Abstract
The Integrative Psychiatric Research (iPSYCH) consortium has established a large Danish population-based Case–Cohort sample (iPSYCH2012) aimed at unravelling the genetic and environmental architecture of severe mental disorders. The iPSYCH2012 sample is nested within the entire Danish population born between 1981 and 2005, including 1 472 762 persons. This paper introduces the iPSYCH2012 sample and outlines key future research directions. Cases were identified as persons with schizophrenia (N=3540), autism (N=16 146), attention-deficit/hyperactivity disorder (N=18 726) and affective disorder (N=26 380), of which 1928 had bipolar affective disorder. Controls were randomly sampled individuals (N=30 000). Within the sample of 86 189 individuals, a total of 57 377 individuals had at least one major mental disorder. DNA was extracted from the neonatal dried blood spot samples obtained from the Danish Neonatal Screening Biobank and genotyped using the Illumina PsychChip. Genotyping was successful for 90% of the sample. The assessments of exome sequencing, methylation profiling, metabolome profiling, vitamin-D, inflammatory and neurotrophic factors are in progress. For each individual, the iPSYCH2012 sample also includes longitudinal information on health, prescribed medicine, social and socioeconomic information, and analogous information among relatives. To the best of our knowledge, the iPSYCH2012 sample is the largest and most comprehensive data source for the combined study of genetic and environmental aetiologies of severe mental disorders.
Introduction
The fundamental nature of mental disorders remains poorly understood, but genetic factors have an important role.1, 2, 3, 4, 5, 6, 7 Considerable progress in psychiatric genetics has been made in recent years, based on large samples and international collaborations, for example, through the pivotal efforts of the Psychiatric Genomics Consortium.8 We can expect that larger samples will reveal new insights to common and rare variants underpinning mental disorders.9
Many environmental factors influencing pre- and postnatal development are associated with schizophrenia, bipolar affective disorder, autism and attention-deficit/hyperactivity disorder, and furthermore, adverse life circumstances increase the risk of mental disorders. Gene–environment synergism contributes to the aetiology of these disorders, but suitable datasets to explore this important field of research have been lacking. To understand the impact of genes and environments over the life course, large and truly population-based longitudinal cohort studies are required.10, 11
As part of the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH: http://iPSYCH.au.dk/), a large case–cohort study has commenced. In most countries, it would not be logistically feasible to compile large, representative population-based samples. In Denmark, the existence of (a) a universal public health care system free of charge, (b) several national longitudinal registers and (c) strict ethical and data protection legislation required to safeguard the privacy of study participants, has provided a remarkable research platform.12 Recent technological developments and a new legal framework for use of bio-banked material for research have created similar possibilities for genetic research.
The vision of iPSYCH was to leverage these combined resources, considering the entire national cohort as our study population. We utilized information on individuals with a diagnosis of selected mental disorders (N=57 377) and a randomly sampled cohort13, 14 of the general population (N=30 000). The sample is known as the iPSYCH Danish case-cohort study (iPSYCH2012). We used neonatal dried blood spots from the Danish Neonatal Screening Biobank to investigate detailed genetic and biomarker information, some of which are markers of environmental exposures. The rich Danish population-based registers were used to add information on all individuals and all their relatives. Thus, we created a comprehensive data source for the combined study of genetic and environmental aetiologies of severe mental disorders. Within the iPSYCH2012 sample, currently around 77 500 individuals have been array genotyped and around 20 000 have been whole exome sequenced. Ten thousand samples have been analysed for ranges of cytokines and neurotrophic factors. Epigenetic and metabolome data from several thousand samples are emerging. For the entire sample and their relatives, detailed longitudinal information related to health, prescribed medicine, social and socioeconomic information exists. This study provides a general overview of the sample design and outlines future research.
The overall design
Individuals diagnosed with schizophrenia, mood disorders, bipolar affective disorder, autism and attention-deficit/hyperactivity disorder were identified through linkage between Danish population-based registers along with a random sample of the same population that supplied the cases.15 Dried blood spots for virtually all individuals were retrieved from the Danish Neonatal Screening Biobank and processed for genotyping. The design includes the ability to efficiently analyse prospectively collected cohort data within the iPSYCH case–cohort sample.15 This particular design provides several advantages: As the cohort is randomly selected from the entire population, we are able to generate unbiased absolute risks and incidence rates and to estimate the effect sizes of genetic markers on risk of mental disorders, which is representative of the entire Danish population. To date, most genetic and epidemiological studies are based on convenient case-control samples, which are prone to biases.15, 16 The iPSYCH2012 sample was preceded by four smaller Danish samples,17, 18, 19, 20, 21, 22, 23, 24 all aiming to investigate the potential interplay between genes and the environment. Collectively, these forerunners informed on the best possible study design to use in the iPSYCH2012 sample (Supplementary Text 1). The following three paragraphs describe the resources and methods used to identify individuals included in the iPSYCH2012 sample.
Selecting the study base
The Danish Civil Registration System was established in 1968,25 where all people alive and living in Denmark were registered. It includes information on the unique personal identification number, sex, date and place of birth, parents’ identifiers and continuously updated information on emigration and death. The personal identification number is used in all national registers enabling accurate linkage within and between registers. The study base included all singleton births with known mothers born between 1 of May 1981 and 31 of December 2005, who were alive and resided in Denmark at their first birthday (N=1 472 762 persons). Selecting births in this period ensures individual samples to be retrieved in the Danish Neonatal Screening Biobank and reasonable distribution of cases and cohort members for all birth years. All residents are registered in the Danish Civil Registration System irrespective of health, income, receipt of social benefits, employment and other socioeconomic characteristics.26
Diagnoses of mental disorders
Persons within the study base were linked via their personal identifier to the Danish Psychiatric Central Research Register27 to obtain information on mental disorders. The Danish Psychiatric Central Research Register was computerized in 1969 and contains data on all admissions to Danish psychiatric in-patient facilities. Information on outpatient visits was included from 1995 onwards. From 1994 onwards, the International Classification of Diseases, 10th revision, Diagnostic Criteria for Research was used for diagnostic classification.28 All persons within the study base, who had a diagnosis of schizophrenia, bipolar disorder, affective disorder, autism and attention-deficit/hyperactivity disorder were included (Table 1). At the time of linkage, the Danish Psychiatric Central Research Register contained all psychiatric contacts until 31 December 2012. Table 1 summarizes the number of individuals across the diagnostic groups.
Table 1. Number of persons included in iPSYCHs population-based sample of the Danish population born 1981–2005.
|
Number of persons |
|||||
|---|---|---|---|---|---|
| Group membershipa | ICD10 diagnosesb | Follow-up period | Initial Sample | Blood spots available in Biobank and sent to Broad | Passed Sample QC |
| Schizophreniac | F20 | 2009–2012 | 3540 | 2830 | 2738 |
| Bipolar disorder | F30–F31 | 1994–2012 | 1928 | 1488 | 1452 |
| Affective disorder | F30–F39 | 1994–2012 | 26 380 | 23 532 | 22 809 |
| Autism | F84.0, F84.1, F84.5, F84.8 or F84.9 | 1994–2012 | 16 146 | 15 418 | 14 812 |
| ADHD | F90.0 | 1994–2012 | 18 726 | 17 835 | 17 249 |
| Any case | All ICD codes listed above | 57 377 | 52 867 | 51 101 | |
| Population-based cohort | Random sampling, i.e., disregarding any diagnostic information | 30 000 | 28 650 | 27 605 | |
| Total number of persons | 86 189 | 80 422 | 77 639 | ||
Abbreviations: ADHD, attention-deficit/hyperactivity disorder; ICD-10, International Classification of Diseases, 10th revision; iPSYCH, Integrative Psychiatric Research; QC, quality control.
Groups are not mutually exclusive.
Initial ICD10 diagnosis used to select case groups. Identification was performed through linkage to the Danish Psychiatric Central Research Register. At the time of linkage, the register contained all contacts up until 31/12/2012.
Include all persons first diagnosed with schizophrenia during the period from 1 January 2009 to 31 December 2012 and persons first diagnosed with schizophrenia before 1 January 2009 that were not already genotyped (N=923). A total of 1887 persons first diagnosed with schizophrenia before 1 January 2009 were previously genotyped (see Supplementary Text 1).
Selecting the population-based cohort
Among the 1 472 762 persons included in the study base, a total of 30 000 persons were chosen uniformly at random (Table 1) corresponding to 2.04% of the study base (=30 000/1 472 762). As the cohort members were chosen randomly, some cohort members may also have the disorders of interest.13, 14 Thus, the cohort selected is representative of the entire Danish population born in the same period.26 In addition, the cohort members are at risk of developing the disorder of interest during follow-up, whereas controls are typically conditioned to be healthy until the study ends.29 We have thereby identified the individuals to be included in the iPSYCH2012 sample. Next, we describe the enrichment with genetic and other biomarker data.
The Danish neonatal screening biobank
Blood spots for individuals included in the iPSYCH2012 sample were retrieved from the Danish Neonatal Screening Biobank within the Danish National Biobank.30 This facility stores dried blood spot samples taken from practically all neonates born in Denmark since 1 May 1981 and stored at −20 °C. These samples were collected primarily for diagnosis of congenital disorders. The samples are stored for follow-up diagnostics, screening, quality control and research. At time of blood sampling (4–7 days after birth), parents are informed in writing about the neonatal screening and that the blood spots are stored in the Danish Neonatal Screening Biobank and can be used for research, pending approval from relevant authorities. The parents are also informed about how to prevent or withdraw the sample from inclusion in research studies.
Genotyping was based on two blood spot punches of 3.2 mm, equivalent to 6 μl of whole blood.30 Biological components are generally very well preserved in neonatal dried blood spot samples, in particular if the samples are stored at −20 °C. However, it may be challenging to analyse the samples due to the very limited amount of biological material available, the nature of dried whole blood on filter paper and decades of storage. In particular, the determination of concentration of biomarkers in dried blood spots is less precise than in serum. This calculation is based on the assumption that one punch 3.2 mm in diameter is equivalent to 3 μl of whole blood, which only applies if the filter paper is fully and evenly saturated. Moreover, measurements are performed on whole blood containing various cell types that may have an influence on the concentration of certain components. The hematocrit, which is usually unknown, is also an important factor for blood components that do not re-distribute into red blood cells. Special high sensitive assays may be required and multi-analyte measurements are preferred to get as much information of the limited samples as possible. The neonatal dried blood spots is suitable for next generation sequencing,31 DNA methylation profiling,32 metabolome profiling, vitamin D,33 multiplex measurements of cytokines,34 antibodies to infectious agents19 and whole transcriptome analysis through microarray35 and RNA-seq.36 Importantly, these measurements are made in samples drawn few days after birth, meaning that case-control differences cannot be ascribed to disease-related confounders as medication, alcohol or substance use, smoking or the disease state itself.
Systematic comparisons of genomic DNA versus whole-genome-amplified DNA37 reveals increased signal noise. Although this has very little impact on genotype calls, it is problematic for Copy Number Variation detection algorithms such as PennCNV.38 Efforts within the iPSYCH community are making progress towards solving the noise issues. Technical reproductions using RNA microarrays reported in Grauholm et al.35 indicated high reproducibility, independently of spot size, and indicated that the critical factor is storage conditions rather than storage length. Ho et al.39 found differences between cerebral palsy cases and matched controls using dried blood spots from the Michigan neonatal screening. Combined these reports strongly indicate that it is possible to do meaningful transcriptome experiments despite prolonged storage at perceived sub-optimal conditions.
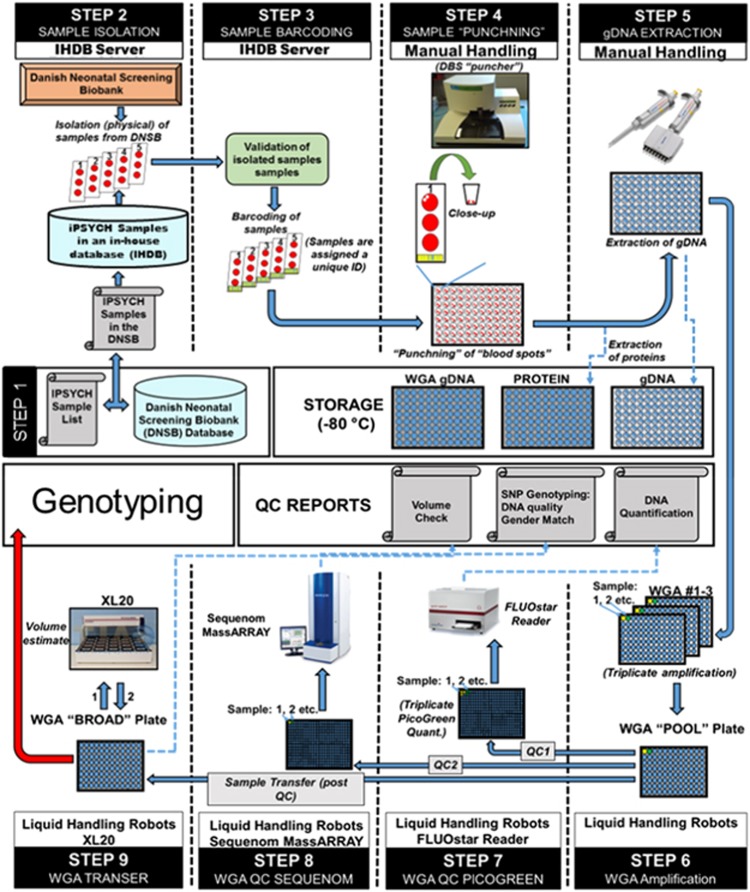
Preparation of samples for genotyping and sequencing from the Danish neonatal screening biobank DNA was extracted and whole genome amplified at the Statens Serum Institut following previously established procedures.40, 41 The sample flow is described in Figure 1.
Figure 1.
The selected samples were correlated with their DNSB identifiers and entered into an in-house developed selection database (Step 1 and 2). Sample identities were then validated and assigned a pseudonymized unique ID (Step 3) before cutting two discs of 3.2 mm of dried blood into a 96-well PCR plate (Step 4). Proteins were washed of the blood spots and stored at −80 °C before DNA was extracted using Extract-N-Amp Blood PCR Kit (Sigma-Aldrich, St Louis, MO, USA) (Step 5). DNA was amplified in triplicates using REPLI-g (Qiagen, Hilden, Germany) and combined to a single sample (Step 6). Finally, concentrations were quantified using Quant-iT picogreen (Invitrogen, Carlsbad, CA, USA) (Step 7) and a genetic fingerprint established using the iPLEX pro Sample ID panel (Agena Bioscience, Hamburg, Germany) (Step 8) before aliquoting a fraction of the sample for genotyping (Step 9).
Array genotyping and quality control
Samples were processed at the Broad Institute (Boston, MA, USA) using the Infinium PsychChip v1.0 array (Illumina, San Diego, CA, USA) in accordance with the manufacturer’s instructions.42 Genotyping was conducted in 25 waves. Variant calls were trained using GenTrain2 (Illumina) on the first wave (4146 samples) using the PsychChip 15048346 B manifest and GenomeStudio version v2011.1. Following autoclustering, loci were manually curated if they had a call frequency below 90%, GenTrain scores below 0.5 or cluster separation below 0.2. During this processing, 3890 loci were excluded and 928 were manually modified. The resulting GenTrain was used to produce GenCall variant calls used for sample level quality control of the entire cohort.43 Samples with call rates below 95% (N=2270) were designated to fail sample quality control (QC). Sex was inferred using heterozygosity on chromosome X; below 20% in males; above 20% in females. Sex obtained from genotyping was compared to the sex recorded in the Danish Civil Registration System and mismatches were excluded. It is extremely unlikely to observe errors in recorded sex in the Danish Civil Registration System.26 About 0.25% (N=224) of the sample did not match the expected sex. Half of the failures (N=119) were due to abnormal structural variation on chromosome X (aneuploidy and loss of heterozygosity). The other half were due to sample mix-ups (N=103). In this study we describe the sample QC only and not the subsequent single-nucleotide polymorphism QC, which vary between studies.
Probe remapping
All probe sequences were queried against an HG19 database using a nucleotide version of the Basic Local Alignment Search Tool. The Basic Local Alignment Search Tool results were compared with the original array manifest, an Illumina update to the array manifest, and the Broad Institute updates to the manifest. The genomic coordinates matched between the Basic Local Alignment Search Tool results and the existing manifests for 95.12% of probes. 2.23% of probes were updated based on the new Basic Local Alignment Search Tool results. 2.11% retained their original mapping. The remaining 0.54% were split between the Broad Institute reference and the Illumina update or the probe was removed from the data set (Supplementary Table 1).
Improving variant calls
GenCall,43 Birdseed44 and zCall45 were used supplementary to improve variant calls. GenCall and Birdseed are genotype calling algorithms best suited for common variants, while zCall is a post-processing step for GenCall to improve genotype calling for rare variants. Approximately half of the probes on the array are common variants (minor allele frequency⩾0.05), while the other half are rare variants (minor allele frequency<0.05). A large subgroup of the rare variants are non-polymorphic within the cohort. A consensus genotype call was made from the three calling algorithms (Supplementary Text 2) using PLINK.46, 47
Ethical framework
The Danish Scientific Ethics Committee, the Danish Health Data Authority, the Danish data protection agency and the Danish Neonatal Screening Biobank Steering Committee approved this study. This is in keeping with the strict ethical framework and the Danish legislation protecting the use of these samples.30, 48 Permission has been granted to study genetic and environmental factors for the development and prognosis of mental disorders. To unravel the foundation of severe mental disorders, it is central that this rich data source is accessible to the international research community to the largest extent possible. It is also paramount to protect the privacy of the individuals included in the study. Owing to the sensitive nature of these data, individual level data can be accessed only through secure servers where download of individual level information is prohibited.49 iPSYCH encourage national and international collaboration. For details, please contact Professor Preben Bo Mortensen, Scientific Director of iPSYCH.
Baseline characteristics
Table 2 shows baseline characteristics of the 86 189 individuals included in the iPSYCH2012 sample. Among these individuals, 77 639 (90%) passed sample QC. In the cohort group, males constituted 51% in both the initial and in the QC’ed sample. The following numbers refer to the initial sample: Overall, 26 380 individuals were included due to suffering from an affective disorder. Among individuals with affective disorder, 543 individuals were incidentally also among the cohort members, that is, the 2.03% random sample of the study base. Overall, 28 812 (96.04%) of the 30 000 cohort members had none of the 5 psychiatric diagnoses until 2012. A total of 49 737 (86.68%) cases and 25 159 (83.86%) cohort members were native Danes. The largest second-generation immigrant group was persons having one or both parents born in Europe followed by one or both parents born in Scandinavia.
Table 2. Baseline characteristics of the iPSYCH2012 case–cohort.
|
Initial sample, N=86 189 |
Passed sample QC, N=77 639 |
|||
|---|---|---|---|---|
| Casesa, N=57 377 | Cohorta, N=30 000 | Casesa, N=51 101 | Cohorta, N=27 605 | |
| Sex | ||||
| Male | 32 243 (56.19%) | 15 308 (51.03%) | 29 048 (56.84%) | 14 090 (51.04%) |
| Female | 25 134 (43.81%) | 14 692 (48.97%) | 22 053 (43.16%) | 13 515 (48.96%) |
| Year of birth | ||||
| 1981–1985 | 11 909 (20.76%) | 4858 (16.19%) | 9409 (18.41%) | 4175 (15.12%) |
| 1986–1990 | 14 506 (25.28%) | 5838 (19.46%) | 12 766 (24.98%) | 5361 (19.42%) |
| 1991–1995 | 14 521 (25.29%) | 6597 (21.99%) | 13 508 (26.43%) | 6190 (22.42%) |
| 1996–2000 | 10 306 (17.96%) | 6461 (21.54%) | 9796 (19.17%) | 6182 (22.39%) |
| 2001–2005 | 6144 (10.71%) | 6246 (20.82%) | 5622 (11.00%) | 5697 (20.64%) |
| Diagnosisb | ||||
| Schizophrenia | 3540 (6.17%) | 90 (0.30%) | 2738 (5.36%) | 65 (0.24%) |
| Bipolar affective disorder | 1928 (3.36%) | 42 (0.14%) | 1452 (2.84%) | 31 (0.11%) |
| Affective disorder | 26 380 (45.98%) | 543 (1.81%) | 22 809 (44.64%) | 467 (1.69%) |
| Autism | 16 146 (28.14%) | 324 (1.08%) | 14 812 (28.99%) | 309 (1.12%) |
| ADHD | 18 726 (32.64%) | 387 (1.29%) | 17 249 (33.75%) | 360 (1.30%) |
| None of the above | 0 (0.00%) | 28812 (96.04%) | 0 (0.00%) | 26 538 (96.13%) |
| Parental originc | ||||
| Denmark | 49 737 (86.68%) | 25 159 (83.86%) | 44 284 (86.66%) | 23 213 (84.09%) |
| Africa | 619 (1.08%) | 446 (1.49%) | 566 (1.11%) | 400 (1.45%) |
| America | 472 (0.82%) | 228 (0.76%) | 419 (0.82%) | 204 (0.74%) |
| Asia | 731 (1.27%) | 672 (2.24%) | 648 (1.27%) | 616 (2.23%) |
| Australia | 51 (0.09%) | 29 (0.10%) | 47 (0.09%) | 29 (0.11%) |
| Europe excl Scandinavia | 2508 (4.37%) | 1557 (5.19%) | 2228 (4.36%) | 1406 (5.09%) |
| Greenland | 341 (0.59%) | 162 (0.54%) | 306 (0.60%) | 148 (0.54%) |
| Middle East | 666 (1.16%) | 625 (2.08%) | 604 (1.18%) | 568 (2.06%) |
| Scandinavia excl Denmark | 1168 (2.04%) | 600 (2.00%) | 1047 (2.05%) | 549 (1.99%) |
| Mixed parentage | 221 (0.39%) | 166 (0.55%) | 190 (0.37%) | 146 (0.53%) |
| Unknown | 863 (1.50%) | 356 (1.19%) | 762 (1.49%) | 326 (1.18%) |
| Birth weight | ||||
| <1500 g | 475 (0.83%) | 145 (0.48%) | 381 (0.75%) | 125 (0.45%) |
| 1500–2499 g | 2613 (4.55%) | 924 (3.08%) | 2197 (4.30%) | 808 (2.93%) |
| 2500–3499 g | 26 735 (46.60%) | 13 302 (44.34%) | 23 758 (46.49%) | 12 245 (44.36%) |
| 3500–4499 g | 25 500 (44.44%) | 14 491 (48.30%) | 22 918 (44.85%) | 13 425 (48.63%) |
| ⩾4500 g | 1739 (3.03%) | 954 (3.18%) | 1600 (3.13%) | 883 (3.20%) |
| Missing information | 315 (0.55%) | 184 (0.61%) | 247 (0.48%) | 119 (0.43%) |
| Gestational age | ||||
| <35 weeks | 1342 (2.34%) | 458 (1.53%) | 1105 (2.16%) | 397 (1.44%) |
| 35–36 weeks | 1907 (3.32%) | 857 (2.86%) | 1648 (3.22%) | 776 (2.81%) |
| 37–38 weeks | 8650 (15.08%) | 4305 (14.35%) | 7754 (15.17%) | 3998 (14.48%) |
| 39–40 weeks | 29 768 (51.88%) | 15 835 (52.78%) | 26 642 (52.14%) | 14 640 (53.03%) |
| ⩾41 weeks | 14 995 (26.13%) | 8239 (27.46%) | 13 440 (26.30%) | 7594 (27.51%) |
| Missing information | 715 (1.25%) | 306 (1.02%) | 512 (1.00%) | 200 (0.72%) |
| Apgar score (5 min) | ||||
| 1–5 | 238 (0.41%) | 113 (0.38%) | 204 (0.40%) | 107 (0.39%) |
| 6–7 | 617 (1.08%) | 224 (0.75%) | 523 (1.02%) | 199 (0.72%) |
| 8–9 | 3460 (6.03%) | 1712 (5.71%) | 3083 (6.03%) | 1573 (5.70%) |
| 10 | 52 284 (91.12%) | 27 590 (91.97%) | 46 666 (91.32%) | 25 445 (92.18%) |
| Missing information | 778 (1.36%) | 361 (1.20%) | 625 (1.22%) | 281 (1.02%) |
Abbreviations: ADHD, attention-deficit/hyperactivity disorder; iPSYCH, Integrative Psychiatric Research; QC, quality control.
Persons who were both selected as cases and cohortees constitute 1188 persons in the initial sample and 1067 persons in the QC’ed sample. These individuals were counted both in the case and cohort group.
Psychiatric diagnosis at time of selection as described in the method section. Some persons may both be selected in the case and cohort group, have more than one diagnoses and may later have additional diagnoses.
Persons with both parents born in Denmark were classified as having Danish parental origin. Persons having a Danish-born parent and a foreign born parent were classified as the foreign-born parents region of birth. Persons having two foreign-born parents born in the same region were classified with that region. Persons having two foreign-born parents born in different regions were classified as mixed.
Comparing the percentage of cases included in the initial sample with the percentage of cases passing the QC revealed no systematic deviations across selected baseline characteristics (Table 2). Comparing the percentage of cohort members included in the initial sample with the percentage of cohort members passing QC also revealed no systematic deviations across baseline characteristics.
Visualization of genetic data by foreign parental origin
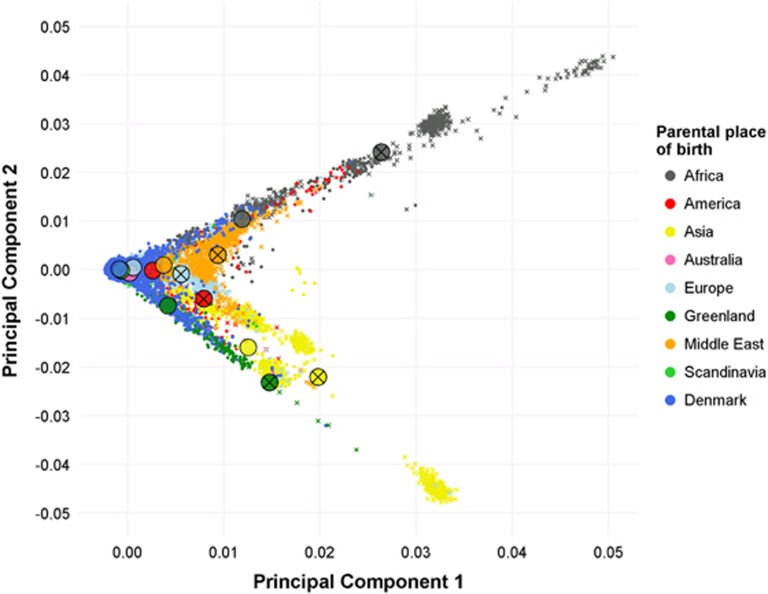
To visualize population substructure for the genetic data, a principle component analysis was conducted (Supplementary Text 3). There was a clear correspondence between the first two principal components based on the genome-wide single-nucleotide polymorphism genotypic data and parental country of birth as registered in the Danish Civil Registration System (Figure 2). Individuals born in Denmark to parents born in other Scandinavian countries clustered together with Danish-born individuals with Danish-born parents, as expected. Within each foreign parental region of birth, individuals with two foreign parents were as anticipated more genetically divergent compared to those having only one foreign-born parent. This finding provides strong evidence of internal validity for processing of individual samples and the ability to link to information in the rich Danish registers on an individual level.
Figure 2.
Scatterplot of the first two principal components colored according to parental region of birth. Big circles indicate mean values for the given parental group. Crosses indicates both parent born abroad within the region indicated by the color. Absence of cross indicate one Danish born parent and one parent born in the region indicated by the color. Persons with unknown information on parental region of birth (N=1088) and mixed parentage are not shown (N=366).
Perspectives
The large iPSYCH2012 sample will provide a solid foundation for a range of studies in decades ahead. We have completed genotyping and plans are advanced for a range of other analyses, including an update and major expansion with cases diagnosed since 2012, as well as including new diagnostic case groups. The sample is thus not only a rich database for research in the current version - it also constitutes a logistic and organizational framework for future studies, although each new study will require relevant ethical permissions. Most other genetic studies are based on samples of convenience rather than utilizing true population-based samples. To our knowledge, no large-scale population-based sample with genome-wide association study data exists elsewhere. In particular, we are confident that the iPSYCH2012 sample provides an important resource to explore novel ways to combine genetic, phenotypic and environmental factors. Phenotypic and environmental factors are readily available through record linkage between the numerous Danish registers or assayed from neonatal dried blood spots.
Access to high-quality, population-based person-linked registers has enabled major contributions to psychiatric epidemiology. For example, researchers have documented key risk factors within psychiatric epidemiology, for example, urban birth,50, 51, 52, 53, 54 paternal age,55, 56, 57 psychiatric family history,58, 59 life-time risk,60 infections,17, 19, 20 neonatal vitamin D deficiency,61 socio-economic adversity,62 treatment resistant schizophrenia,63 pharmacological treatment,64 suicide65 and excess mortality.66 Key features such as the avoidance of selection bias and control of multiple confounders have been important aspects of these studies. However, genetic studies have traditionally not had access to population-based samples, with cases often recruited from multiplex families, or convenient samples of prevalent cases in contact with mental health services. The iPSYCH2012 sample includes a large representative sample of severe mental disorders from a representative sampling frame. The possibility to link the iPSYCH2012 sample to the comprehensive and high quality Danish population-based registers offers researchers unique possibilities to study the interplay between the genetic factors, and variables from the environment, and variables related to health,27, 67, 68, 69 mortality, income and social and socioeconomic characteristics.70, 71, 72 Genetic association studies are by default observational studies, subject to many of the same sources of bias and confounding as other epidemiological studies.16 Therefore, we believe our samples can assist the assessment of the potential impact of such biases and especially lack thereof, and point toward new avenues of research. For example, it has been shown that the genetic associations with schizophrenia identified in the seminal Psychiatric Genomics Consortium paper3 were stronger in more chronic cases than in first episode cases.73 This may suggest that, in future studies, the genetic architecture of schizophrenia could perhaps be refined to identify genes particularly associated with the risk of developing disease, and genes particularly predicting a chronic course, something that could have important preventive and clinical implications. Such future studies will benefit from the continued dialogue between epidemiological studies as iPSYCH and the large-scale studies available only through collaboration in international consortia.
The iPSYCH2012 sample will be able to leverage single-nucleotide polymorphism -derived, genome-wide metrics such as disease-specific polygenic risk scores.74, 75, 76 These provide a continuous measure of liability (rather than a categorical measure of family history), which will greatly enhance our ability to combine genetic, environmental and phenotypic data in disease prediction. We have found higher polygenic loading for schizophrenia in both cases and controls with family histories of mental disorders.77 Also 48% of the effect associated with family history of psychoses was mediated through the polygenic risk score for schizophrenia.78 To further explore the association between the risk of schizophrenia and the polygenetic risk score for schizophrenia, we have investigated the interplay with infections,79 treatment resistant schizophrenia,80 chronicity of schizophrenia,73 and mortality and suicidal behaviour.81
Since the initiation of the iPSYCH2012 sample, other related Danish projects have built on the same framework as that used within iPSYCH, for example, anorexia (5703 cases), obsessive-compulsive disorder (7747 cases), conduct disorder (4205 cases), hyperkinetic conduct disorder (3690 cases) and 1546 twin pairs. All samples gain power in utilizing cohort members within the iPSYCH2012 sample, while also contributing to the unique possibilities of the iPSYCH2012 sample.
Strengths and limitations
Identification of cases within the iPSYCH2012 sample is based on contacts to in- and out-patient psychiatric departments and visits to psychiatric emergency care units in a nation where treatment is provided through the government healthcare system free of charge, and where no private psychiatric hospitals exist. Financial factors are thus less likely to influence pathways to healthcare in Denmark compared to many other nations.82 Unlike samples of convenience, the iPSYCH2012 sample is representative of the Danish population irrespectively of (a) recall bias, (b) emigration or death before sampling, (c) institutional care, (d) imprisonment, (e) being homeless, (f) health and (g) socioeconomic status.26 In contrast to most genetic studies, the iPSYCH2012 sample also provides the unique possibility to explore the potential impact of the longitudinal trajectory on causes and outcomes of mental disorders.
Register-based studies like the current study cannot identify persons with untreated disorders or disorders treated in primary health care only. Most cases with mild to moderate mental disorders, for example, mild or moderate depression and anxiety disorders are thus not registered in the Danish Psychiatric Central Research Register.27 The major strength of the iPSYCH2012 sample approach is the comprehensive clinical assessment of all mental disorders treated in secondary healthcare in a nationwide population. Validation of the Clinician-derived key diagnoses (schizophrenia, single depressive episode, affective disorder, attention-deficit/hyperactivity disorder and autism) has been carried out with good results.83, 84, 85, 86, 87, 88
Limitations include that, from an ethical point of view, we are not allowed to re-contact individuals for any reason. At present, it is also unclear to which extend it will be possible to enrich the iPSYCH2012 sample with information from cohorts including more detailed information on study participants (for example, see refs 89, 90, 91, 92).
We believe that the iPSYCH2012 sample will aid in accelerating psychiatric research in preventing and treating severe mental disorders for the benefit of patients, their families and friends, and the society.
Acknowledgments
This study was supported by The Lundbeck Foundation (grant numbers R102-A9118 and R155-2014-1724), Denmark, the Stanley Medical Research Institute, an Advanced Grant from the European Research Council (project number 294838) and the Stanley Center for Psychiatric Research at Broad Institute and Centre for Integrated Register-based Research at Aarhus University. This research has been conducted using the Danish National Biobank resource, supported by the Novo Nordisk Foundation. Professor John J McGrath is supported by grant APP1056929 from the John Cade Fellowship from the National Health and Medical Research Council and the Danish National Research Foundation (Niels Bohr Professorship). We thank Betina Trabjerg, National Centre for Register-Based Research, Aarhus University, School of Business and Social Sciences, Aarhus, Denmark, for technical help in producing the principal component plot. We are indebted to the late Mads Vilhelm Hollegaard for his contribution to make sample material accessible for analysis from the Danish Neonatal Screening Biobank. Mads’ pioneering work will be used in this and future studies.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E et al. Discovery of the first genome-wide significant risk loci For ADHD. bioRxiv 2017; http://www.biorxiv.org/content/early/2017/06/03/145581. [DOI] [PMC free article] [PubMed]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB et al. Most genetic risk for autism resides with common variation. Nat Genet 2014; 46: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Direk N, Williams S, Smith JA, Ripke S, Air T, Amare AT et al. An analysis of two genome-wide association meta-analyses identifies a new locus for broad depression phenotype. Biol Psychiatry 2016; 82: 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group C. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Owen MJ. The implications of the shared genetics of psychiatric disorders. Nat Med 2016; 22: 1214–1219. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Agrawal A, Bulik C, Andreassen OA, Borglum A, Breen G et al. Psychiatric genomics: an update and an agenda. bioRxiv 2017; http://www.biorxiv.org/content/early/2017/03/10/115600. [DOI] [PMC free article] [PubMed]
- McGrath JJ, Mortensen PB, Visscher PM, Wray NR. Where GWAS and epidemiology meet: opportunities for the simultaneous study of genetic and environmental risk factors in schizophrenia. Schizophrenia Bull 2013; 39: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank L. Epidemiology. The epidemiologist's dream: Denmark. Science 2003; 301: 163. [DOI] [PubMed] [Google Scholar]
- Frank L. Epidemiology. When an entire country is a cohort. Science 2000; 287: 2398–2399. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Susser E. The use of well controls: an unhealthy practice in psychiatric research. Psychol Med 2011; 41: 1127–1131. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Susser E. Genome-wide association studies: does only size matter? Am J Psychiatry 2010; 167: 741–744. [DOI] [PubMed] [Google Scholar]
- Borgan O, Langholz B, Samuelsen SO, Goldstein L, Pogoda J. Exposure stratified case-cohort designs. Lifetime Data Anal 2000; 6: 39–58. [DOI] [PubMed] [Google Scholar]
- Clayton D, Hills M. Statistical Models in Epidemiology. Oxford University Press: Oxford, New York, Tokyo, 1993. [Google Scholar]
- Mortensen PB, Norgaard-Pedersen B, Waltoft BL, Sorensen TL, Hougaard D, Torrey EF et al. Toxoplasma gondii as a risk factor for early-onset schizophrenia: analysis of filter paper blood samples obtained at birth. Biol Psychiatry 2007; 61: 688–693. [DOI] [PubMed] [Google Scholar]
- Nyegaard M, Demontis D, Foldager L, Hedemand A, Flint TJ, Sorensen KM et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry 2010; 15: 119–121. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen CB, McGrath JJ, Hougaard DM, Norgaard-Petersen B, Mors O et al. Neonatal antibodies to infectious agents and risk of bipolar disorder: a population-based case-control study. Bipolar Disord 2011; 13: 624–629. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen CB, Hougaard DM, Norgaard-Petersen B, Mors O, Borglum AD et al. A Danish National Birth Cohort study of maternal HSV-2 antibodies as a risk factor for schizophrenia in their offspring. Schizophrenia Res 2010; 122: 257–263. [DOI] [PubMed] [Google Scholar]
- Demontis D, Nyegaard M, Buttenschon HN, Hedemand A, Pedersen CB, Grove J et al. Association of GRIN1 and GRIN2A-D with schizophrenia and genetic interaction with maternal herpes simplex virus-2 infection affecting disease risk. Am J Med Genet B Neuropsychiatr Genet 2011; 156B: 913–922. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Demontis D, Pedersen MS, Agerbo E, Mortensen PB, Borglum AD et al. Risk of schizophrenia in relation to parental origin and genome-wide divergence. Psychol Med 2012; 42: 1515–1521. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen CB, Hougaard DM, Norgaard-Petersen B, Mors O, Borglum A et al. Maternal antibodies to cytomegalovirus and schizophrenia risk. Schizophrenia Bull 2011; 37: 58. [Google Scholar]
- Borglum AD, Demontis D, Grove J, Pallesen J, Hollegaard MV, Pedersen CB et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol Psychiatry 2014; 19: 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen CB. The Danish Civil Registration System. Scand J Public Health 2011; 39: 22–25. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Gotzsche H, Moller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Danish Med Bull 2006; 53: 441–449. [PubMed] [Google Scholar]
- Mors O, Perto GP, Mortensen PB. The Danish Psychiatric Central Research Register. Scand J Public Health 2011; 39(7 Suppl): 54–57. [DOI] [PubMed] [Google Scholar]
- Organization WHWHO ICD-10: Psykiske lidelser og adfærdsmæssige forstyrrelser. Klassifikation og diagnosekriterier [WHO ICD-10: Mental and Behavioural Disorders. Classification and Diagnostic Criteria]. Copenhagen: Munksgaard Danmark, 1994. [Google Scholar]
- Waltoft BL, Pedersen CB, Nyegaard M, Hobolth A. The importance of distinguishing between the odds ratio and the incidence rate ratio in GWAS. BMC Med Genet 2015; 16: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgaard-Pedersen B, Hougaard DM. Storage policies and use of the Danish Newborn Screening Biobank. J Inherit Metab Dis 2007; 30: 530–536. [DOI] [PubMed] [Google Scholar]
- Poulsen JB, Lescai F, Grove J, Baekvad-Hansen M, Christiansen M, Hagen CM et al. High-quality exome sequencing of whole-genome amplified neonatal dried blood spot DNA. PLoS ONE 2016; 11: e0153253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollegaard MV, Grauholm J, Norgaard-Pedersen B, Hougaard DM. DNA methylome profiling using neonatal dried blood spot samples: a proof-of-principle study. Mol Genet Metab 2013; 108: 225–231. [DOI] [PubMed] [Google Scholar]
- Eyles DW, Morley R, Anderson C, Ko P, Burne T, Permezel M et al. The utility of neonatal dried blood spots for the assessment of neonatal vitamin D status. Paediatr Perinat Epidemiol 2010; 24: 303–308. [DOI] [PubMed] [Google Scholar]
- Skogstrand K, Thorsen P, Norgaard-Pedersen B, Schendel DE, Sorensen LC, Hougaard DM. Simultaneous measurement of 25 inflammatory markers and neurotrophins in neonatal dried blood spots by immunoassay with xMAP technology. Clin Chem 2005; 51: 1854–1866. [DOI] [PubMed] [Google Scholar]
- Grauholm J, Khoo SK, Nickolov RZ, Poulsen JB, Baekvad-Hansen M, Hansen CS et al. Gene expression profiling of archived dried blood spot samples from the Danish Neonatal Screening Biobank. Mol Genet Metab 2015; 116: 119–124. [DOI] [PubMed] [Google Scholar]
- Bybjerg-Grauholm J, Hagen CM, Khoo SK, Johannesen ML, Hansen CS, Baekvad-Hansen M et al. RNA sequencing of archived neonatal dried blood spots. Mol Genet Metab Rep 2017; 10: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekvad-Hansen M, Bybjerg-Grauholm J, Poulsen JB, Hansen CS, Hougaard DM, Hollegaard MV. Evaluation of whole genome amplified DNA to decrease material expenditure and increase quality. Mol Genet Metab Rep 2017; 11: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007; 17: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho NT, Furge K, Fu W, Busik J, Khoo SK, Lu Q et al. Gene expression in archived newborn blood spots distinguishes infants who will later develop cerebral palsy from matched controls. Pediatric Res 2013; 73(4 Pt 1): 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollegaard MV, Grauholm J, Borglum A, Nyegaard M, Norgaard-Pedersen B, Orntoft T et al. Genome-wide scans using archived neonatal dried blood spot samples. BMC Genomics 2009; 10: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollegaard MV, Grove J, Grauholm J, Kreiner-Moller E, Bonnelykke K, Norgaard M et al. Robustness of genome-wide scanning using archived dried blood spot samples as a DNA source. BMC Genet 2011; 12: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C et al. Whole‐Genome Genotyping 2006; 410: 359–376. [DOI] [PubMed]
- Illumina. Illumina GenCall Data Analysis Software. Illumina Techinal Note 2005. Available fromhttps://www.illumina.com/Documents/products/technotes/technote_gencall_data_analysis_software.pdf.
- Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008; 40: 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JI, Crenshaw A, Carey J, Grant GB, Maguire J, Fromer M et al. zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics 2012; 28: 2543–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlev M. Genomic databases and biobanks in Denmark. J Law Med Ethics 2015; 43: 743–753. [DOI] [PubMed] [Google Scholar]
- Statistics D. Guidelines for Transferring Aggregated Results from Statistics Denmark’s Research Services 2017 (cited 25 January 2017). Available fromhttp://dst.dk/ext/8804839804/0/forskning/Guidelines-for-Transferring-Aggregated-Results-from-Statistics-Denmark-s-Research-Services—pdf.
- Pedersen CB. No evidence of time trends in the urban-rural differences in schizophrenia risk among five million people born in Denmark from 1910 to 1986. Psychol Med 2006; 36: 211–219. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen CB, Westergaard T, Wohlfahrt J, Ewald H, Mors O et al. Familial and non-familial risk factors for schizophrenia: a population-based study. Schizophrenia Res 1998; 29: 13. [Google Scholar]
- Pedersen CB, Mortensen PB. Are the cause(s) responsible for urban-rural differences in schizophrenia risk rooted in families or in individuals? Am J Epidemiol 2006; 163: 971–978. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Mortensen PB. Evidence of a dose-response relationship between urbanicity during upbringing and Schizophrenia risk. Arch Gen Psychiatry 2001; 58: 1039–1046. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Mortensen PB. Why factors rooted in the family may solely explain the urban-rural differences in schizophrenia risk estimates. Epidemiol Psichiatr Soc 2006; 15: 247–251. [PubMed] [Google Scholar]
- McGrath JJ, Petersen L, Agerbo E, Mors O, Mortensen PB, Pedersen CB. A comprehensive assessment of parental age and psychiatric disorders. JAMA Psychiatry 2014; 71: 301–309. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, McGrath J, Mortensen PB, Petersen L. The importance of father's age to schizophrenia risk. Mol Psychiatry 2014; 19: 530–531. [DOI] [PubMed] [Google Scholar]
- Petersen L, Mortensen PB, Pedersen CB. Paternal age at birth of first child and risk of schizophrenia. Am J Psychiatry 2011; 168: 82–88. [DOI] [PubMed] [Google Scholar]
- Dean K, Stevens H, Mortensen PB, Murray RM, Walsh E, Pedersen CB. Full spectrum of psychiatric outcomes among offspring with parental history of mental disorder. Arch Gen Psychiatry 2010; 67: 822–829. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen CB, Westergaard T, Wohlfahrt J, Ewald H, Mors O et al. Effects of family history and place and season of birth on the risk of schizophrenia. N Engl J Med 1999; 340: 603–608. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Mors O, Bertelsen A, Waltoft BL, Agerbo E, McGrath JJ et al. A comprehensive nationwide study of the incidence rate and lifetime risk for treated mental disorders. JAMA Psychiatry 2014; 71: 573–581. [DOI] [PubMed] [Google Scholar]
- McGrath JJ, Eyles DW, Pedersen CB, Anderson C, Ko P, Burne TH et al. Neonatal vitamin D status and risk of schizophrenia: a population-based case-control study. Arch Gen Psychiatry 2010; 67: 889–894. [DOI] [PubMed] [Google Scholar]
- Ostergaard SD, Larsen JT, Dalsgaard S, Wilens TE, Mortensen PB, Agerbo E et al. Predicting ADHD by assessment of Rutter's indicators of adversity in infancy. PLoS ONE 2016; 11; doi: 10.1371/journal.pone.0157352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberley T, Stovring H, Sorensen HJ, Horsdal HT, MacCabe JH, Gasse C. Predictors of treatment resistance in patients with schizophrenia: a population-based cohort study. Lancet Psychiatry 2016; 3: 358–366. [DOI] [PubMed] [Google Scholar]
- Dalsgaard S, Leckman JF, Mortensen PB, Nielsen HS, Simonsen M. Effect of drugs on the risk of injuries in children with attention deficit hyperactivity disorder: a prospective cohort study. Lancet Psychiatry 2015; 2: 702–709. [DOI] [PubMed] [Google Scholar]
- Nordentoft M, Mortensen PB, Pedersen CB. Absolute risk of suicide after first hospital contact in mental disorder. Arch Gen Psychiatry 2011; 68: 1058–1064. [DOI] [PubMed] [Google Scholar]
- Dalsgaard S, Ostergaard SD, Leckman JF, Mortensen PB, Pedersen MG. Mortality in children, adolescents, and adults with attention deficit hyperactivity disorder: a nationwide cohort study. Lancet 2015; 385: 2190–2196. [DOI] [PubMed] [Google Scholar]
- Gjerstorff ML. The Danish Cancer Registry. Scand J Public Health 2011; 39(7 Suppl): 42–45. [DOI] [PubMed] [Google Scholar]
- Kildemoes HW, Sorensen HT, Hallas J. The Danish National Prescription Registry. Scand J Public Health 2011; 39(7 Suppl): 38–41. [DOI] [PubMed] [Google Scholar]
- Lynge E, Sandegaard JL, Rebolj M. The Danish National Patient Register. Scand J Public Health 2011; 39(7 Suppl): 30–33. [DOI] [PubMed] [Google Scholar]
- Baadsgaard M, Quitzau J. Danish registers on personal income and transfer payments. Scand J Public Health 2011; 39(7 Suppl): 103–105. [DOI] [PubMed] [Google Scholar]
- Jensen VM, Rasmussen AW. Danish Education Registers. Scand J Public Health 2011; 39(7 Suppl): 91–94. [DOI] [PubMed] [Google Scholar]
- Petersson F, Baadsgaard M, Thygesen LC. Danish registers on personal labour market affiliation. Scand J Public Health 2011; 39(7 Suppl): 95–98. [DOI] [PubMed] [Google Scholar]
- Meier SM, Agerbo E, Maier R, Pedersen CB, Lang M, Grove J et al. High loading of polygenic risk in cases with chronic schizophrenia. Mol Psychiatry 2015. [DOI] [PubMed]
- Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res 2007; 17: 1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Polygenic epidemiology. Genet Epidemiol 2016; 40: 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerbo E, Mortensen PB, Wiuf C, Pedersen MS, McGrath J, Hollegaard MV et al. Modelling the contribution of family history and variation in single nucleotide polymorphisms to risk of schizophrenia: a Danish national birth cohort-based study. Schizophrenia Res 2012; 134: 246–252. [DOI] [PubMed] [Google Scholar]
- Agerbo E, Sullivan PF, Vilhjalmsson BJ, Pedersen CB, Mors O, Borglum AD et al. Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiatry 2015; 72: 635–641. [DOI] [PubMed] [Google Scholar]
- Benros ME, Trabjerg BB, Meier S, Mattheisen M, Mortensen PB, Mors O et al. Influence of polygenic risk scores on the association between infections and schizophrenia. Biol Psychiatry 2016; 80: 609–616. [DOI] [PubMed] [Google Scholar]
- Wimberley T, Gasse C, Meier SM, Agerbo E, MacCabe JH, Horsdal HT. Polygenic risk score for schizophrenia and treatment-resistant schizophrenia. Schizophr Bull 2017; 43: 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen TM, Trabjerg BB, Mors O, Borglum AD, Hougaard DM, Mattheisen M et al. Association of the polygenic risk score for schizophrenia with mortality and suicidal behavior - a Danish population-based study. Schizophrenia Res 2016; 184: 122–127. [DOI] [PubMed] [Google Scholar]
- Demyttenaere K, Bruffaerts R, Posada-Villa J, Gasquet I, Kovess V, Lepine JP et al. Prevalence, severity, and unmet need for treatment of mental disorders in the World Health Organization World Mental Health Surveys. JAMA 2004; 291: 2581–2590. [DOI] [PubMed] [Google Scholar]
- Bock C, Bukh JD, Vinberg M, Gether U, Kessing LV. Validity of the diagnosis of a single depressive episode in a case register. Clin Pract Epidemiol Ment Health 2009; 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessing LV. Validity of diagnoses and other clinical register data in patients with affective disorder. Eur Psychiatry 1998; 13: 392–398. [DOI] [PubMed] [Google Scholar]
- Lauritsen MB, Jorgensen M, Madsen KM, Lemcke S, Toft S, Grove J et al. Validity of childhood autism in the Danish Psychiatric Central Register: findings from a cohort sample born 1990-1999. J Autism Dev Disord 2010; 40: 139–148. [DOI] [PubMed] [Google Scholar]
- Uggerby P, Ostergaard SD, Roge R, Correll CU, Nielsen J. The validity of the schizophrenia diagnosis in the Danish Psychiatric Central Research Register is good. Dan Med J 2013; 60: A4578. [PubMed] [Google Scholar]
- Jakobsen KD, Frederiksen JN, Hansen T, Jansson LB, Parnas J, Werge T. Reliability of clinical ICD-10 schizophrenia diagnoses. Nord J Psychiatry 2005; 59: 209–212. [DOI] [PubMed] [Google Scholar]
- Mohr-Jensen C, Vinkel Koch S, Briciet Lauritsen M, Steinhausen HC. The validity and reliability of the diagnosis of hyperkinetic disorders in the Danish Psychiatric Central Research Registry. Eur Psychiatry 2016; 35: 16–24. [DOI] [PubMed] [Google Scholar]
- Olsen J, Melbye M, Olsen SF, Sorensen TI, Aaby P, Andersen AM et al. The Danish National Birth Cohort—its background, structure and aim. Scand J Public Health 2001; 29: 300–307. [DOI] [PubMed] [Google Scholar]
- Hundrup YA, Simonsen MK, Jørgensen T, Obel EB. Cohort profile: the Danish nurse cohort. Int J Epidemiol 2012; 41: 1241–1247. [DOI] [PubMed] [Google Scholar]
- Tjonneland A, Olsen A, Boll K, Stripp C, Christensen J, Engholm G et al. Study design, exposure variables, and socioeconomic determinants of participation in Diet, Cancer and Health: a population-based prospective cohort study of 57,053 men and women in Denmark. Scand J Public Health 2007; 35: 432–441. [DOI] [PubMed] [Google Scholar]
- Sorensen CJ, Pedersen OB, Petersen MS, Sorensen E, Kotze S, Thorner LW et al. Combined oral contraception and obesity are strong predictors of low-grade inflammation in healthy individuals: results from the Danish Blood Donor Study (DBDS). PLoS ONE 2014; 9: e88196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.