolation and Quantitative Reverse Transcription–Polymerase Chain Reaction
Total cellular RNA was isolated, and relative expression of ACTA2 was quantified using the comparative cycle threshold method as previously reported (11).
Polyacrylamide Hydrogel Preparation
Culture of NHLFs on hydrogels constructed of varying stiffness (26) is described in the online supplement.
Statistical Analysis
Data distribution was assessed using the D’Agostino-Pearson omnibus test or the Shapiro-Wilk test on the basis of sample size. For continuous data, parametric comparisons were made using Student’s t test, and nonparametric data were compared using the Mann-Whitney U test. Categorical data were compared with one-way analysis of variance and Fisher’s exact test. Receiver operating characteristic curve analysis of the Pittsburgh IPF cohort was used to determine the mtDNA cutoff. Kaplan-Meier analysis was performed to determine survival associations. Multivariate Cox regression hazard ratios (HRs) were used to model associations with relevant covariates. These evaluations were performed using Prism 7.0 (GraphPad Software, La Jolla, CA) or MedCalc (MedCalc Software, Ostend, Belgium) software.
Results
TGF-β1–stimulated NHLFs Show Altered Cellular Metabolism
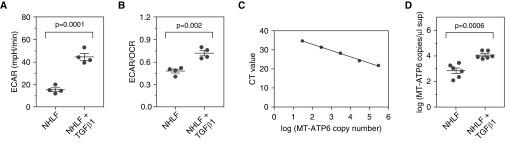
To determine whether the metabolic changes described in NHLFs exposed to fibrotic stimuli (27) were accompanied by extracellular accumulation of mitochondria-related DAMPS, we characterized the bioenergetic response of NHLFs to TGF-β1. NHLFs obtained from three independent donors used between passages 3 and 7 were cultured in the absence or presence of 5 ng/ml of TGF-β1 and assessed for various metabolic parameters. After 24 hours, transmission electron microscopic examination of these cells revealed the elongated mitochondrial morphology that has previously been described following stimulation with TGF-β1 (see Figure E1 in the online supplement). By 7 days, evaluation of mitochondrial substructure by structured illumination microscopy–based immunofluorescence of the mitochondrial outer membrane marker TOM20 and the major mtDNA packaging protein mitochondrial transcription factor A showed that, in contrast to unstimulated cells, which displayed the typical findings of narrow mitochondria (28), TGF-β1–treated cells adopted a strikingly altered and enlarged mitochondrial appearance (Figure E2A). Conventional fluorescence microscopy performed at the same time point revealed that TGF-β1–stimulated cells showed the linear organization of α-SMA consistent with myofibroblast transformation (Figure E2B). Despite this phenotypic change, mitochondrial membrane potential was similar between groups (mean fluorescence intensity ratio of MitoTracker Red to MitoTracker Green, 1.709 ± 0.929 vs. 1.857 ± 1.274; n = 4 for both groups; P > 0.05) (Figure E2C). However, bioenergetic measurements with the Seahorse XFe96 Analyzer identified that, relative to naive cells, TGF-β1–treated NHLFs displayed enhanced glycolysis as measured by substantial increases in the mean ECAR (15.39 ± 1.684 mpH/min vs. 44.63 ± 2.978 mpH/min; P = 0.0001; n = 4 for both groups) (Figure 1A) and the ratio of ECAR to OCR (mean ECAR/OCR ratio, 0.4804 ± 0.026 vs. 0.7195 ± 0.036; P = 0.002; n = 4 for both groups) (Figure 1B). These data confirm that TGF-β1 alters mitochondrial morphology and glycolytic reprogramming in previously normal lung fibroblasts.
Figure 1.
Transforming growth factor-β1 (TGF-β1)–stimulated normal human lung fibroblasts (NHLFs) show altered metabolism and high concentrations of extracellular mitochondrial DNA. (A and B) NHLFs stimulated with 5 ng/ml of TGF-β1 for 7 days (right) demonstrated the previously reported increase in glycolysis relative to unstimulated cells (left) as measured by significant elevations in (A) extracellular acidification rate (ECAR) and (B) ratio of ECAR to oxygen consumption rate (OCR). Data are presented as mean ECAR (mpH/min) and mean (±SEM) ratio of ECAR to OCR, respectively. (C) A standard curve was developed from serial dilutions of a commercially available plasmid containing the sequence of the human MT-ATP6 gene. (D) Relative to supernatants obtained from NHLFs cultured with normal medium (left), the mean MT-ATP6 copy number is significantly increased in the supernatant of NHLFs stimulated with 5 ng/ml of TGF-β1 for 7 days (right). Data are presented graphically as log base 10 of the raw values (MT-ATP6 copies per microliter of supernatant) with mean ± SEM. A graph including the raw values is presented in Figure E2D. CT = cycle threshold.
TGF-β1–stimulated NHLFs Generate Extracellular mtDNA
To determine whether these mitochondrial changes were accompanied by the extracellular accumulation of a mitochondria-related DAMP, we employed a well-described method of polymerase chain reaction (PCR)-based quantification of MT-ATP6 copy number in the supernatants of these cells (Figure 1C) (17, 22). We found that, relative to unstimulated samples, supernatants obtained from TGF-β1–exposed NHLFs displayed a robust increase in the mean MT-ATP6 copy numbers (2.838 ± 0.214 log copies per microliter vs. 4.055 ± 0.118 log copies per microliter; P = 0.0006; n = 6 for both groups) (Figures 1D and E2D). These studies show that the extracellular milieu of TGF-β1–exposed NHLFs contains high concentrations of mtDNA.
TGF-β1–stimulated NHLFs Show Reduced Mitochondrial Mass
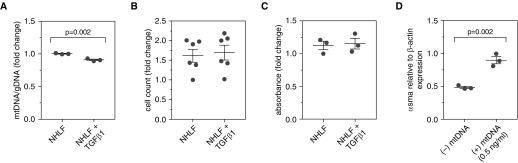
The finding of increased extracellular mtDNA in these studies suggested that prolonged TGF-β1 exposure might result in the loss of mitochondria from the intracellular space. To test this hypothesis, NHLFs from three separate donors were subjected to stimulation with 5 ng/ml of TGF-β1 for 7 days, at which point mitochondrial mass was determined using PCR-based comparison of DNA derived from mitochondria (assessed by the MT-ATP6 gene) and the genome (measured by the ACTB gene). This approach identified a decline in the mean ratio of mtDNA to genomic DNA (1.003 ± 0.005 vs. 0.909 ± 0.012; n = 3 for both groups; P = 0.002) (Figure 2A), showing that the extracellular accumulation of mtDNA in this context is paralleled by a reduction in intracellular mitochondrial mass.
Figure 2.
Transforming growth factor-β1 (TGF-β1) stimulation of normal human lung fibroblasts (NHLFs) reduces mitochondrial mass without affecting cell viability. (A) NHLFs were stimulated with 5 ng/ml of TGF-β1 for 7 days, at which point mitochondrial mass was determined using polymerase chain reaction–based comparison of DNA derived from mitochondria (assessed by the MT-ATP6 gene) and the genome (measured by the β-actin gene). Relative to unstimulated cells (left), there was a significant decline in the mean ratio of mitochondrial DNA (mtDNA) to genomic DNA (gDNA) in TGF-β1–treated cells (right). Data are presented as the mean (±SEM) fold change in ratio of mtDNA to gDNA. (B) Cell counts in NHLF cultures stimulated with (right) and without (left) 5 ng/ml of TGF-β1 for 7 days were unchanged across both conditions. Data are presented as mean (±SEM) fold change in cell count. (C) Assessment of viability with the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay revealed similar fold changes in absorbance at a wavelength of 540 nm between NHLFs stimulated with (right) and without (left) 5 ng/ml of TGF-β1 for 7 days. Data are presented as the mean (±SEM) fold change in absorbance. (D) Stimulation of NHLFs with 0.5 ng/ml of mtDNA for 48 hours significantly increased α-smooth muscle actin (α-SMA) expression, relative to β-actin, by NHLFs. Data are presented as mean (±SEM) α-smooth muscle actin expression relative to β-actin.
TGF-β1–stimulated NHLFs Are Viable
The finding of reduced intracellular mitochondrial mass combined with increased extracellular mtDNA suggested that, as has been reported in other scenarios, mitochondria may be actively released by stimulated cells (29). However, an alternative explanation is that the mtDNA stems from the lysis of necroptotic cells. To explore the latter concept, we compared pre- and postculture cell counts from these experiments and found unchanged numbers (P > 0.05 for all comparisons; n = 6 for both groups) (Figure 2B). In addition, viability assessment performed on a subset of these cells with the MTT assay was similar in both conditions (P > 0.05 for all comparisons; n = 3 for both groups) (Figure 2C). These data show that stimulation with TGF-β1 does not influence the quantity or viability of NHLFs, supporting the contention that the presence of extracellular mtDNA is unlikely to result from the death of nonviable cells.
mtDNA Stimulation Induces α-SMA Expression in NHLFs
A variety of immune and stromal cells can internalize and respond to CpG-rich DNA (14, 18, 30). To determine whether fibroblasts exhibit similar responses to mtDNA, NHLFs from three separate donors were grown to confluence, stimulated for 48 hours with mtDNA isolated from a parallel fibroblast culture, and assessed for expression of α-SMA via reverse transcription–PCR. As compared with unstimulated NHLFs, cells exposed to mtDNA developed increased expression of ACTA2 relative to ACTB (0.483 ± 0.016 vs. 0.897 ± 0.054; n = 3 for both groups; P = 0.002) (Figure 2D). These data demonstrate a functional ramification of mtDNA exposure by showing it augments α-SMA expression in otherwise normal lung fibroblasts.
Direct Contact with Stiff Surfaces Phenocopies the Effect of TGF-β1
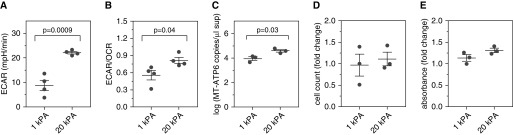
The data presented above reveal a relationship between fibroblast activation and extracellular mtDNA in the presence of soluble fibrotic mediators. However, given the pivotal contribution of mechanotransductive signaling to fibroblast activation (31), we thought biophysical influences might also be involved. Thus, NHLFs were cultured for 7 days on tunable hydrogels constructed to approximate the mean stiffness of the normal (1 kPa) and fibrotic (20 kPa) human lung (26). Unlike the glass culture substrates, which are bioinert, the hydrogel allows standardization of coating matrix (in this case, collagen) between the soft and stiff conditions so that the only independent variable is the stiffness of the culture substrate. Increased expression of α-SMA in the 20-kPa samples was confirmed via immunofluorescence (Figure E3A). Measurements of aerobic glycolysis emulated those caused by TGF-β1; relative to NHLFs cultured on the low-stiffness hydrogels, cells grown on the 20-kPa gels displayed substantially elevated ECAR (8.793 ± 2.116 mpH/min vs. 22.13 ± 0.497 mpH/min; P = 0.0009; n = 4 for both groups) (Figure 3A) and ECAR/OCR (0.556 ± 0.083 vs. 0.815 ± 0.052; n = 4 for both groups; P = 0.04) (Figure 3B). When culture supernatants were assessed for mtDNA content, a greater than fourfold increase in the detection of MT-ATP6 copies was observed (mean value, 3.95 ± 0.153 log copies per microliter vs. 4.59 ± 0.11 log copies per microliter; P = 0.03; n = 3 for both groups) (Figures 3C and E3B). Limitations in cell number precluded measurement of mitochondrial mass, but postculture cell counts and MTT-based assessments were stable across conditions (Figures 3D and 3E). These findings show that the NHLF response to increased local stiffness phenocopies the glycolytic reprogramming and extracellular mtDNA increases caused by TGF-β1.
Figure 3.
Direct contact with stiff surfaces phenocopies exposure to transforming growth factor-β1 in normal human lung fibroblasts (NHLFs). Measurements of cellular metabolism revealed enhanced aerobic glycolysis in NHLFs grown on the 20-kPa hydrogels for 7 days (right) compared with cells grown on the 1-kPa hydrogels (left), as evidenced by significantly elevated (A) extracellular acidification rate (ECAR) and (B) ECAR/oxygen consumption rate (OCR) ratio. Data are shown as mean (±SEM) ECAR (mpH/min) and mean (±SEM) ratio of ECAR/OCR, respectively. (C) After 7 days, relative to supernatant obtained from NHLFs seeded on the 1-kPa hydrogel (left), there was a significant increase in MT-ATP6 concentration in the supernatant of NHLFs seeded on the 20-kPa hydrogel (right). Data are presented graphically as log base 10 of the raw values (MT-ATP6 copies per microliter of supernatant) with mean (±SEM). A graph including the raw values is presented in Figure E3B. NHLFs grown on the 20-kPa hydrogel (right) demonstrated no significant change in (D) cell counts or (E) viability based on 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay compared with cells grown on the 1-kPa hydrogel (left) for 7 days. Data are presented as mean (±SEM) fold change in cell count and mean (±SEM) fold change in absorbance, respectively.
IPF Fibroblasts Exhibit High Concentrations of Extracellular mtDNA
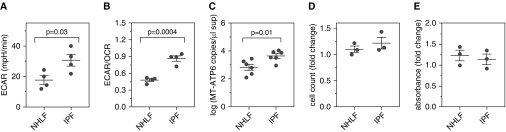
The data above show increased mtDNA levels in the microenvironment of NHLFs exposed to multiple fibrogenic factors. To determine the clinical relevance of these findings, we evaluated these endpoints in fibroblasts obtained from IPF lungs. Immunostaining confirmed the increased expression of α-SMA (Figure E4A), and bioenergetic evaluation revealed the robustly increased mean ECAR (17.69 ± 2.773 mpH/min vs. 30.68 ± 3.868 mpH/min; n = 4 for both groups; P = 0.03) (Figure 4A) and ECAR/OCR ratio (0.48 ± 0.026 vs. 0.864 ± 0.048; n = 4 for both groups; P = 0.0004) (Figure 4B) that have previously been reported for these cells (9, 27). Furthermore, relative to NHLFs, IPF fibroblasts developed a significant increase in extracellular MT-ATP6 concentrations (mean value, 2.838 ± 0.214 log copies per microliter vs. 3.646 ± 0.162 log copies per microliter; P = 0.01; n = 6 for both groups) (Figures 4C and E4B). Importantly, postculture cell counts and MTT-based assessment revealed stable numbers and viability, respectively (Figures 4D and 4E). These data show that the α-SMA–expressing, glycolytically reprogrammed phenotype of IPF fibroblasts is accompanied by a microenvironment that is enriched for mtDNA.
Figure 4.
Idiopathic pulmonary fibrosis (IPF) fibroblasts exhibit enhanced glycolysis and increased extracellular mitochondrial DNA. Compared with normal human lung fibroblasts (NHLFs) (left), IPF fibroblasts (right) displayed enhanced aerobic glycolysis, as measured by a significantly elevated (A) extracellular acidification rate (ECAR) and (B) ECAR/oxygen consumption rate (OCR) ratio. Data are presented as mean (±SEM) ECAR (mpH/min) and mean (±SEM) ratio of ECAR/OCR, respectively. (C) Relative to samples obtained from NHLFs (left), a significant increase in MT-ATP6 concentration was detected in supernatants from IPF fibroblasts (right). Data are presented graphically as log base 10 of the raw values (MT-ATP6 copies per microliter of supernatant) with mean (±SEM). A graph including the raw values is presented in Figure E4B. (D) Cell counts were unchanged between NHLFs (left) and IPF fibroblasts (right). Data are presented as mean (±SEM) fold change in cell count. (E) 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay demonstrated no significant differences in the viability of NHLFs (left) and IPF fibroblasts (right). Data are presented as mean (±SEM) fold change in absorbance.
mtDNA Is Elevated in the BAL Fluid of Patients with IPF
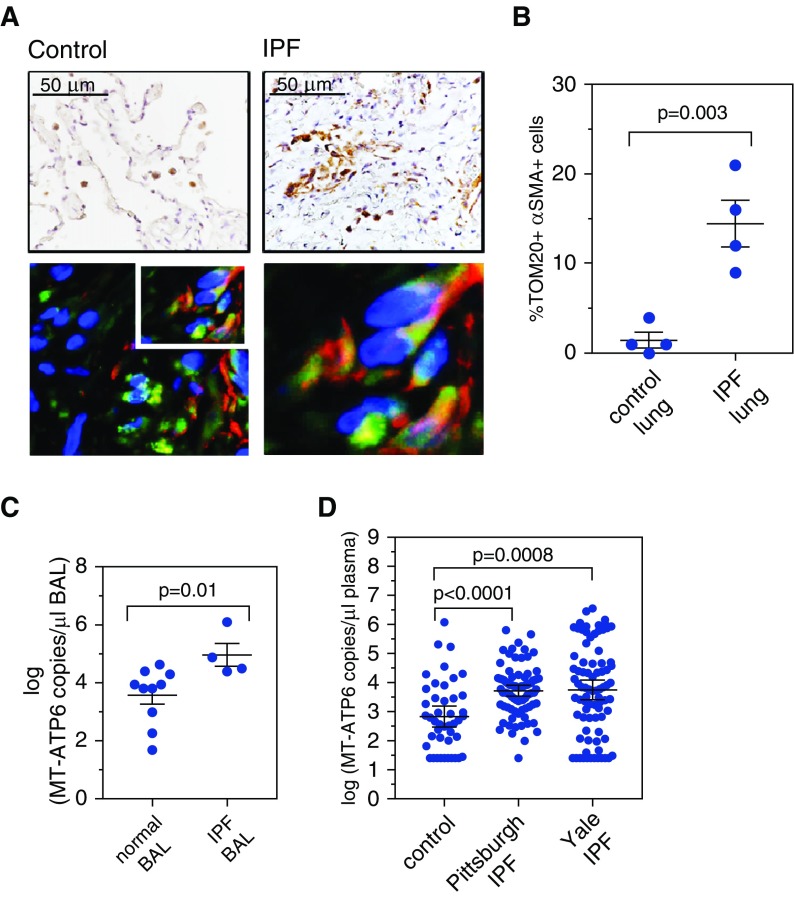
We then explored the above findings in vivo. Immunohistochemistry-based detection of TOM20 in IPF lung tissue revealed the previously reported expression in alveolar epithelial cells and macrophages (32) and also in α-SMA–expressing cells located in areas of fibrosis (Figures 5A and 5B). We did not attempt to quantify the extracellular MT-ATP6 content of lung tissue, owing to the technical impossibility of creating a cell-free preparation via tissue dissociation without lysing cells. Although there are differences in recovery with BAL samples obtained from healthy subjects versus those with fibrotic lung disease, multiple studies have demonstrated the useful role of BAL in IPF (33), and we accepted it as an additional surrogate of the IPF lung microenvironment. Thus, we analyzed the cell-free supernatant of archived BAL samples from subjects with newly diagnosed IPF (n = 4) and demographically matched control subjects lacking known interstitial lung disease (n = 10). Relative to samples from control subjects, BAL samples from IPF subjects demonstrated a robust increase in the median MT-ATP6 concentration (3.87 log copies per microliter vs. 4.68 log copies per microliter; P = 0.01) (Figures 5C and E5A). Due to the theoretical concern that the MT-ATP6 primers might amplify bacteria present in the BAL, we confirmed specificity by showing that these primers do not amplify DNA derived from E. coli (Figure E5B). These data show that mtDNA not only accumulates in the supernatants of NHLFs exposed to fibrogenic stimuli but also exists in high concentrations in the IPF lung.
Figure 5.
Mitochondrial DNA is elevated in the lungs and plasma of patients with idiopathic pulmonary fibrosis (IPF). (A) Immunohistochemistry-based detection of TOM20 in IPF lung tissues (relative to control samples) revealed strong detection in fibrotic regions of lung in cells that also expressed α-smooth muscle actin (α-SMA). Top left: Normal lung stained with TOM20 shows detection in macrophages (brown). Top right: IPF lung stained with TOM20 shows detection in fibrotic regions (brown). Images are counterstained with hematoxylin, and scale bar is 50 μm. Bottom left: Combined immunofluorescence for α-SMA (red) and TOM20 (green) on IPF lung tissue. Slides are counterstained with 4′,6-diamidino-2-phenylindole (blue nuclear stain), and images include a 50-μm scale bar. Bottom right: Enlargement of the boxed area shows codetection of TOM20 and α-SMA in spindle-shaped cells with the appearance of fibroblasts. (B) Relative to control lung tissue, there were a significantly higher percentage of TOM20-positive cells expressing α-SMA in the IPF lung. Data are presented as mean (±SEM) percentage of TOM20 cells that also express α-SMA. (C) Relative to bronchoalveolar lavage (BAL) samples obtained from subjects without parenchymal lung disease (left), the median concentration of MT-ATP6 was significantly increased in BAL samples obtained from IPF subjects (right). Data are presented graphically as log base 10 of the raw values of MT-ATP6 copies per microliter of BAL with median value and interquartile range. A graph including the raw values is presented in Figure E5A. (D) Relative to plasma specimens obtained from aged-matched control subjects (left), median concentration of MT-ATP6 in both the Pittsburgh (middle) and Yale (right) IPF cohorts was significantly increased. Data are presented graphically as log base 10 of the raw values of MT-ATP6 copies per microliter of plasma with median value and interquartile range. A graph including the raw values is presented in Figure E6A.
Plasma mtDNA Is Elevated in Two Independent IPF Cohorts
It has become increasingly clear that molecular events in the IPF lung are detectable in the circulation (34). To evaluate whether mtDNA might display such properties, we obtained plasma from a cohort of aged but otherwise healthy individuals lacking known inflammatory or fibrotic disease, as well as from two large, well-characterized IPF cohorts. Baseline characteristics are described in Table 1. Both cohorts were mostly white (96.30% and 97.56%, respectively) and male (77.78% and 86.59%, respectively). Baseline FVC percent predicted values were 65.38% and 75.43%, respectively, and diffusing capacity of the lung for carbon monoxide (DlCO) percent predicted values were 50.41% and 42.89%, respectively. Disease severity was assessed with the GAP (gender, age, and physiology) index, with mean scores of 3.88 and 4.26, respectively. Relative to plasma from control subjects (n = 44), plasma obtained from the Pittsburgh IPF cohort (n = 81) contained substantial increases in median MT-ATP6 copy numbers (2.68 log copies per microliter vs. 3.77 log copies per microliter; P < 0.0001) (Figures 5D and Figure E6A). These findings were subsequently validated in the Yale IPF cohort (2.68 log copies per microliter vs. 3.765 log copies per microliter; P = 0.0008) (Figures 5D and Figure E6A), and median MT-ATP6 copy numbers did not differ between the two cohorts (P = 0.8354). These data demonstrate that, in addition to being present in the supernatants of fibrotically reprogrammed NHLFs, mtDNA is present in high concentrations in the lungs and blood of patients with IPF.
Table 1.
Baseline Characteristics of Study Populations
| Control Subjects | Pittsburgh IPF | Yale IPF | |
|---|---|---|---|
| n | 44 | 81 | 82 |
| Age, yr | |||
| Mean ± SD | 70.23 ± 7.13 | 67.18 ± 8.27 | 71.72 ± 7.93 |
| Sex, n (%) | |||
| Male | 31 (70.45) | 63 (77.78) | 71 (86.59) |
| Female | 13 (29.55) | 18 (22.22) | 11 (13.41) |
| Race n (%) | |||
| White | 39 (88.64) | 78 (96.30) | 80 (97.56) |
| Other | 5 (11.36) | 3 (3.70) | 2 (2.50) |
| Smoking status, n (%) | |||
| Ever/current | 9 (20.45) | 38 (46.91) | 66 (80.49) |
| Never | 35 (79.55) | 26 (32.10) | 16 (19.51) |
| Unknown | 0 (0) | 17 (20.99) | 0 (0) |
| FVC, % predicted, mean ± SD | 65.38 ± 18.44 | 75.43 ± 17.60 | |
| DlCO, % predicted, mean ± SD | 50.41 ± 18.19 | 42.89 ± 14.94 | |
| GAP index, mean ± SD | 3.88 ± 1.56 | 4.26 ± 1.36 |
Definition of abbreviations: DlCO = diffusing capacity of the lung for carbon monoxide; IPF = idiopathic pulmonary fibrosis.
Excessive Plasma mtDNA Is Predictive of All-Cause Mortality
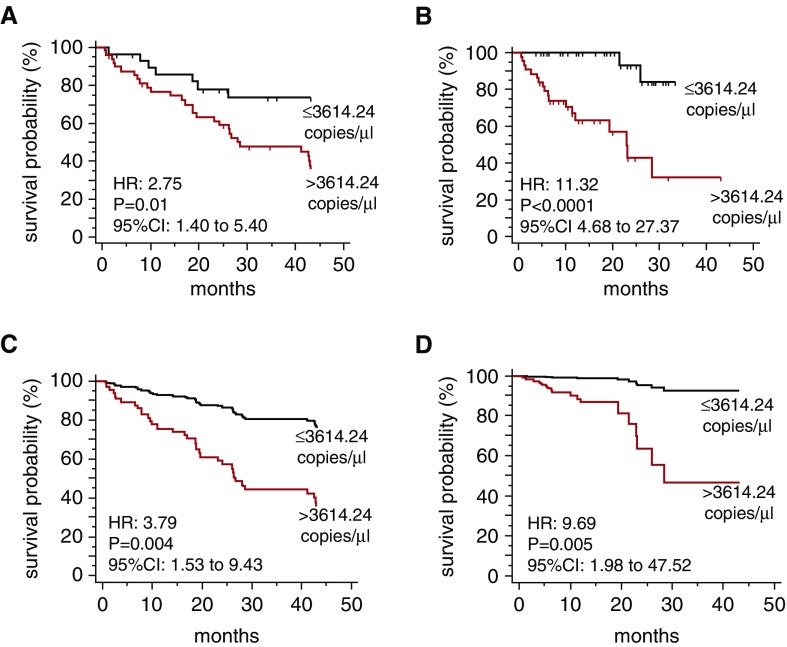
Emerging evidence suggests that substances found in the circulation might convey important prognostic information in IPF (34). We examined the predictive value of plasma mtDNA for all-cause mortality during a 43-month longitudinal follow-up, which is within the median survival length for patients with IPF. Receiver operating characteristic curve analysis of the Pittsburgh IPF cohort revealed that a plasma MT-ATP6 copy number of 3,614.24 copies per microliter can be reliably used as a dichotomous variable to stratify subjects as being at low risk (≤3,614.24 copies per microliter) or high risk (>3,614.24 copies per microliter) for all-cause mortality with a sensitivity of 80.0% and specificity of 54.3% (area under the curve, 0.62; P = 0.045) (Figure E6B). Clinical characteristics of subjects with low and high MT-ATP6 concentrations, based on this threshold value, are shown in Table E1. Although there was no detectable difference between the subjects with MT-ATP6 copy numbers above or below 3,614.24, individuals with high MT-ATP6 concentrations trended toward lower FVC percent predicted and DlCO percent predicted values and higher GAP index scores that did not reach statistical significance. Kaplan-Meier analysis of the Pittsburgh IPF cohort showed that values exceeding this cutoff were significantly associated with all-cause mortality (unadjusted HR, 2.75; 95% confidence interval [CI], 1.40–5.40; P = 0.012) (Figure 6A). Following covariate adjustments for age, sex, race, FVC percent predicted, DlCO percent predicted, and GAP index score, the 3,614.24 copies per microliter cutoff was an even stronger predictor of all-cause mortality (adjusted HR, 3.79; 95% CI, 1.53–9.43; P = 0.004) (Figure 6C and Table E2). Because of missing data, an effect of smoking could not be assessed. The same cutoff value was then validated in the Yale IPF cohort (unadjusted HR, 11.32; 95% CI, 4.68–27.37; P < 0.0001) (Figure 6B); following adjustment for covariates, it remained a robust predictor of all-cause mortality (adjusted HR, 9.69; 95% CI, 1.98–47.52; P = 0.005) (Figure 6D).
Figure 6.
Excessive plasma mitochondrial DNA is predictive of all-cause mortality in two independent cohorts. Kaplan-Meier plot for all-cause mortality reveals a significant survival benefit for subjects with a plasma MT-ATP6 concentration greater than or equal to 3,614.24 copies per microliter (top line) relative to those with a plasma MT-ATP6 concentration greater than 3,614.24 copies per microliter (bottom line). These findings were (A) derived from the Pittsburgh idiopathic pulmonary fibrosis (IPF) cohort and (B) validated in the Yale IPF cohort. Following adjustments for covariates of age, sex, race, FVC percent predicted, adjusted diffusion capacity for carbon monoxide percent predicted, and GAP index score, the cutpoint of 3,614.24 copies per microliter remained a strong predictor of all-cause mortality in (C) the Pittsburgh IPF cohort and (D) the Yale IPF cohort. CI = confidence interval; HR = hazard ratio.
Post-treatment Plasma mtDNA Concentrations Are Reduced in Patients Who Respond to Pirfenidone
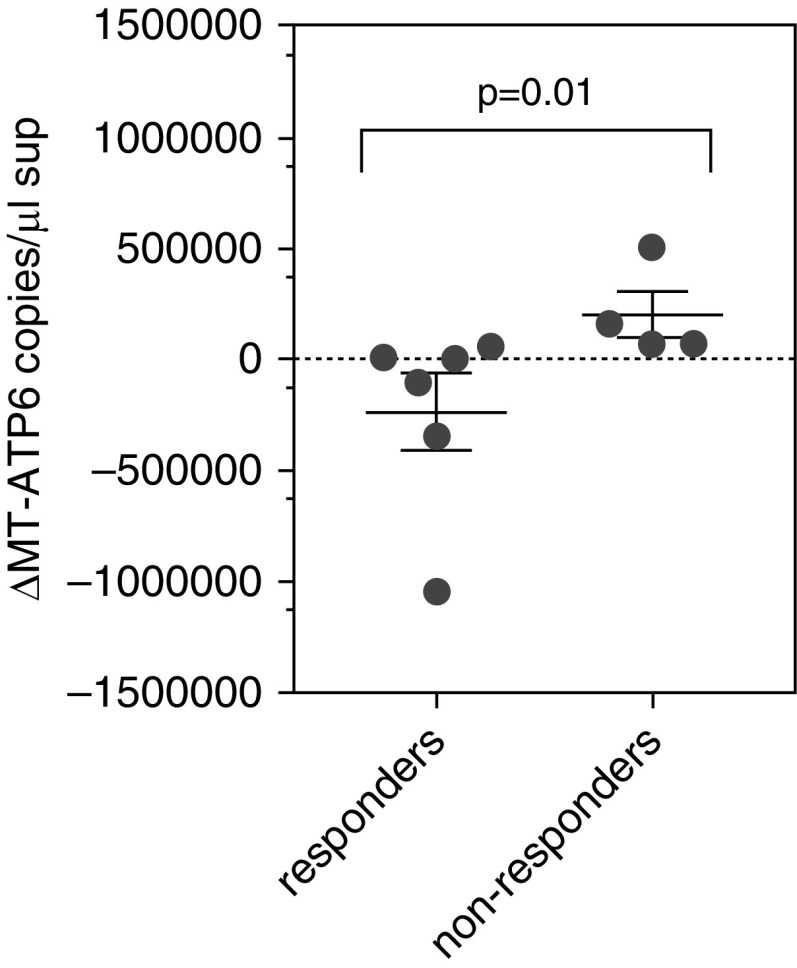
Having found that elevations in circulating mtDNA are predictive of reduced event-free survival, we assessed temporal changes in plasma mtDNA following treatment with the antifibrotic agent pirfenidone, a pyridone derivative that improves mitochondrial function in experimentally induced injury models (35) and enacts modest stabilizing effects on ventilatory decline as well as, potentially, survival in a subset of patients with IPF (1). To this end, we evaluated longitudinal changes in mtDNA during a 3-month follow-up in subjects who responded to pirfenidone by retaining stable FVC percent predicted value after 1 year of treatment (responders, n = 6), and we compared these changes with those of patients who demonstrated greater than 10% reduction in FVC percent predicted (nonresponders, n = 4) despite 1 year of pirfenidone therapy. Plasma MT-ATP6 concentrations were obtained prior to the initiation of therapy and 3 months after starting therapy. Relative to nonresponders, in whom median MT-ATP6 copy numbers increased by 114,284 copies per microliter, responders experienced a reduction in MT-ATP6 copy number on the order of 50,997 copies per microliter (P = 0.01) (Figure 7). Although these numbers are small, they do suggest that serial MT-ATP6 measurements might be a useful predictor of treatment efficacy and clinical outcomes in IPF.
Figure 7.
Post-treatment plasma mitochondrial DNA concentrations are reduced in patients who respond to pirfenidone. Relative to subjects who did not respond to pirfenidone (nonresponders; right), as demonstrated by a greater than 10% reduction in FVC percent predicted after 1 year of treatment, subjects who responded to pirfenidone by retaining stable FVC after 1 year of treatment (responders; left) showed a significant decline in their change in median plasma MT-ATP6 concentration, as compared with their baseline MT-ATP6 concentration, after 3 months of therapy. Data are presented graphically as change in median with interquartile range of MT-ATP6 copies per microliter of plasma.
Discussion
This study lends new insight into the molecular factors linking fibroblast activation with innate immunity and clinical progression in the IPF disease state. Using a combination of standard cell culture approaches and a mechanotransductive tissue culture platform, we show that interactions with biochemical stimuli (TGF-β1), as well as biophysical factors such as local substrate stiffness, induce a state of glycolytic reprogramming that is accompanied by the accumulation of mtDNA in the extracellular milieu. These observations replicate the spontaneous phenotype of IPF fibroblasts. They are also supported by the finding of increased mtDNA concentrations in the lungs and blood of patients with IPF, where, in the latter compartment, they display an impressive association with disease progression and reduced event-free survival. When viewed in combination, this constellation of findings demonstrates a previously unrecognized and highly novel connection between biochemical and biophysical interactions, mitochondrial bioenergetics, detection of mitochondrial DAMPS, and fibroblast activation that carries great ramifications for IPF.
Our finding that NHLFs exposed to fibrotic stimuli generate high levels of extracellular mtDNA expands the role of mitochondria in IPF pathogenesis. Previous studies in this area have been focused largely on how metabolic reprogramming influences the phenotype of epithelium (32) and fibroblasts (27). Our study extends this work to show that fibroblasts adapt to local cues by adopting mitochondrial changes and excessive quantities of extracellular mtDNA, which engenders autocrine or paracrine induction of α-SMA expression (36–41). This finding is especially intriguing because mtDNA functions as a DAMP for TLR9, a pattern recognition receptor that mediates TGF-β1–induced fibroblast activation and is associated with a particularly aggressive disease course in IPF (37, 42–47). However, mtDNA might contribute to disease via additional mechanisms, such as by directing activation of the macrophages which exist at the interface of injury and repair or by contributing to the formation of neutrophil extracellular traps that have recently been described in association with the pathologic remodeling conditions in the human lung (29, 48). It is also possible that fibroblasts sense mtDNA as a danger signal indicative of injury and, as such, assume a repair program characterized by α-SMA expression to enact repair. Understanding these aspects will require evaluation in ex vivo modeling systems and in dedicated animal studies. It will also require a thorough understanding of the nature of extracellular mtDNA, its oxidation state, how it traffics to the extracellular space, and whether canonical or noncanonical TGF-β1 signaling pathways account for these observations.
One particularly novel aspect of our work is the relationship between mtDNA and mechanotransduction. Cellular responses to local tissue stiffness are believed to drive the fibroblast activation in a variety of settings, including IPF (49). In our study, culture of otherwise normal lung fibroblasts on hydrogels approximating the stiffness of the fibrotic lung that have previously been shown to induce α-SMA expression (31, 50) led to substantial increases in the detection of extracellular mtDNA. Although cultured endothelial cells subject to cyclical stress demonstrate degraded mitochondrial integrity (51), to our knowledge, ours are the first data demonstrating a direct link between the biophysical aspects of the lung microenvironment and metabolic remodeling in fibroblasts. Because stiffness-induced α-SMA expression is known to be at least partially TGF-β1 dependent (52), our work suggests that changes in mitochondrial function could occur via a similar mechanism through additional mechanotransductive signaling pathways, such as Rho-associated kinase 1, YAP/TAZ, and Hippo (11).
In a direct link to the IPF disease state, the finding that mtDNA demonstrates robust outcome predictive capacity in two large IPF cohorts suggests a real clinical impact of our findings. Additional investigations relating mtDNA to clinical predictors of mortality and FVC percent predicted decline, which is currently difficult to predict on the basis of clinical factors alone (53), might improve disease stratification in IPF. However, substantial work is required to better understand the mechanistic basis for our observations, how it might relate to patient-specific factors such as smoking and comorbid conditions, and how it might relate to currently available management strategies. We have not determined whether plasma mtDNA stems from intact mitochondria present in circulating exosomes, whether this DNA is oxidized as has recently been shown for nonalcoholic steatohepatitis (17), or whether these findings are directly related to the TLR9 activation that has been proposed as a contributor to poor outcomes in IPF. We have not determined the cell of origin, though our ex vivo modeling suggests that production by fibroblasts might be at least partially involved. We have not determined whether other mitochondrial DAMPS such as formyl peptides, as well as other DAMPS such as extracellular ATP, demonstrate similar properties, and we have not evaluated the relationship of mtDNA to other molecular biomarkers of disease progression, such as matrix metalloproteinase 7, surfactant protein D, chemokine (C-C motif) ligand 18, or Krebs von den Lungen-6, or to genomic biomarkers of stability, such as mucin 5B (34). We also have not fully determined whether mtDNA functions as a molecular predictor of therapeutic responses, which will require access to larger cohorts of treated patients with serial sampling. Despite these limitations, our work delivers evidence supporting examination of circulating mtDNA as a mechanism-based prognostic biomarker of IPF. Further investigation of these findings could yield new management approaches for this enigmatic disease.
Acknowledgments
Acknowledgment
The authors are extremely grateful to all our patients with IPF and the control subjects who generously donated their time and specimens for our studies. The authors also thank Dr. Gerald S. Shadel and Zheng Wu for sharing their expertise in mitochondrial biology. The authors additionally thank Dr. Brian J. Murphy for contributing his editorial expertise with graphics in the preparation of the figures.
Footnotes
Supported by National Institutes of Health (NIH) grant T32HL007778-21 (C.R.); a senior scholar award from the Ellison Medical Foundation (D.F.B.); European Respiratory Society/European Union RESPIRE 2 Marie Skłodowska-Curie fellowship grant 8860/2015 (A.T.); NIH grants K24 AR060297, P30 AR061271, and UL1TR000005 (C.F.-B.); NIH grants R01 DK101984 and P30 DK045735 (R.I.H.); NIH grant R01 HL126094 (C.S.D.C.); a VA merit award and NIH grant 2R56DK0 5U01AA021912-02 (W.M.); NIH grants U01 HL122626, UH3 HL123886, and R01 HL127349 (N.K.); NIH grants HL109233, HL125850, and HL112702 as well as grants from the Gabriel and Alma Elias Research Fund and the Greenfield Foundation (E.L.H.); and a faculty startup package provided by the Department of Pathology at Stony Brook University School of Medicine (G.T.). Support for Nikon structured illumination microscopy was provided by grant 1S10OD016405-01 to Stony Brook University.
Author Contributions: C.R. and H.S.: performed experiments and analyzed data; M.G.: recruited subjects, procured biospecimens, and analyzed data; J.D.H.-M., M.S., D.F.B., K.G., K.L., R.I.H., C.S.D.C., W.M., and N.K.: analyzed data; Y.C., A.O.-A., C. Brandsdorfer, J.W., C. Blaul, J.F., H.P., T.W., A.T., and V.K.: performed experiments; D.E.A.-O.: assisted with enrollment and clinical characterization of subjects; J.T.P.: procured biospecimens; A.L.G.: assisted with hydrogel studies; C.F.-B.: procured lung tissue and analyzed data; E.L.H. and G.T.: conceived of the experimental design and analyzed data. All authors participated in manuscript preparation and provided final approval of the submitted work.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201612-2480OC on August 7, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Blackwell TS, Tager AM, Borok Z, Moore BB, Schwartz DA, Anstrom KJ, Bar-Joseph Z, Bitterman P, Blackburn MR, Bradford W, et al. Future directions in idiopathic pulmonary fibrosis research: an NHLBI workshop report. Am J Respir Crit Care Med. 2014;189:214–222. doi: 10.1164/rccm.201306-1141WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley B, Collard HR, King TE., Jr Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183:431–440. doi: 10.1164/rccm.201006-0894CI. [DOI] [PubMed] [Google Scholar]
- 3.Herzog EL, Mathur A, Tager AM, Feghali-Bostwick C, Schneider F, Varga J. Review: interstitial lung disease associated with systemic sclerosis and idiopathic pulmonary fibrosis: how similar and distinct? Arthritis Rheumatol. 2014;66:1967–1978. doi: 10.1002/art.38702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley B, Ryerson CJ, Vittinghoff E, Ryu JH, Tomassetti S, Lee JS, Poletti V, Buccioli M, Elicker BM, Jones KD, et al. A multidimensional index and staging system for idiopathic pulmonary fibrosis. Ann Intern Med. 2012;156:684–691. doi: 10.7326/0003-4819-156-10-201205150-00004. [DOI] [PubMed] [Google Scholar]
- 5.Herazo-Maya JD, Noth I, Duncan SR, Kim S, Ma SF, Tseng GC, Feingold E, Juan-Guardela BM, Richards TJ, Lussier Y, et al. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Sci Transl Med. 2013;5:205ra136. doi: 10.1126/scitranslmed.3005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards TJ, Kaminski N, Baribaud F, Flavin S, Brodmerkel C, Horowitz D, Li K, Choi J, Vuga LJ, Lindell KO, et al. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;185:67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore BB, Lawson WE, Oury TD, Sisson TH, Raghavendran K, Hogaboam CM. Animal models of fibrotic lung disease. Am J Respir Cell Mol Biol. 2013;49:167–179. doi: 10.1165/rcmb.2013-0094TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Negmadjanov U, Godic Z, Rizvi F, Emelyanova L, Ross G, Richards J, Holmuhamedov EL, Jahangir A. TGF-β1-mediated differentiation of fibroblasts is associated with increased mitochondrial content and cellular respiration. PLoS One. 2015;10:e0123046. doi: 10.1371/journal.pone.0123046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie N, Tan Z, Banerjee S, Cui H, Ge J, Liu RM, Bernard K, Thannickal VJ, Liu G. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am J Respir Crit Care Med. 2015;192:1462–1474. doi: 10.1164/rccm.201504-0780OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez IE, Eickelberg O. The impact of TGF-β on lung fibrosis: from targeting to biomarkers. Proc Am Thorac Soc. 2012;9:111–116. doi: 10.1513/pats.201203-023AW. [DOI] [PubMed] [Google Scholar]
- 11.Thannickal VJ, Henke CA, Horowitz JC, Noble PW, Roman J, Sime PJ, Zhou Y, Wells RG, White ES, Tschumperlin DJ. Matrix biology of idiopathic pulmonary fibrosis: a workshop report of the National Heart, Lung, and Blood Institute. Am J Pathol. 2014;184:1643–1651. doi: 10.1016/j.ajpath.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, Broderick SM, Wade MS, Hysi P, Scuirba J, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trujillo G, Meneghin A, Flaherty KR, Sholl LM, Myers JL, Kazerooni EA, Gross BH, Oak SR, Coelho AL, Evanoff H, et al. TLR9 differentiates rapidly from slowly progressing forms of idiopathic pulmonary fibrosis. Sci Transl Med. 2010;2:57ra82. doi: 10.1126/scitranslmed.3001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirillov V, Siler JT, Ramadass M, Ge L, Davis J, Grant G, Nathan SD, Jarai G, Trujillo G. Sustained activation of Toll-like receptor 9 induces an invasive phenotype in lung fibroblasts: possible implications in idiopathic pulmonary fibrosis. Am J Pathol. 2015;185:943–957. doi: 10.1016/j.ajpath.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Ellson CD, Dunmore R, Hogaboam CM, Sleeman MA, Murray LA. Danger-associated molecular patterns and danger signals in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2014;51:163–168. doi: 10.1165/rcmb.2013-0366TR. [DOI] [PubMed] [Google Scholar]
- 16.Maeda A, Fadeel B. Mitochondria released by cells undergoing TNF-α-induced necroptosis act as danger signals. Cell Death Dis. 2014;5:e1312. doi: 10.1038/cddis.2014.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Martinez I, Santoro N, Chen Y, Hoque R, Ouyang X, Caprio S, Shlomchik MJ, Coffman RL, Candia A, Mehal WZ. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest. 2016;126:859–864. doi: 10.1172/JCI83885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu X, Wu G, Yao Y, Zeng J, Shi D, Lv T, Luo L, Song Y. Intratracheal administration of mitochondrial DNA directly provokes lung inflammation through the TLR9-p38 MAPK pathway. Free Radic Biol Med. 2015;83:149–158. doi: 10.1016/j.freeradbiomed.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 19.Chrysanthopoulou A, Mitroulis I, Apostolidou E, Arelaki S, Mikroulis D, Konstantinidis T, Sivridis E, Koffa M, Giatromanolaki A, Boumpas DT, et al. Neutrophil extracellular traps promote differentiation and function of fibroblasts. J Pathol. 2014;233:294–307. doi: 10.1002/path.4359. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi K, Araya J, Minagawa S, Hara H, Saito N, Kadota T, Sato N, Yoshida M, Tsubouchi K, Kurita Y, et al. Involvement of PARK2-mediated mitophagy in idiopathic pulmonary fibrosis pathogenesis. J Immunol. 2016;197:504–516. doi: 10.4049/jimmunol.1600265. [DOI] [PubMed] [Google Scholar]
- 21.Krychtiuk KA, Ruhittel S, Hohensinner PJ, Koller L, Kaun C, Lenz M, Bauer B, Wutzlhofer L, Draxler DF, Maurer G, et al. Mitochondrial DNA and Toll-like receptor-9 are associated with mortality in critically ill patients. Crit Care Med. 2015;43:2633–2641. doi: 10.1097/CCM.0000000000001311. [DOI] [PubMed] [Google Scholar]
- 22.Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, Quintana C, Osorio JC, Wang Z, Zhao Y, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577. doi: 10.1371/journal.pmed.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu C, Sun H, Gulati M, Herazo-Maya J, Osafo-Addo A, Brandsdorfer C, Blaul C, Faunce J, Pan H, Slade M, et al. Mitochondrial DNA is released from lung fibroblasts in response to the fibrotic lung microenvironment and is predictive of mortality in idiopathic pulmonary fibrosis [abstract] Am J Respir Crit Care Med. 2017;195:A6917. [Google Scholar]
- 24.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.American Thoracic Society; European Respiratory Society. American Thoracic Society/European Respiratory Society international multidisciplinary consensus classification of the idiopathic interstitial pneumonias. Am J Respir Crit Care Med. 2002;165:277–304. doi: 10.1164/ajrccm.165.2.ats01. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Zhu Y, Pan H, Chen X, Balestrini JL, Lam TT, Kanyo JE, Eichmann A, Gulati M, Fares WH, et al. Netrin-1 regulates fibrocyte accumulation in the decellularized fibrotic sclerodermatous lung microenvironment and in bleomycin-induced pulmonary fibrosis. Arthritis Rheumatol. 2016;68:1251–1261. doi: 10.1002/art.39575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie N, Tan Z, Banerjee S, Cui H, Ge J, Liu RM, Bernard K, Thannickal VJ, Liu G. Glycolytic Reprogramming in myofibroblast differentiation and lung fibrosis. Am J Respir Crit Care Med. 2015;192:1462–1474. doi: 10.1164/rccm.201504-0780OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta. 2012;1819:914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Yousefi S, Mihalache C, Kozlowski E, Schmid I, Simon HU. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 2009;16:1438–1444. doi: 10.1038/cdd.2009.96. [DOI] [PubMed] [Google Scholar]
- 30.Behboudi S, Chao D, Klenerman P, Austyn J. The effects of DNA containing CpG motif on dendritic cells. Immunology. 2000;99:361–366. doi: 10.1046/j.1365-2567.2000.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, Tschumperlin DJ. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol. 2010;190:693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, Corey C, Herazo-Maya JD, Sembrat J, Lee JS, et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest. 2015;125:521–538. doi: 10.1172/JCI74942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagai S, Handa T, Ito Y, Takeuchi M, Izumi T. Bronchoalveolar lavage in idiopathic interstitial lung diseases. Semin Respir Crit Care Med. 2007;28:496–503. doi: 10.1055/s-2007-991522. [DOI] [PubMed] [Google Scholar]
- 34.Ley B, Brown KK, Collard HR. Molecular biomarkers in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;307:L681–L691. doi: 10.1152/ajplung.00014.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JF, Liu H, Ni HF, Lv LL, Zhang MH, Zhang AH, Tang RN, Chen PS, Liu BC. Improved mitochondrial function underlies the protective effect of pirfenidone against tubulointerstitial fibrosis in 5/6 nephrectomized rats. PLoS One. 2013;8:e83593. doi: 10.1371/journal.pone.0083593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou B, Liu Y, Kahn M, Ann DK, Han A, Wang H, Nguyen C, Flodby P, Zhong Q, Krishnaveni MS, et al. Interactions between β-catenin and transforming growth factor-β signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP) J Biol Chem. 2012;287:7026–7038. doi: 10.1074/jbc.M111.276311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julian MW, Shao G, Bao S, Knoell DL, Papenfuss TL, VanGundy ZC, Crouser ED. Mitochondrial transcription factor A serves as a danger signal by augmenting plasmacytoid dendritic cell responses to DNA. J Immunol. 2012;189:433–443. doi: 10.4049/jimmunol.1101375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanjuan MA, Rao N, Lai KT, Gu Y, Sun S, Fuchs A, Fung-Leung WP, Colonna M, Karlsson L. CpG-induced tyrosine phosphorylation occurs via a TLR9-independent mechanism and is required for cytokine secretion. J Cell Biol. 2006;172:1057–1068. doi: 10.1083/jcb.200508058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holm CK, Paludan SR, Fitzgerald KA. DNA recognition in immunity and disease. Curr Opin Immunol. 2013;25:13–18. doi: 10.1016/j.coi.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang JZ, Liu Z, Liu J, Ren JX, Sun TS. Mitochondrial DNA induces inflammation and increases TLR9/NF-κB expression in lung tissue. Int J Mol Med. 2014;33:817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 45.Lamphier MS, Sirois CM, Verma A, Golenbock DT, Latz E. TLR9 and the recognition of self and non-self nucleic acids. Ann N Y Acad Sci. 2006;1082:31–43. doi: 10.1196/annals.1348.005. [DOI] [PubMed] [Google Scholar]
- 46.Bao W, Xia H, Liang Y, Ye Y, Lu Y, Xu X, Duan A, He J, Chen Z, Wu Y, et al. Toll-like receptor 9 can be activated by endogenous mitochondrial DNA to induce podocyte apoptosis. Sci Rep. 2016;6:22579. doi: 10.1038/srep22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zauner L, Nadal D. Understanding TLR9 action in Epstein-Barr virus infection. Front Biosci (Landmark Ed) 2012;17:1219–1231. doi: 10.2741/3982. [DOI] [PubMed] [Google Scholar]
- 48.Porto BN, Stein RT. Neutrophil extracellular traps in pulmonary diseases: too much of a good thing? Front Immunol. 2016;7:311. doi: 10.3389/fimmu.2016.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Southern BD, Grove LM, Rahaman SO, Abraham S, Scheraga RG, Niese KA, Sun H, Herzog EL, Liu F, Tschumperlin DJ, et al. Matrix-driven myosin II mediates the pro-fibrotic fibroblast phenotype. J Biol Chem. 2016;291:6083–6095. doi: 10.1074/jbc.M115.712380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med. 2012;186:866–876. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartolák-Suki E, Imsirovic J, Parameswaran H, Wellman TJ, Martinez N, Allen PG, Frey U, Suki B. Fluctuation-driven mechanotransduction regulates mitochondrial-network structure and function. Nat Mater. 2015;14:1049–1057. doi: 10.1038/nmat4358. [DOI] [PubMed] [Google Scholar]
- 52.Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol Biol Cell. 2001;12:2730–2741. doi: 10.1091/mbc.12.9.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ley B, Bradford WZ, Vittinghoff E, Weycker D, du Bois RM, Collard HR. Predictors of mortality poorly predict common measures of disease progression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2016;194:711–718. doi: 10.1164/rccm.201508-1546OC. [DOI] [PMC free article] [PubMed] [Google Scholar]