Abstract
Objective
To characterize the phenotypic spectrum, molecular genetic findings, and functional consequences of pathogenic variants in early-onset KCNT1 epilepsy.
Methods
We identified a cohort of 31 patients with epilepsy of infancy with migrating focal seizures (EIMFS) and screened for variants in KCNT1 using direct Sanger sequencing, a multiple-gene next-generation sequencing panel, and whole-exome sequencing. Additional patients with non-EIMFS early-onset epilepsy in whom we identified KCNT1 variants on local diagnostic multiple gene panel testing were also included. When possible, we performed homology modeling to predict the putative effects of variants on protein structure and function. We undertook electrophysiologic assessment of mutant KCNT1 channels in a xenopus oocyte model system.
Results
We identified pathogenic variants in KCNT1 in 12 patients, 4 of which are novel. Most variants occurred de novo. Ten patients had a clinical diagnosis of EIMFS, and the other 2 presented with early-onset severe nocturnal frontal lobe seizures. Three patients had a trial of quinidine with good clinical response in 1 patient. Computational modeling analysis implicates abnormal pore function (F346L) and impaired tetramer formation (F502V) as putative disease mechanisms. All evaluated KCNT1 variants resulted in marked gain of function with significantly increased channel amplitude and variable blockade by quinidine.
Conclusions
Gain-of-function KCNT1 pathogenic variants cause a spectrum of severe focal epilepsies with onset in early infancy. Currently, genotype-phenotype correlations are unclear, although clinical outcome is poor for the majority of cases. Further elucidation of disease mechanisms may facilitate the development of targeted treatments, much needed for this pharmacoresistant genetic epilepsy.
Autosomal dominant pathogenic variants in KCNT1, encoding the sodium-activated potassium channel, are identified in a wide spectrum of epileptic disorders with variable age at onset and cognitive outcome. These include severe early-onset epileptic encephalopathies such as Ohtahara and West syndromes1,2 and epilepsy of infancy with migrating focal seizures (EIMFS),3–14 as well as autosomal dominant and sporadic severe nocturnal frontal lobe epilepsies (ADNFLE and NFLE),10,15,16 but the genotype-phenotype relationship appears to be unclear. We undertook detailed clinical, molecular genetic, and functional characterization of a cohort of patients with KCNT1-related epilepsy.
Methods
Patient recruitment
We recruited patients with EIMFS (n = 31) to a research study investigating the genetic basis of early-onset epileptic encephalopathy (EOEE) between 2011 and 2016, following an earlier national surveillance study.4 Inclusion criteria were epilepsy with onset at <2 years and unknown etiology. Diagnostic criteria for EIMFS were as described in the previous study.4 Patients were recruited at Great Ormond Street Hospital, London, UK, and by referral from other centers in the United Kingdom and internationally. Two patients who had routine local diagnostic multiple gene panel testing revealing KCNT1 variants were also included.
Standard protocol approvals, registrations, and patient consents
We obtained written informed consent from families in whom research genetic investigations were undertaken. The study was approved by the National Research Ethics Service (London-Bloomsbury, Research Ethics Committee reference 13/LO/0168, Integrated Research Application System project identifier 95005). We collected anonymized data from patients tested on the diagnostic next-generation sequencing panel (n = 3) as part of an approved case note review project (Great Ormond Street Hospital Research and Development Department, 16NM11).
Genetic testing
We used a variety of different methods (table e-1, http://links.lww.com/WNL/A6), including direct Sanger sequencing, multiple gene panel testing with the TruSeq Custom Amplicon panel and SureSelect panel, exome sequencing (e-Methods, http://links.lww.com/WNL/A8; tables e-1 and e-2, http://links.lww.com/WNL/A6), and diagnostic chromosomal microarray.
Homology modeling
HMMscan17 against Pfam (database of sequence-based domain families)18 identified 2 domains in the sequence of human KCNT1 (isoform 1): ion channel (PF07885, at position 278–346) and calcium-activated BK potassium channel alpha-subunit family (PF03493, at position 495–598) (e-Methods).
Electrophysiologic assessment of mutant KCNT1 in xenopus oocyte model
We introduced variants into a wild-type (WT) human KCNT1 expression construct19 using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). cDNAs were transcribed in vitro (mMessage mMachine; Ambion, Austin, TX). Oocytes were prepared, and 2-electrode voltage clamp recording was performed after 14 to 24 hours of expression. We also recorded currents before and after the application of Quinidine (e-Methods).
Results
Clinical and molecular genetic features of KCNT1 mutation–positive patients
Clinical presentation
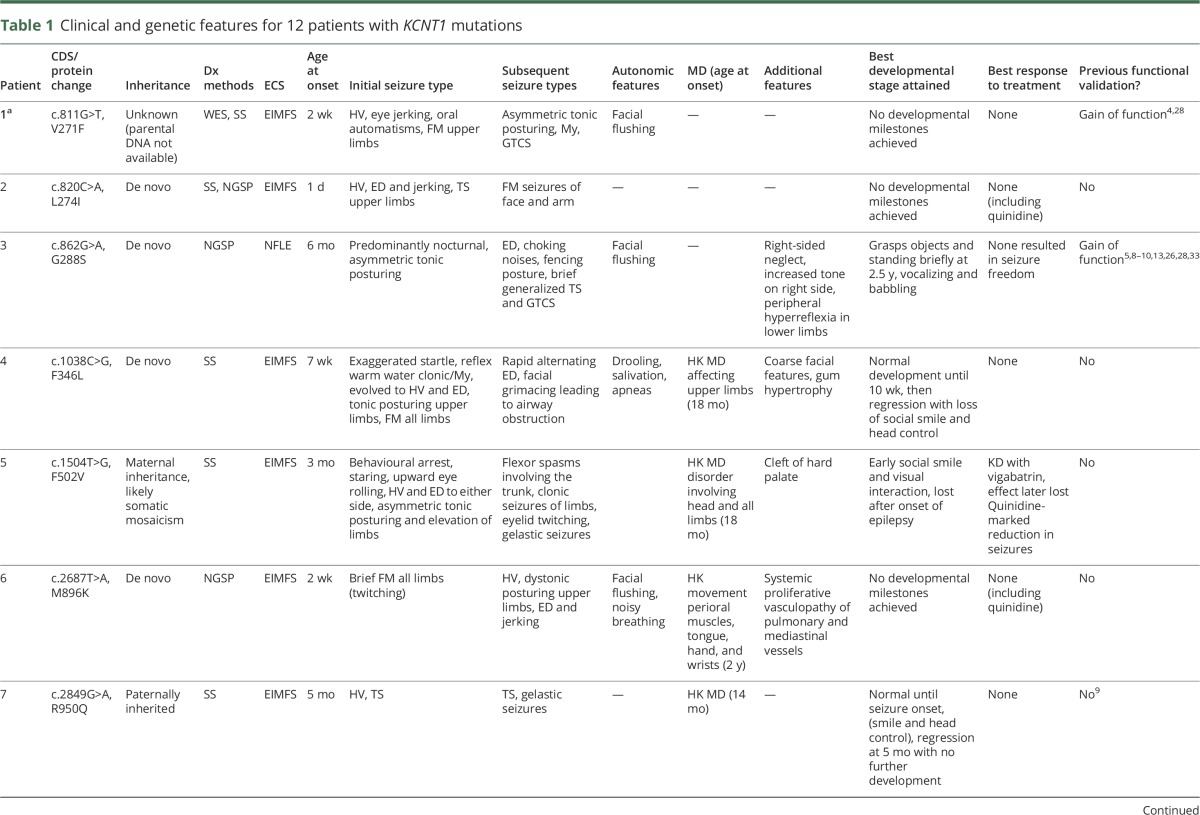
We identified pathogenic variants in KCNT1 in 12 patients, 5 through direct Sanger sequencing, 2 from whole-exome sequencing, and 5 from the Great Ormond Street Hospital diagnostic panel (5 of 800 tested patients with EOEE/developmental delay).
Clinical features are summarized in table 1. Median age at seizure onset was 3.5 weeks (range 1 day–6 months). Most patients developed seizures consistent with EIMFS. Two patients (patients 3 and 11) presented with severe, early-onset NFLE, characterized by asymmetric tonic posturing and later fencing posture. We noted similar frontal seizure semiology in patients with EIMFS (e.g., patient 12). All patients developed axial hypotonia, and upper motor neuron signs emerged in 3 patients. Four patients had a choreiform movement disorder (onset 14–24 months); 1 patient developed generalized dystonia at 18 months. Onset of hyperkinesia was not related to medication (including vigabatrin) nor triggered by intercurrent illness. Initial age at presentation, disease course, response to medication, brain MRI, and EEG findings were similar for both KCNT1 pathogenic variant–positive and –negative patients from the cohort. However, 5 of 12 pathogenic variant–positive patients with EIFMS presented with a severe movement disorder compared to 2 of 19 KCNT1-negative cases. Most had extensive uninformative laboratory metabolic and genetic investigations. Abnormal muscle respiratory chain enzyme activity for complex I and/or II was detected in patients 4 and 8 of uncertain significance (table e-3, http://links.lww.com/WNL/A6). For patient 4, the muscle biopsy was taken during an intercurrent illness and repeated after clinical recovery, revealing a more borderline result. In patient 8, borderline abnormalities in complex I and II ratios were found. Neither patient had other systemic, biochemical, or radiologic features of mitochondrial disease or concurrent sodium valproate treatment.
Table 1.
Clinical and genetic features for 12 patients with KCNT1 mutations
In general, neurodevelopmental outcome was markedly impaired in all patients with EIMFS. All patients had a trial of at least 5 different medications. Response to treatment was, in general, poor (table 1). Three of 8 patients who received the ketogenic diet in combination with other antiepileptic drugs responded with ≈75% seizure reduction. Three patients were treated with quinidine. Patient 2 received 40 mg/kg/d without adverse events but with no effect on seizure burden. Patient 5 was treated with quinidine at 40 mg/kg/d, leading to a marked reduction in seizure frequency. Patient 6 showed some initial transient reduction in seizure frequency at 30 mg/kg/d. For this patient, the unexpected development of a severe proliferative pulmonary and mediastinal vasculopathy resulted in life-threatening pulmonary hemorrhage. Investigations failed to identify an underlying vasculitis, and quinidine was subsequently withdrawn. The patient later died despite initial successful pulmonary embolization.
EEG features
All patients with an EIMFS phenotype had a “migrating” ictal focus with discrete ictal involvement of differing cortical areas within the same EEG (table e-4, http://links.lww.com/WNL/A6). Although not always evident at initial presentation, it developed by 7 months of age in most patients. Periods of EEG suppression or burst suppression were noted in 8 of 12 patients; 6 of these patients had seizure onset in the first 4 weeks of life. Further atypical EEG features included a generalized electrodecremental response in 5 patients and hypsarrhythmia in 1 patient.
Radiologic features
Neuroimaging was available for review in 11 of 12 patients. The majority developed predominantly frontal cerebral atrophy by 3 years of age (figures e-1A and e-1B, http://links.lww.com/WNL/A7; table e-5, http://links.lww.com/WNL/A6). Cerebellar atrophy was also evident in 4 patients (figure e-1C). We noted an open operculum in the first 6 months of life in patient 4 (figure e-1B). Delayed myelination was evident in 9 of 11 patients who had imaging after 3 months of age. In some patients, early brain imaging was normal. Magnetic resonance spectroscopy was abnormal with a relatively reduced N-acetylcholine peak in 3 of 4 patients.
Molecular genetic findings
We identified 12 patients with pathogenic variants in KCNT1 (table 1 and table e-6, http://links.lww.com/WNL/A6); 8 have been previously reported and 4 are unpublished.4,5,9,10,16 Eight of 12 patients had C-terminus variants, of whom 5 had the commonly reported variant A934T. We have identified 4 (including 2 unpublished) pathogenic variants causing EIMFS, namely V271F, L274I, G288S, and F346L located in or between transmembranes 5 and 6. All are missense variants that are predicted to be pathogenic (table e-6), affecting highly conserved amino acid residues (figure e-2, http://links.lww.com/WNL/A7), and are not reported in 1000 Genomes, the ExAC database, or the Exome Variant Server.20–24 For 9 of 12 cases, variants occurred de novo. Parental DNA was not available for patient 1. In patient 5, we found the same KCNT1 variant in an asymptomatic mother and her affected child. We noted a lower heterozygous peak on Sanger sequencing of both salivary and blood-derived maternal genomic DNA (figure e-3), which may reflect somatic mosaicism. In patient 7, the variant was inherited from the unaffected father with no difference in peak size on Sanger sequencing (figure e-4). The recurrent A934T variant was identified in 5 patients, 4 with an EIMFS presentation and 1 (patient 11) with an NFLE phenotype.
Protein homology modeling of mutant KCNT1
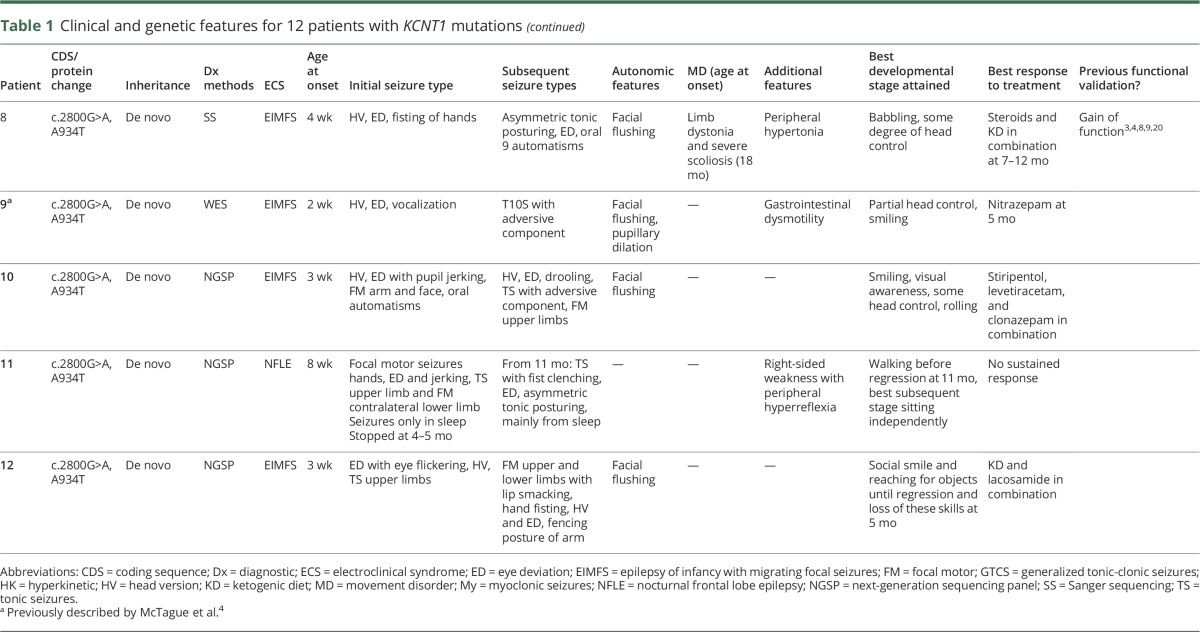
Homology modeling was performed for 2 novel mutations: F346L, located in the ion channel domain (residues 270–353), and F502V, located in the gating region (residues 373–1174, although residues 1,045–1,174 could not be modeled). F346 is located on the inner helix of the transmembrane pore (figure 1, A and B). It is part of the hydrophobic cavity, which mediates interactions between the inner-membrane helices of 2 adjacent subunits (figure 1C) and is thus responsible for maintaining the stability of the open conformation. In the modeled closed-state conformation, the helix containing F346 and the inner helix from the other protomer undergo conformational changes (figure e-5, http://links.lww.com/WNL/A7). Therefore, mutation to leucine (F346L) is likely to destabilize the open state by perturbing the hydrophobic interactions because the side chain of leucine is smaller (figure 1D), affecting the equilibrium between the closed and open states. In addition, the packing arrangement in the K+ channels involving the pore and the inner helix is known to be critical for the stability of the tetrameric assembly, ion conduction function, and cation selectivity. Thus, F346L might be detrimental to these functions.25
Figure 1. Modeling the ion channel and gating apparatus of KCNT1.
(A) Side view of the homology model of the KCNT1 ion channel (residues 278–346) as a tetramer. F346 is present on the edge of the inner helix (in gold) and interacts with the inner helix of the adjacent subunit in the tetrameric arrangement. Membrane position is shown in spheres. (B) Top view of the tetramer arrangement of the ion channel and location of F346 on the inner helix. (C) F346 is part of the hydrophobic cavity (shown as surface), which mediates interactions between the inner membrane helices of the 2 subunits. F346 is shown in green; the surrounding hydrophobic residues are shown in red. (D) On mutation to leucine (F346L, in green), the hydrophobic interactions between the 2 subunits are likely to be reduced (black circle) because the side chain of leucine is much shorter than phenylalanine. (E) Model of a dimer of the gating ring (residues 373–1,044; residues 1,045–1,174 could not be modeled), which is a tetramer (dimer of the modeled dimer). Each subunit possesses 2 RCK domains: RCK1 (in blue) and RCK2 (in gold). F502 (in green) is present in the RCK1 domain, near the intersubunit interface (assembly interface). The RCK1-RCK2 intrasubunit interface is purple (residues from RCK1) and orange (residues from RCK2). The dimer interfaces formed by both RCK-1 and RCK-2 are indicated by an arrow. (F) F502 (green) and its neighboring hydrophobic residues (red), including W476, with which it could potentially form a pi-pi interaction. Distance between the centroid (spheres) of the 2 rings (F502 and W476) is 4.7 Å, and the angle between the ring planes is 27.3°. (G) F502V could abolish the formation of the potential pi-pi interaction with W476 and is likely to reduce the hydrophobic interactions (black circle) because the side chain of valine is smaller than that of phenylalanine.
Within each protomer of the KCNT1 gating region, there are 2 tandem RCK domains (RCK1 and RCK2) that serve as regulators of potassium conductance (figure 1E). These form flexible intrasubunit and intersubunit (figure 1E) interfaces that facilitate functional tetramer formation.26 F502 is located in RCK1 and predicted to form a pi-pi interaction with W476 from αD (figure 1F). F502 is also surrounded by a number of hydrophobic residues (I472, L473, A475, V500, and A503), which may play a role in stabilizing the gating ring (figure 1F). The amino acid substitution F502V is predicted to result in destabilization of these hydrophobic interactions, given the smaller valine side chain (figure 1G), and abolition of potential pi-stacking with resultant disruption of the stable assembly interface.
Electrophysiologic assessment of mutant KCNT1
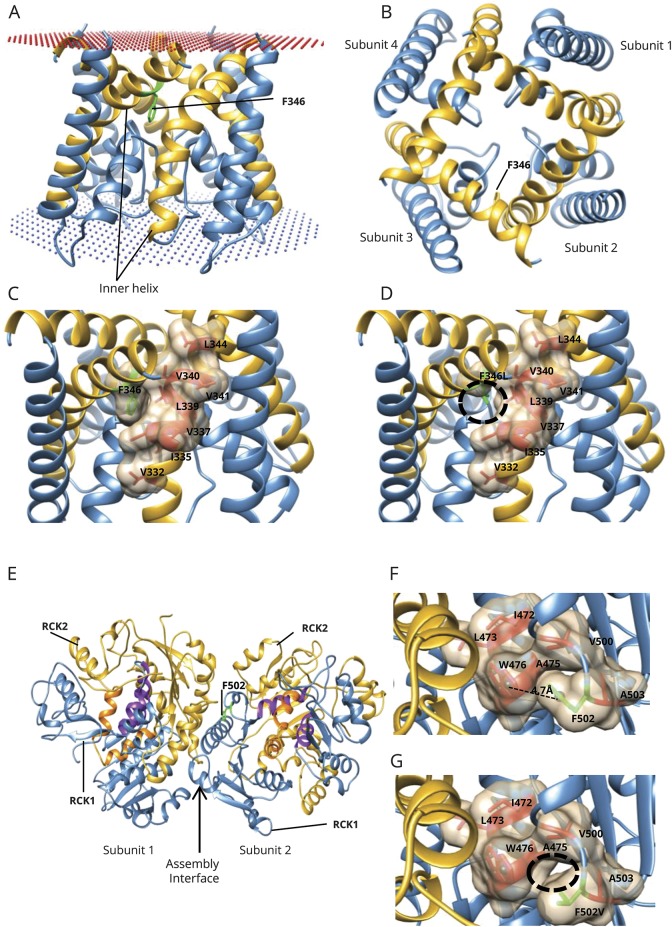
We evaluated the 4 previously unpublished variants and V271F, which we previously described4 and was recently studied in a xenopus oocyte system.27 All mutations resulted in an increased current magnitude compared to WT (figure 2A). We noted that for variants V271F and F346L, the rate of activation was slowed at higher voltages compared to WT, and in others (M896K, F502V and L274I), the activation rates were generally faster than WT (figure 2A). Investigation of the current-voltage relationship showed that mutant channels were very weakly voltage dependent, and in some cases, voltage dependence of steady-state activation was essentially absent (figure 2, B and C) with only a residual Goldman-Hodgkin-Katz rectification. Assessment of average peak currents at 10 mV revealed a significant difference between both individual mutant channels and summated data compared to WT (figure 2, D and E).
Figure 2. Functional investigation of KCNT1 mutations in a xenopus oocyte model.
(A) Representative current traces obtained from oocytes expressing WT and EIMFS mutants (M896K, F502V, V271F, F346L, and L274I). Oocytes were held at −90 mV and stepped from −80 to 80 mV for 600 milliseconds every 5 seconds. Scale bars apply to all traces. (B) Current-voltage relationships for WT (n = 32), M896K (n = 15), F502V (n = 13), V271F (n = 9), F346L (n = 11), and L274I (n = 12). Currents were averaged and then normalized to the value at a test potential of 80 mV (Imax). (C) Comparison of current-voltage relationships between WT (solid circles, n = 32) and EIMFS mutations (M896K [squares, n = 15], F502V [triangles, n = 13], V271F [hexagons, n = 9], F346L [diamonds, n = 11], and L274I [inverted triangles, n = 12]). Currents were averaged and then normalized to the value at a test potential of 80 mV (Imax). (D) Average peak currents at 10 mV for WT (n = 44), M896K (n = 19), F502V (n = 16), V271F (n = 10), F346L (n = 11), and L274I (n = 12) channels. Peak currents for each mutant channel at 10 mV were compared to the peak currents for the WT channel at 10 mV. ***p < 0.001, ****p < 0.0001. (E) Comparison of pooled WT (n = 44) and EIMFS (n = 68) currents at 10 mV. ****p < 0.0001. EIMFS = epilepsy of infancy with migrating focal seizures; WT = wild-type.
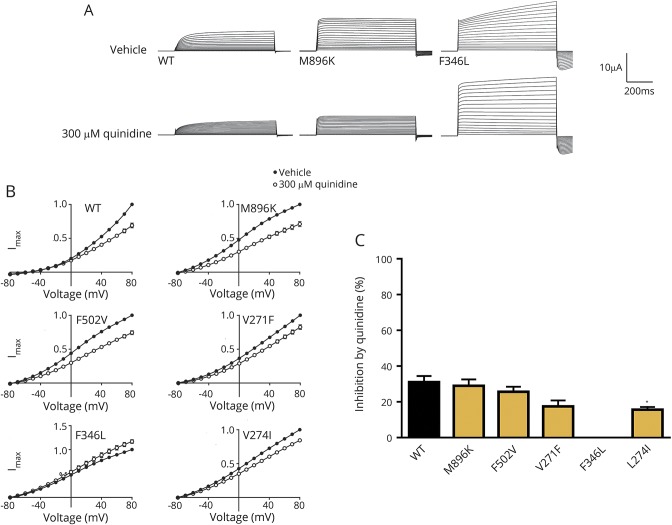
Effect of 300 μmol/L quinidine on mutant KCNT1
Quinidine 300 µmol/L had variable current-blocking effects in different mutant channels. For F346L, peak current was completely insensitive to quinidine, although it had some effect on activation kinetics (figure 3A). The differential sensitivity of KCNT1 mutants to quinidine was clearly shown in the current-voltage relationship (figure 3B) and percentage of inhibition at maximum current, 80 mV (figure 3C). There is some correlation between the in vitro studies and clinical response in patient 5 (figure 3). M896K had the most marked in vitro blockade by quinidine, and patient 6 showed some initial clinical response. F346L showed no quinidine response at all, and the patient harboring this mutation was not treated with quinidine.
Figure 3. Effect of quinidine on xenopus oocytes expressing hKCNT1 channels.
(A) Representative current traces obtained from oocytes expressing WT and EIMFS mutants (M896K and F346L) with application of vehicle (ND96) and 300 μmol/L quinidine. Oocytes were held at −90 mV and stepped from −80 to 80 mV for 600 milliseconds every 5 seconds. Scale bars apply to all traces. (B) Current-voltage relationships for WT (n = 32), M896K (n = 15), F502V (n = 13), V271F (n = 9), F346L (n = 11), and L274I (n = 12) hKCNT1 channels in the presence of vehicle (ND96) and 300 μmol/L quinidine. Currents were averaged and then normalized to the value at a test potential of 80 mV (Imax). (C) Average percent inhibition at 80 mV of WT (n = 31) and EIMFS (M896K, n = 15; F502V, n = 13; V271F, n = 9; F346L, n = 11; and; L274I, n = 12) hKCNT1 channels by quinidine (300 μmol/L) depicting the variable degree of block by 300 μmol/L quinidine (1-way analysis of variance followed by Bonferroni post hoc analysis). *p < 0.1. EIMFS = epilepsy of infancy with migrating focal seizures; WT = wild-type.
Discussion
We report a cohort of patients with early-onset epilepsy associated with pathogenic variants in KCNT1, which encodes the sodium-activated potassium channel KCa4.1 (sequence like a calcium-dependent potassium channel [SLACK], Slo2.2). KCNT1 is widely expressed throughout the brain, as well as in the dorsal root ganglia, kidney, and heart, and is responsible for slow hyperpolarization after bursts of action potentials.28,29 KCNT1 also has direct interactions with Fragile X-related protein.29 Compared with other potassium channels, KCNT1 is involved in a highly extensive protein network, suggesting a putative role in cognitive developmental processes.3,8,28,30
To date, KCNT1 variants have been reported in a wide range of epilepsies (table e-7, http://links.lww.com/WNL/A6).1–16,31,32 We identified patients with the same variant associated with varying electroclinical phenotypes (table 1). Phenotypic variability has been reported within single families in which different individuals may present with either ADNFLE or EIMFS.10 Such intrafamilial variation in phenotype is also described in SCN1A kindreds; Dravet syndrome, febrile seizures, and a variety of other generalized epilepsies may be reported in the same family.33 Furthermore, while the majority of variants in our cohort occurred de novo, 2 patients inherited variants from an unaffected parent. The mechanisms underlying phenotypic variability and true/apparent nonpenetrance are unclear but may be related to somatic mosaicism, variant type, other genetic/epigenetic factors, or differential expression of alternative KCNT1 transcripts.9,10,29,34,35
The majority of patients with pathogenic KCNT1 variants in our cohort had electroclinical EIFMS, although this is likely to reflect ascertainment bias. Indeed, 2 of the 5 KCNT1-positive patients identified by the diagnostic panel from a larger cohort of 800 patients with EOEE/developmental delay had an NFLE-like presentation. Although movement disorders are increasingly reported in other severe early-onset genetic epilepsies, they appear to be rare in KCNT1 epilepsy.36 We describe several atypical EEG features. Generalized electrodecrement and hypsarrhythmia, more classically associated with infantile spasms, have been previously described in EIMFS.2,4,9,10,31 EEG suppression, classically seen in Ohtahara syndrome,37 has been only rarely described in EIMFS.4,9 Extensive diagnostic investigations undertaken in patients with KCNT1 mutations were unyielding other than abnormal respiratory chain enzyme analysis of muscle tissue in 2 patients. The relevance of these findings is not clear, but secondary mitochondrial effects may be evident in KCNT1 epilepsy, as often reported in other severe drug-resistant epilepsies.38 Other genetic and environmental influences on mitochondrial function may also play a role.
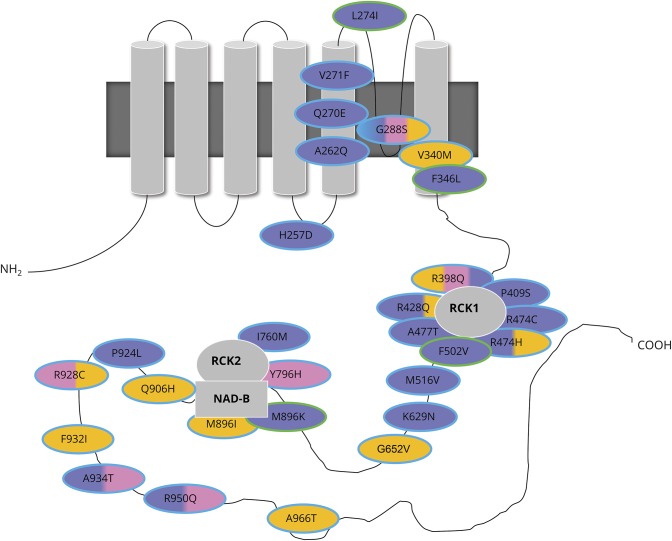
KCNT1 tetramers form a transmembrane sodium-activated potassium channel. Each subunit consists of 6 transmembrane domains with an extended cytoplasmic carboxy (C-) terminus (figure 4). The majority of reported pathogenic variants (table e-7, http://links.lww.com/WNL/A6), as seen in this study, are located in the C-terminus with clustering around the RCK and nicotinamide adenine dinucleotide–binding domains (figure 4). More recently, several variants have been identified within transmembrane domain 5 and in the pore-forming regions between transmembrane domains 4 and 54,5,8–10 (table e-7), and this study also demonstrates epilepsy-associated mutations in transmembrane domains.
Figure 4. Schematic diagram of the location of mutations in KCNT1 in this and previously published studies.
KCNT1 encodes sequence like a calcium-dependent potassium channel (SLACK), which forms tetramers (top left) or heteromers with KCNT2 or sequence like an intermediate conductance K channel (SLICK). The structure comprises 6 transmembrane domains with a pore-forming region, regulator of potassium conductance (RCK), and nicotinamide adenine dinucleotide–binding (NAD-B) domains. EIMFS phenotypes are shaded in purple, ADNFLE or NFLE in pink, others (Ohtahara syndrome, leukoencephalopathy, focal epilepsy, EOEE, West syndrome, unaffected) in orange. Mutations giving rise to >1 phenotype are shaded with a combination of the corresponding colors. Novel mutations identified in this study are outlined in green, those identified in previous studies in turquoise. ADNFLE = autosomal dominant nocturnal frontal lobe epilepsy; EIMFS = epilepsy of infancy with migrating focal seizures; EOEE = early-onset epileptic encephalopathy; NFLE = nocturnal frontal lobe epilepsy.
To date, different model systems have been used to determine the functional effects of KCNT1 variants.1,5,7,10,13,19,27 Our protein homology structural modeling data predict abnormal gating or protein instability within the pore-forming region as a putative disease mechanism. In silico modeling of G288S has predicted similar detrimental effects,5 while Y775H is predicted to affect sodium sensitivity of the channel.27 KCNT1 variants may therefore alter structural properties of the protein, contributing to altered channel function. Consistent with previous reports1,3,13,19,27 (table e-7, http://links.lww.com/WNL/A6), our xenopus oocyte model demonstrated that KCNT1 pathogenic variants display a gain-of-function effect with increased current amplitude (figure 2). Previous studies have sought to correlate disease severity with the degree of gain of function.1,19 However, in keeping with more recent studies,8 such correlation was not evident in our study. KCNT1 variants result in an increased Po (probability of the channel being open), which may be due to increased mutant channel cooperativity or altered sodium sensitivity.8,27 In a recent study, sodium removal from the pipette solution had a less negative effect on G288S channel amplitude than WT, suggesting reduced sodium sensitivity in the mutant.13 Heterotetramer formation may be of importance in vivo. In 1 study, mutant KCNT1 homomers revealed a more marked gain of function than mutant WT heteromers.13 A significant remaining question is how KCNT1 gain-of-function variants with predicted effects on neuronal hyperpolarization result in epilepsy.28 Altered voltage sensitivity may result in KCNT1 channels opening at more depolarized potentials, allowing a persistent hyperpolarizing current, with resultant interneuronal disinhibition as reported in SCN1A-related epilepsy.39 Conversely, increased repolarization permitting more frequent and rapid action potentials may also play a role.27,34
Recently, quinidine has been identified as a novel therapy for patients with KCNT1-related epilepsy. In in vitro models, quinidine has been shown to reduce the abnormal increase in mutant KCNT1 channel amplitude.19 For 1 patient with EIMFS with the KCNT1 variant R428Q, in vitro testing showed quinidine sensitivity, and treatment resulted in a dramatic improvement in seizure control with neurodevelopmental gains.3,7 However, in more recent studies, patient response has been variable and not always as predicted by in vitro studies.11 Indeed, another patient with the same variant (R428Q) but different epilepsy phenotype (unclassified EOEE) failed to respond to quinidine, albeit at a later stage in the disease course.14 Most recently, a patient with West syndrome had a good response but only with a higher dose of 60 mg/kg/d.2 Clinical response may possibly be determined by the specific variant, other genetic factors, epilepsy phenotype, and drug timing within a therapeutic window. In our series, we treated 3 patients with quinidine, and 1 patient showed a clinical response. One patient developed a severe pulmonary vasculopathy, after which quinidine was discontinued. Systemic vasculitis has been reported with quinidine treatment.40 While investigations in our patient did not reveal overt evidence of vasculitis, the observed pulmonary dysfunction may represent an adverse drug-related event. The precise mechanism of KCNT1 blockade by quinidine is unclear, and it is possible that the disease mechanism for F346L, perhaps involving abnormal channel-opening dynamics as suggested by the modeling data, is not modifiable by quinidine. Our data suggest that quinidine should be considered as a therapeutic option for patients with KCNT1 variants, but used with caution. Larger studies will provide further guidance about clinical utility, patient selection, optimum age at administration, and dose. Other KCNT1 modulators, including bepridil and clofililum, have been identified as possible alternative therapies.28 Like quinidine, bepridil has been shown in vitro to reversibly block mutant KCNT1 channels at a lower concentration than WT channels.13 However, similar to quinidine, potential cardiac effects and lack of specificity may limit use in patients.
Pathogenic variants in KCNT1 cause a wide spectrum of severe epilepsies typically associated with impaired neurologic development and significant disease burden. As demonstrated, in vitro model systems may be useful to validate putative variants and to confirm pathogenicity, although genotype-phenotype correlations remain unclear. Evaluation of new therapies, including KCNT1-specific blockers, remains a research priority for this devastating pharmacoresistant group of epilepsies.
Acknowledgment
The authors thank the patients and their families for their participation in this study. They thank all clinicians referring patients with EIMFS for inclusion in the research study (Dr. Mike Pike, Dr. Stefan Spinty, Dr. Ailsa McLellan, Dr. Mary King, Dr. Andrew Curran, Dr. Hans Randby, Dr. Linda de Meirleir, Dr. Jost Richter, Dr. Elaine King, Dr. Martin Piepkorn, Dr. Mary O'Regan, Dr. Ariane Biebl, Dr. Gary McCullagh, and Dr. Siobhan West). They thank Erin Heinzen, PharmD, PhD, for whole-exome sequencing of 5 patients at the Duke Center for Genomic Medicine, Durham, NC.
Glossary
- ADNFLE
autosomal dominant nocturnal frontal lobe epilepsy
- EIMFS
epilepsy of infancy with migrating focal seizures
- EOEE
early-onset epileptic encephalopathy
- NFLE
nocturnal frontal lobe epilepsy
- RCK
regulator of potassium conductance
- SLACK
sequence like a calcium-dependent potassium channel
- WT
wild-type
Note added in proof
Recently, 3 patients with de novo KCNT1 mutations and massive systemic to pulmonary collateral artery formation, presenting with pulmonary hemorrhage requiring embolization, were described. These patients had not been treated with quinidine. However, the mechanism remains unclear and further investigation of the expression and role of KCNT1 in the cardiovascular system is required.41
Author contributions
Amy McTague, Umesh Nair, Sony Malhotra, and Esther Meyer have contributed to drafting/revising the manuscript for content, including medical writing for content, study concept or design, acquisition of data, analysis or interpretation of data. Natalie Trump has contributed to drafting/revising the manuscript for content, including medical writing for content, acquisition of data, analysis or interpretation of data. Elena V. Gazina, Apostolos Papandreou, and Adeline Ngoh have contributed to drafting/revising the manuscript for content, acquisition of data, analysis or interpretation of data. Sally Ackermann and Gautam Ambegaonkar have contributed to drafting/revising the manuscript for content, acquisition of data. Richard Appleton has contributed to drafting/revising the manuscript for content, including medical writing for content, acquisition of data, analysis or interpretation of data. Archana Desurkar has contributed to drafting/revising the manuscript for content, acquisition of data. Christin Eltze and Rachel Kneen have contributed to drafting/revising the manuscript for content, including medical writing for content, acquisition of data, analysis or interpretation of data. Ajith V. Kumar and Karine Lascelles have contributed to drafting/revising the manuscript for content, acquisition of data. Tara Montgomery has contributed to drafting/revising the manuscript for content, acquisition of data, analysis or interpretation of data. Venkateswaran Ramesh and Rajib Samanta have contributed to drafting/revising the manuscript for content, acquisition of data. Richard H. Scott has contributed to drafting/revising the manuscript for content, acquisition of data, analysis or interpretation of data. Jeen Tan has contributed to drafting/revising the manuscript for content, acquisition of data. William Whitehouse and Annapurna Poduri have contributed to drafting/revising the manuscript for content, including medical writing for content, acquisition of data. Ingrid E. Scheffer, W.K. “Kling” Chong, and J. Helen Cross have contributed to drafting/revising the manuscript for content, including medical writing for content, acquisition of data, analysis or interpretation of data. Maya Topf, Steven Petrou, and Manju A. Kurian have contributed to drafting/revising the manuscript for content, including medical writing for content, study concept or design, acquisition of data, analysis or interpretation of data.
Study funding
This project was supported by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London.
Disclosure
A. McTague was funded by a Medical Research Council Clinical Research Training Fellowship (MR/L001497/1), the Medical Research Foundation for travel and conference fees (MRF-007-0003-STD-MCTAG), a Grass Foundation Travel Bursary from the American Epilepsy Society, and a Junior Investigator travel scholarship from the University of South California. U. Nair and S. Malhotra report no disclosures relevant to the manuscript. E. Meyer is funded by SPARKS. N. Trump reports no disclosures relevant to the manuscript. E.V. Gazina is funded by the Australian Research Council (APP1106027). A. Papandreou is funded by a joint Action Medical Research and British Paediatric Neurology Association research training fellowship. A. Ngoh is funded by Guarantors of Brain, Friends of Landau Kleffner Syndrome and Action Medical Research. S. Ackermann and G. Ambegaonkar report no disclosures relevant to the manuscript. R. Appleton served on the scientific committees for and received a single honorarium from Novartis and GW Pharma and is the Chief Investigator for the EcLiPSE Study funded by an National Institute for Health Research Health Technology Assessment grant (12/127/134). A. Desurkar, C. Eltze, R. Kneen, A.V. Kumar, K. Lascelles, T. Montgomery, V. Ramesh, R. Samanta, and R.H. Scott report no disclosures relevant to the manuscript. J. Tan received an industry-funded bursary for conference travel (£300) from UCB Pharma. W. Whitehouse serves on the editorial board of NeuroEducation. A. Poduri is on the Scientific Advisory Board of the Dravet Syndrome Foundation, serves as an associate editor for Epilepsia and contributing editor for Epilepsy Currents, and is on the editorial board for Annals of Neurology. She receives research support from the National Institute of Neurological Disorders and Stroke (NINDS), Citizens United for Research in Epilepsy, and the Boston Children's Hospital Translational Research Program. I.E. Scheffer serves on the editorial boards of Neurology and Epileptic Disorders; may accrue future revenue on a pending patent WO61/010176: Therapeutic Compound that relates to discovery of PCDH19 gene; is one of the inventors listed on a patent held by Bionomics Inc on diagnostic testing of using the SCN1A gene, WO2006/133508; has received speaker honoraria or scientific board consulting fees from Athena Diagnostics, UCB, GSK, Eisai, and Transgenomics; has received funding for travel from Athena Diagnostics, UCB, Eisai, and GSK; and has received research support from the National Health and Medical Research Council of Australia, NINDS, Health Research Council of New Zealand, US Department of Defense, and March of Dimes. W.K. Chong reports no disclosures relevant to the manuscript. J.H. Cross is on the advisory boards of Shire, GSK, Eisai, Vitaflo, UCB, and Takeda for which remuneration has been made to her department; has a patent in application with Vitaflo; and is the chief investigator for clinical trials with Vitaflo, GW Pharma, and Zogenix for which remuneration has been made to the department. M. Topf reports no disclosures relevant to the manuscript. S. Petrou is scientific founder of Praxis Precision Medicine, Boston, and a Scientific Advisory Board Member of Pairnomix, Minneapolis, and has received shares for both. Dr. Petrou's research is supported by the Lulu Foundation, Vitaflo/Nestle, Citizens United for Research in Epilepsy, Mallinckrodt Pharmaceuticals, Clarus Ventures, DHB Foundation, Australian Research Council, National Health and Medical Research Council, and SCN2A Research Foundation. M.A. Kurian receives research/grant support from a Wellcome Trust Intermediate Clinical Fellowship (WT098524MA). Go to Neurology.org/N for full disclosures.
References
- 1.Martin HC, Kim GE, Pagnamenta AT, et al. Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum Mol Genet 2014;23:3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukuoka M, Kuki I, Kawawaki H, et al. Quinidine therapy for West syndrome with KCNT1 mutation: a case report. Brain Dev 2017;39:80–83. [DOI] [PubMed] [Google Scholar]
- 3.Barcia G, Fleming MR, Deligniere A, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet 2012;44:1255–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McTague A, Appleton R, Avula S, et al. Migrating partial seizures of infancy: expansion of the electroclinical, radiological and pathological disease spectrum. Brain 2013;136:1578–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishii A, Shioda M, Okumura A, et al. A recurrent KCNT1 mutation in two sporadic cases with malignant migrating partial seizures in infancy. Gene 2013;531:467–471. [DOI] [PubMed] [Google Scholar]
- 6.Vanderver A, Simons C, Schmidt JL, et al. Identification of a novel de novo p.Phe932Ile KCNT1 mutation in a patient with leukoencephalopathy and severe epilepsy. Pediatr Neurol 2014;50:112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bearden D, Strong A, Ehnot J, DiGiovine M, Dlugos D, Goldberg EM. Targeted treatment of migrating partial seizures of infancy with quinidine. Ann Neurol 2014;76:457–461. [DOI] [PubMed] [Google Scholar]
- 8.Kim GE, Kronengold J, Barcia G, et al. Human slack potassium channel mutations increase positive cooperativity between individual channels. Cell Rep 2014;9:1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohba C, Kato M, Takahashi N, et al. De novo KCNT1 mutations in early-onset epileptic encephalopathy. Epilepsia 2015;56:e121–e128. [DOI] [PubMed] [Google Scholar]
- 10.Møller RS, Heron SE, Larsen LHG, et al. Mutations in KCNT1 cause a spectrum of focal epilepsies. Epilepsia 2015;56:e114–e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikati MA, Jiang Y-H, Carboni M, et al. Quinidine in the treatment of KCNT1 positive epilepsies. Ann Neurol 2015;78:995–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen NM, Conroy J, Shahwan A, et al. Unexplained early onset epileptic encephalopathy: exome screening and phenotype expansion. Epilepsia 2015;57:e12–e17. [DOI] [PubMed] [Google Scholar]
- 13.Rizzo F, Ambrosino P, Guacci A, et al. Characterization of two de novo KCNT1 mutations in children with malignant migrating partial seizures in infancy. Mol Cell Neurosci 2016;72:54–63. [DOI] [PubMed] [Google Scholar]
- 14.Chong PF, Nakamura R, Saitsu H, Matsumoto N, Kira R. Ineffective quinidine therapy in early-onset epileptic encephalopathy with KCNT1 mutation. Ann Neurol 2016;79:502–503. [DOI] [PubMed] [Google Scholar]
- 15.Heron SE, Smith KR, Bahlo M, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet 2012;44:1188–1190. [DOI] [PubMed] [Google Scholar]
- 16.Hildebrand MS, Myers CT, Carvill GL, et al. A targeted resequencing gene panel for focal epilepsy. Neurology 2016;86:1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol 2011;7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res 2009;38:D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milligan CJ, Li M, Gazina EV, et al. KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidine. Ann Neurol 2014;75:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes. Available at: http://browser.1000genomes.org/index.html. Accessed June 14, 2016.
- 21.ExAC Browser (beta)|Exome Aggregation Consortium. Available at: http://exac.broadinstitute.org/. Accessed June 14, 2016.
- 22.NHLBI Exome Sequencing Project (ESP). Available at: http://evs.gs.washington.edu/EVS/. Accessed June 14, 2016.
- 23.PolyPhen-2. Available at: http://genetics.bwh.harvard.edu/pph2/. Accessed June 14, 2016.
- 24.Available at: http://provean.jcvi.org/genome_submit_2.php. Accessed June 14, 2016.
- 25.Doyle DA, Morais Cabral J, Pfuetzner RA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 1998;280:69–77. [DOI] [PubMed] [Google Scholar]
- 26.Yuan P, Leonetti MD, Pico AR, Hsiung Y, MacKinnon R. Structure of the human BK channel Ca2+-activation apparatus at 3.0 Å resolution. Science 2010;329:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Q-Y, Zhang F-F, Xu J, et al. Epilepsy-related slack channel mutants lead to channel over-activity by two different mechanisms. Cell Rep 2016;14:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaczmarek LK. Slack, slick, and sodium-activated potassium channels. ISRN Neurosci 2013;2013:354262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown MR, Kronengold J, Gazula V-R, et al. Amino-termini isoforms of the slack K+ channel, regulated by alternative promoters, differentially modulate rhythmic firing and adaptation. J Physiol 2008;586:5161–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim GE, Kaczmarek LK. Emerging role of the KCNT1 slack channel in intellectual disability. Front Cell Neurosci 2014;8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen AS, Berkovic SF, Cossette P, et al. De novo mutations in epileptic encephalopathies. Nature 2013;501, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arai-Ichinoi N, Uematsu M, Sato R, et al. Genetic heterogeneity in 26 infants with a hypomyelinating leukodystrophy. Hum Genet 2015;135:89–98. [DOI] [PubMed] [Google Scholar]
- 33.Scheffer IE, Zhang Y-HH, Jansen FE, Dibbens L. Dravet syndrome or genetic (generalized) epilepsy with febrile seizures plus? Brain Dev 2009;31:394–400. [DOI] [PubMed] [Google Scholar]
- 34.Lim CX, Ricos MG, Dibbens LM, Heron SE. KCNT1 mutations in seizure disorders: the phenotypic spectrum and functional effects. J Med Genet 2016;53:217–225. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Kronengold J, Yan Y, et al. The N-terminal domain of slack determines the formation and trafficking of slick/slack heteromeric sodium-activated potassium channels. J Neurosci 2009;29:5654–5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howell KB, McMahon JM, Carvill GL, et al. SCN2A encephalopathy: a major cause of epilepsy of infancy with migrating focal seizures. Neurology 2015;85:958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtahara S. A study on the age dependent epileptic encephalopathy. No To Hattatsu 1977;9:2–21. [PubMed] [Google Scholar]
- 38.Folbergrová J, Kunz WS. Mitochondrial dysfunction in epilepsy. Mitochondrion 2012;12:35–40. [DOI] [PubMed] [Google Scholar]
- 39.Yu FH, Mantegazza M, Westenbroek RE, et al. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci 2006;9:1142–1149. [DOI] [PubMed] [Google Scholar]
- 40.Lipsker D, Walther S, Schulz R, Navé S, Cribier B. Life-threatening vasculitis related to quinidine occurring in a healthy volunteer during a clinical trial. Eur J Clin Pharmacol 1998;54:815. [DOI] [PubMed] [Google Scholar]
- 41.Kawasaki Y, Kuki I, Ehara E, et al. Three cases of KCNT1 mutations: malignant migrating partial seizures in infancy with massive systemic to pulmonary collateral arteries. J Pediatr Epub 2017 October 5. [DOI] [PubMed] [Google Scholar]