Significance
We demonstrate a technology that we believe could transform the way nutritional deficiency testing is done worldwide. Micronutrient deficiencies such as vitamin A and iron affect a third of the world’s population with consequences such as blindness, higher child mortality, anemia, poor pregnancy outcomes, and reduced work capacity. Our testing platform simultaneously quantifies concentrations of ferritin, retinol-binding protein, and C-reactive protein, allowing one to diagnose iron and vitamin A deficiency and inflammation status (to better inform and help with the interpretation of the result) in about 15 min. Our approach combines multiple florescent markers and immunoassay approaches on a single test strip, allowing us to span the five orders of magnitude difference in the physiological range of these markers.
Keywords: micronutrient deficiency diagnostics, smartphone, point-of-care diagnostics, multicolor lateral flow, fluorescence immunoassay
Abstract
Micronutrient deficiencies such as those of vitamin A and iron affect a third of the world’s population with consequences such as night blindness, higher child mortality, anemia, poor pregnancy outcomes, and reduced work capacity. Many efforts to prevent or treat these deficiencies are hampered by the lack of adequate, accessible, and affordable diagnostic methods that can enable better targeting of interventions. In this work, we demonstrate a rapid diagnostic test and mobile enabled platform for simultaneously quantifying iron (ferritin), vitamin A (retinol-binding protein), and inflammation (C-reactive protein) status. Our approach, enabled by combining multiple florescent markers and immunoassay approaches on a single test, allows us to provide accurate quantification in 15 min even though the physiological range of the markers of interest varies over five orders of magnitude. We report sensitivities of 88%, 100%, and 80% and specificities of 97%, 100%, and 97% for iron deficiency (ferritin <15 ng/mL or 32 pmol/L), vitamin A deficiency (retinol-binding protein <14.7 μg/mL or 0.70 μmol/L) and inflammation status (C-reactive protein >3.0 μg/mL or 120 nmol/L), respectively. This technology is suitable for point-of-care use in both resource-rich and resource-limited settings and can be read either by a standard laptop computer or through our previously developed NutriPhone technology. If implemented as either a population-level screening or clinical diagnostic tool, we believe this platform can transform nutritional status assessment and monitoring globally.
Iron deficiency (ID) and vitamin A deficiency (VAD) are two of the most prevalent micronutrient deficiencies worldwide. ID affects 2 billion people and is a common cause of anemia, which may reduce physical work capacity in adults or lead to impaired brain development in children (1). VAD causes night blindness, mostly among children and pregnant women, affecting about 29% of the population in low- and mid-income countries (2). VAD also negatively affects the immune system and results in lowered erythropoiesis (3). Moreover, ID and VAD often coexist (4) and the interaction between ID and VAD may exacerbate each deficiency (5–7). Therefore, simultaneous access to iron and vitamin A status among populations at risk is important. ID and VAD due to dietary inadequacy can be potentially effectively treated at early stages by changing diet and/or taking supplements (8–10), although it can be challenging at the population level. Measuring ID and VAD status represents an important first step in managing these deficiencies.
Ferritin and retinol-binding protein (RBP) in serum are common biomarkers used for assessment of iron and vitamin A status. Ferritin serves to store and transport iron in blood, an indicator of ID (11). The normal range of ferritin in serum is between 15 ng/mL (32 pmol/L) and 150 ng/mL (316 pmol/L) for females or 200 ng/mL (421 pmol/L) for males, while the cutoff for depleted iron storage in children less than 5 y old is <12 ng/mL (26 pmol/L) and <30 ng/mL (63 pmol/L) if inflammation presents. Nearly all retinol, the circulating form of vitamin A in serum, is bound to RBP, so RBP concentrations in blood can be used as an indicator of vitamin A status (12). However, different RBP cutoffs such as 14.5 μg/mL (0.69 μmol/L), 14.7 μg/mL (0.70 μmol/L), and 17.4 μg/mL (0.83 μmol/L) for a diagnostic of VAD have been proposed (13–16), because the molar ratio between retinol and RBP depends on many factors. For the purposes of this paper, we consider a serum RBP concentration lower than 14.7 μg/mL (0.70 μmol/L) as vitamin A deficient. This cutoff corresponds to the diagnostic standard for VAD at 0.70 μmol/L of serum retinol concentration, assuming a 1:1 retinol to RBP ratio in circulation (17), which has also been demonstrated in previous research (15, 18). A challenge to diagnosing ID and VAD, however, is that RBP and ferritin are both acute phase proteins. This means that RBP concentrations in blood can be temporarily reduced and ferritin concentrations can be temporarily increased by acute infection and inflammation (19–21). Therefore, readings taken during acute infection or inflammation periods can result in a false deficiency/sufficiency diagnosis. As such, C-reactive protein (CRP) or similar markers of inflammation should also be measured, to correctly interpret iron and vitamin A status. For people with moderate infection, serum CRP concentration is usually higher than 5.0 μg/mL (200 nmol/L), while for healthy people, serum CRP concentration is usually lower than 3.0 μg/mL (120 nmol/L) (22).
In this paper we introduce a rapid, portable diagnostic test for ID and VAD and demonstrate its performance compared with standard reference measurements in human samples. Incorporation of sandwich-type ferritin and CRP assays with a competitive RBP assay onto a single multiplexed test allowed us to not only detect ferritin concentrations as low as 15 ng/mL (32 pmol/L), but also measure RBP concentrations, which are over times higher than ferritin concentrations, at ∼70 μg/mL (3.3 μmol/L). We show that the test requires around 15 min to accurately quantify ferritin, RBP, and CRP concentrations in serum samples through evaluation of the fluorescent signal intensity on each test line. The device yielded sensitivities of 88%, 100%, and 80% and specificities of 97%, 100%, and 97% for ID, VAD, and inflammation status. We also demonstrate how multicolor fluorescence allows us to ensure that cross-binding does not significantly affect the results. Like other smartphone-based diagnostic devices, functions such as tracking users’ diagnostic records and communicating the result via email are inherited (23–29). Common variabilities observed in other devices, caused by such things as auto-color correction performed by the camera (30), have been avoided with our raw data subtraction algorithm.
Results
Multiplexed Micronutrient Deficiency Test-Strip Architecture.
Our platform consists of a reusable stand-alone reader, which we call the TIDBIT and expand on later, and a disposable custom multiplexed fluorescence test strip that measures ferritin, RBP, and CRP concentrations. The multiplex nutrition assay was optimized in two ways. First, because serum RBP concentrations (>20 μg/mL or 0.95 μmol/L in a healthy group) can be ∼10 times higher than serum CRP concentrations (<3 μg/mL or 0.12 μmol/L for healthy people) and times higher than serum ferritin concentrations (<15 ng/mL or 32 pmol/L for ID), we combined a sandwich ferritin assay, a competitive RBP assay, and a sandwich CRP assay on one single test strip, allowing us to cover the entire physical range of the three biomarkers with one assay. Second, we labeled R-phycoerythrin (RPE), fluorescein (FITC), and phycoerythrin/Cyaine5 (PE/Cy5) on detection antibodies for ferritin, RBP, and CRP fluorescence assays, respectively. The combination of RPE, FITC, and PE/Cy5 is suitable for a three-color fluorescence test because they have similar excitation wavelengths (∼488 nm) but different emission spectra, which peak at 564 nm (RPE), 532 nm (FITC), and 649 nm (PE/Cy5), leading to distinguishable colors: orange, green, and red, respectively. As we demonstrate later, the major benefits of using the fluorescence tags with different colors for different markers are that potential cross-binding between antibodies can be determined by observing whether a fluorescence color appears in the wrong detection area. Also using a higher quantum-yield fluorescence tag on the marker with lower concentration may balance the brightness of each test line and reduce the dynamic range required.
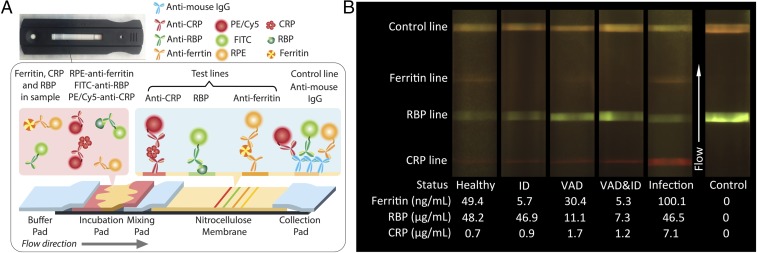
Fig. 1A shows a test-strip schematic. The test strip consists of a buffer pad that accepts running buffer; an incubation pad that incubates the sample with labeled antibodies; a mixing pad; a nitrocellulose membrane with immobilized anti-CRP, RBP, antiferritin, and secondary antibodies sequentially in the direction of flow; and another cellulose fiber pad to collect the waste sample at the end. To simplify operation we incorporated an incubation pad in our test strip, as a substitute for the sample and conjugation pads traditionally used in lateral flow test strips. The incubation pad is preloaded with labeled antibodies, allowing preincubation of sample and labeled antibodies immediately as soon as the sample is added.
Fig. 1.
Diagnostic test schematic and fluorescence image of sample cases. (A) Scheme of the multiplexed test strip. The sandwich lateral flow assay used for ferritin and CRP results in higher fluorescence signal intensity with increasing concentration, while higher RBP concentrations result in lower signal intensity due to the use of a competitive assay. (B) Sample results from different humans with known ferritin, RBP, and CRP concentrations in serum, showing the colorimetric change of detection lines in the range of interest. The infection sample shows an elevated CRP concentration. As a result, even though the ferritin and RBP concentration remained in normal range, no valid diagnostic conclusion can be made with this test.
To perform a test, first 15 μL of human serum was added on the incubation pad and the test strip was left in a light-free environment for 3 min. Then 60 μL running buffer was added to the buffer pad to initiate the flow. The flow front first reaches the CRP test line, then the RBP test line, and then the ferritin test line on the nitrocellulose membrane. All conjugation antibodies are mouse monoclonal antibodies, so goat antimouse secondary antibodies are dispensed as a control line for all three markers. After a test is performed, fluorescence tags are bound on the test lines and the intensities of fluorescence light are related to the concentration of the marker in the sample. Those conjugation antibodies that are not captured by the test lines are captured by the secondary antibodies in the control line. Therefore, the color on the control line is a mix of green, orange, and red emitted light. The control line demonstrates that the test worked properly and helps locate the position of test lines in the image because the distance between lines on the image is fixed.
The TIDBIT reader automatically images the test after 15 min (Fig. S1). In Fig. 1B, we present a set of typical fluorescence images acquired by the reader from human serum samples with previously characterized concentrations of CRP, ferritin, and RBP by ELISA. The health status of the participants was determined based on the ELISA measurements of ferritin, RBP, and CRP concentrations and is listed underneath each of the cases shown in Fig. 1B. All of the electronic components and optical filters are commercially available, and the TIDBIT reader can cost as little as $95 to manufacture, and each multiplex test for three biomarkers costs around $1.50.
TIDBIT Reader and Fluorescence Imaging System.
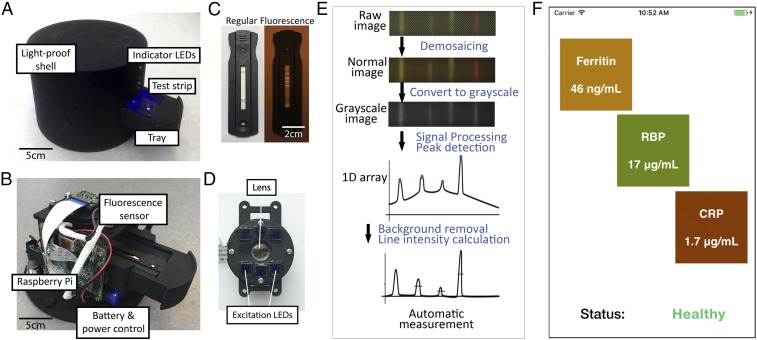
Fig. 2 A and B shows the design of the TIDBIT reader. The reader links to a standard laptop or our previously developed NutriPhone (26) technology to interpret the results and display them to the user. In the reader, a tray is built to accept test-strip cartridges with a variety of shapes. As shown in Fig. 2C, fluorescence signals appear on the test strip only during the fluorescence imaging mode. Fig. 2D shows the design of the fluorescence detection system. The sensor was developed using a Raspberry Pi camera module. It excites the fluorescence signal on the test strip, using six blue LEDs covered by band-pass optical filters with a center wavelength at 458 nm. A focusing lens with f = 25 mm is aligned to the optical path of the camera. A 535-nm long-pass optical filter covers the camera to eliminate excitation light (Fig. S2). The detection area on the test strip is aligned to the optical path of the sensor to maximize image quality.
Fig. 2.
Multicolored VAD and ID diagnostic platform. (A and B) TIDBIT reader overview showing components and internal structures of the device. (C) Pictures of test strips. (C, Left to Right) In normal indoor ambient light (Left) and fluorescence signal of the test strip (Right). (D) Assembled integrated fluorescence sensor. The LED lights were filtered by a band-pass optical filter to reduce background noise in the fluorescence images. (E) Image-processing algorithm used by the system. (F) Screenshot of the smartphone at the result page.
To avoid variability introduced by the camera autocorrection, we developed image-processing software that measures biomarker concentrations with unprocessed raw data directly from the complementary metal–oxide–semiconductor (CMOS) sensor. Fig. 2E shows the algorithm whereby the captured image is processed and the results are quantified. Briefly, binary data directly extracted from the camera are transformed to a raw image, followed by cropping dark edges so that only the test strip in the image remains. Then the image is converted to grayscale and integration of grayscale value in the direction vertical to the flow is performed, to reduce the 2D image to a 1D array. Then locations of test and control lines are determined. At intervals between each of these locations, a polynomial fitting of points is performed to find the brightness of background. The background is then subtracted from the original brightness profile to find the true brightness of the control and test lines, which represents the intensity of the fluorescence signal on the test strip. The average brightness value of each test line is then stored. Further details of the image-processing algorithm are in SI Materials and Methods and Fig. S3. Finally, the result is displayed as shown in Fig. 2F.
If concentration of a given marker falls within our physiologically relevant dynamic range, the TIDBIT reader provides quantitative analysis for all three biomarkers. Otherwise it tells whether the concentration of biomarkers is greater than the upper bound, or less than the lower bound of the test range. Details on the physiological range of all three biomarkers and the way we present the results are included in SI Materials and Methods and Fig. S10). The TIDBIT reader has a 16-GB SD card as storage. Results are stored in both the TIDBIT reader and the mobile device after each test, and all previous results can be read at any time.
Ferritin, RBP, and CRP Assay Quantification.
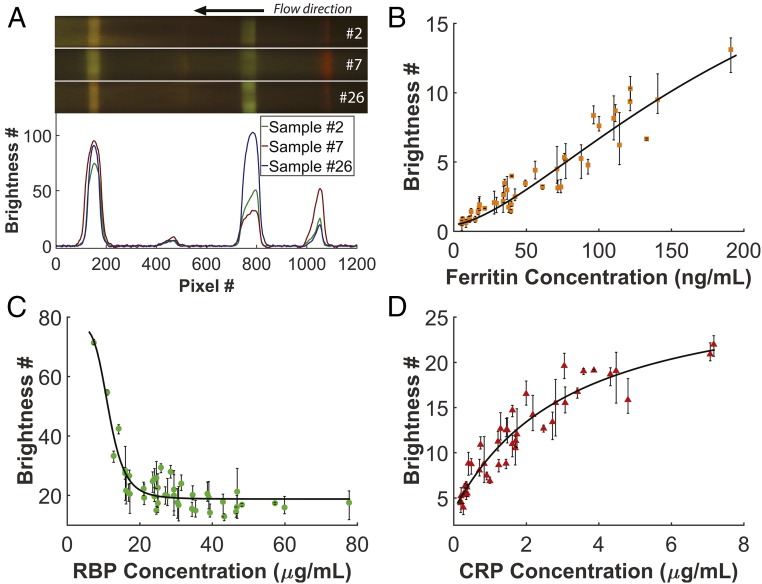
Forty-three human whole blood samples from different participants were used to quantify the TIDBIT assay. The blood samples were purchased from a commercial source (Research Blood Components, LLC) and were all from US adult donors with no appearance of infectious disease. Concentrations of ferritin, CRP, and RBP in the samples were characterized with commercial ELISA kits (Abcam, Inc.). No data were excluded. Four batches of test strips were manufactured and randomly selected for each test. The test strips were stored in a light-free environment at room temperature until used. No significant batch-to-batch variability between test strips was observed, and storage up to 6 wk had no noticeable effect on the test result, as shown in Figs. S4 and S5. Human serum samples were separated with a portable centrifuge from whole blood and then used as direct input into the test. Fig. 3A show the colorimetric variation of test lines from three different human serum samples with known ferritin, RBP, and CRP concentration and their brightness profile acquired by the imaging-processing algorithm. The images of strips were rescaled to 40% along the direction vertical to the flow.
Fig. 3.
Calibration of the biomarkers. (A) Colorimetric variation of the multiplex test strip from three different human serum samples: #2 (with ferritin = 34 ng/mL or 75 pmol/L, RBP = 16.0 μg/mL or 0.76 μmol/L and CRP = 0.37 μg/mL or 15 nmol/L), #7 (with ferritin = 42 ng/mL or 93 pmol/L, RBP = 31.5 μg/mL or 1.48 μmol/L, and CRP = 3.41 μg/mL or 136 nmol/L), and #26 (with ferritin = 40 ng/mL or 89 pmol/L, RBP = 11.0 μg/mL or 0.52 μmol/L, and CRP = 0.27 μg/mL or 11 nmol/L). (B–D) Data points showing the average intensity of the fluorescence signal for each marker at different concentration and calibration of each marker: . Error bars show the range of values obtained from three test strips with the same sample. Values of parameters can be found in Table S1.
Brightness values of test lines were then correlated to the readout of commercial ELISA kits, as shown in Fig. 3 B–D. For each of the 43 human samples, three test strips were used. The brightness values are averaged for the three test strips and range of the brightness values is shown as error bars. According to the ELISA results, 9 of 43 (20.9%) participants were ID, 4 of 43 (9.3%) participants were VAD, and 10 of 43 (23.2%) participants were subject to minimal or moderate inflammation. We then fitted four-parameter logistic curves on each marker such that , and the calibration functions were stored to predict concentration of each marker in the microcontroller software. Four-parameter curve-fitting results show () for ferritin, () for RBP, and () for CRP. The system shows high accuracy in predicting biomarker concentration based on the fitting curve. Detailed information about calibration functions can be found in Table S1. The fitting curve for ferritin shows good linearity within the whole physiological range (15∼200 ng/mL or 32∼421 pmol/L). The fitting curve for CRP indicates a moderate saturation effect at higher concentration (>3 μg/mL or 120 nmol/L); however, no hook effect was observed in this study. For the RBP assay we optimized it to maximize its capability to distinguish VAD (RBP <14.7 μg/mL or <0.70 μmol/L) and thus compromised on its performance in quantifying RBP concentrations higher than 25 μg/mL (1.19 μmol/L), which still falls in the healthy range, as we expand upon in the following section.
TIDBIT System Performance Evaluation.
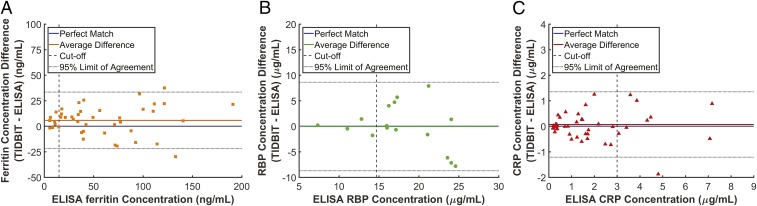
Performance of the system was evaluated by comparing the concentration of the biomarkers acquired by the TIDBIT system and concentration of biomarkers determined with laboratory standard ELISA kits, as shown in Fig. 4 A–C, in Bland–Altman plots. Linear regression (Figs. S6–S8) is also applied for each biomarker. Predicted results from the TIDBIT system are highly correlated with results from the standard method. Compared with a perfect match with regression coefficient (RC) equal to 1, the ferritin assay shows RC close to a perfect match at +1.06 ( = 0.03, P < 0.0001), with root-mean-square error (rmse) at 14.4 ng/mL (32 pmol/L) and at 0.92, while the CRP assay has RC at +1.03 ( = 0.04, P < 0.0001), with rmse at 0.65 μg/mL (26 nmol/L) and at 0.88. For the RBP test, because it is a competitive assay and we optimized the RBP assay to maximize its accuracy around the diagnostic threshold (14.7 μg/mL or 0.70 μmol/L), the test line intensity remains low at higher RBP concentration (>25 μg/mL or 1.19 μmol/L) as expected. For samples with actual RBP concentration less than 25 μg/mL (1.19 μmol/L), the RBP assay has RC at +0.97 ( = 0.05, P < 0.0001), with rmse at 4.34 μg/mL (0.21 μmol/L) and at 0.56. Samples with RBP values above the quantitative range are excluded from Fig. 4B. The TIDBIT system yielded a sensitivity and specificity at 88% (95% CI 47.3–99.6) and 97% (95% CI 85.0–99.9) for ferritin, 100% (95% CI 39.7–100.0) and 100% (95% CI 90.9–100.0) for RBP, and 80% (95% CI 55.5–99.7) and 97% (95% CI 84.2–99.9) for CRP. Moreover, to maximize the overall diagnostic accuracy of the system, the cutoff for ID, VAD, and inflammation can be set to ferritin concentration less than 27 ng/mL (60 pmol/L), RBP concentration less than 14.7 μg/mL (0.70 μmol/L), and CRP concentration greater than 2.8 μg/mL (112 nmol/L). Under these conditions the TIDBIT system then yielded a 100% (95% CI 59.0–100.0) sensitivity and 95% (95% CI 81.3–99.3) specificity for ferritin, 100% (95% CI 39.7–100.0) sensitivity and 100% (95% CI 90.9–100.0) specificity for RBP, and a 100% (95% CI 66.3–100.0) sensitivity and 94% (95% CI 80.3–99.2) specificity for CRP. More information about TIDBIT’s performance at other cutoffs can be found in receiver operating characteristics (ROC) curves in Figs. S6–S8.
Fig. 4.
Comparison between ELISA- and TIDBIT-characterized concentrations of biomarkers. (A) Bland–Altman plot for the ferritin test. The orange line indicates a bias at +5.7 ng/mL (+12.7 pmol/L). Dashed line indicates cutoff at 15 ng/mL (32 pmol/L). (B) Bland–Altman plot for the RBP test. Since the assay gives only a quantitative result for RBP <25 μg/mL (1.19 μmol/L), only sample RBP <25 μg/mL (1.19 μmol/L) was compared. Green line indicates a bias at −0.05 μg/mL (−2.3 nmol/L). Dashed line indicates cutoff at 14.7 μg/mL (0.70 μmol/L). (C) Bland–Altman plot for the CRP test. Red line indicates a bias at +0.06 μg/mL (+2.4 nmol/L). Dashed line indicates cutoff at 3 μg/mL (120 nmol/L).
Cross-Binding and Limit of Detection Quantification.
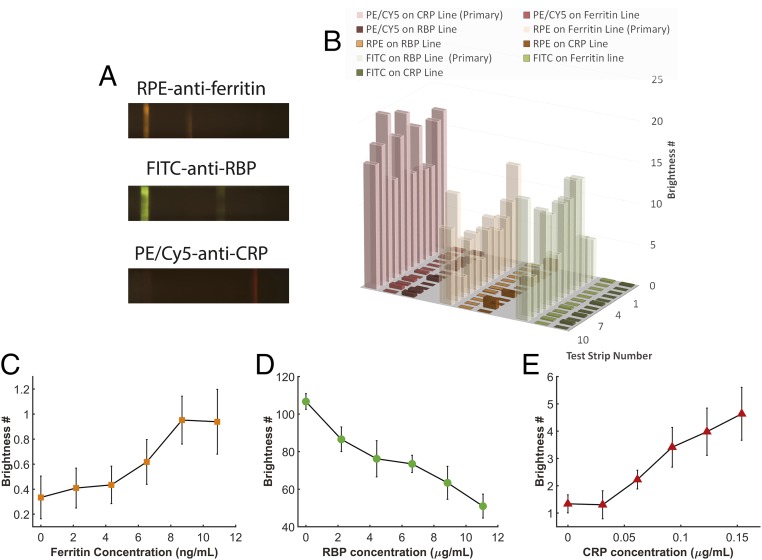
Cross-binding is a factor that can cause potential error in multiplexed lateral flow assays (30). With our multicolored fluorescent test strips, it can be easily accounted for since the incorrect fluorescence signal can be detected at the improper detection site. To demonstrate the level of cross-binding in the TIDBIT system we ran human serum tests with only one type of antibody conjugation loaded on the incubation pad. The result is shown in Fig. 5A. It shows that test lines capture only their target biomarkers, proving that nonspecific cross-binding between antibodies and markers is limited. Furthermore, to demonstrate that cross-binding has only a small effect on readout, we tested levels of cross-binding in 12 human serum samples, as shown in Fig. 5B. For each sample, only one type of antibody conjugation was loaded, and the level of cross-binding was evaluated as the ratio of the brightness value on the incorrect test lines to the brightness value of the correct test line. Error bars indicate the SD of the cross-binding level. As is shown, cross-binding between all biomarker/test line pairs was limited to less than 2% (Fig. S9).
Fig. 5.
Cross-binding test and limit of detection quantification. (A) Fluorescence image series for an assay with only one type of conjugation antibodies preloaded. (B) Evaluation of cross-binding level in 12 human serum samples. No significant cross-binding between biomarkers was found. (C–E) Limit of detection for each marker in multiplex tests. Eight tests were performed at each data point and error bars show the SD at each data point. The assay has a limit of detection lower than the diagnostics cutoff for all three biomarkers.
We also evaluated the limit of detection for each biomarker, as shown in Fig. 5 C–E. Since serum samples with very low levels of ferritin, RBP, and CRP are hard to obtain, we used resuspended standard dried serum (Siemens, Inc.) to perform the test. For each data point, eight test strips were used and the error bar shows SD of the result. The nonzero readout for the ferritin and CRP test line intensity at zero concentration indicates there was some nonspecific binding on the corresponding test lines. Based on these results we determined that the TIDBIT system has limits of detection lower than 10.9 ng/mL (24 pmol/L), 2.2 μg/mL (0.10 μmol/L), and 0.092 μg/mL (3.7 nmol/L) for ferritin, RBP, and CRP. The limit of detection for all of the biomarkers was lower than the diagnostic threshold for both adults and children. For children, an alternative form of the assay could also be developed to optimize sensitivity in their relevant range.
Discussion
Rapid immunoassay tests for multiple targets are challenging. Current rapid diagnostic tests usually label multiple types of conjugation antibodies with the same optical tags (latex beads or colloidal gold nanoparticles), and thus cross-binding can be difficult to distinguish. The adoption of a three-color fluorescent assay in our device offers a direct advantage in that cross-binding can be relatively easily spotted by observing the incorrect fluorescence signal at a given test site.
In this paper we demonstrated a rapid point-of-care test for ID and VAD by quantitatively measuring ferritin, RBP, and CRP concentrations. The test is enabled by our TIDBIT system and provides results in around 15 min. The system could have significant impact in areas of the world with high instances of micronutrient deficiencies, by providing a rapid, easy-to-operate tool for population-level micronutrient status surveys in situations where both ID and VAD need to be diagnosed (1, 17, 31). If implemented with on-strip blood separation, the system’s high sensitivity and specificity will allow it to be further applied to individual-level assessment at point of care.
Materials and Methods
Antibody-fluorescence signal conjugation was prepared using Lightening-Link Conjugation Kits (Innova Bioscience Ltd.). Test strips are based on HF180 cards (EMD Millipore). Serum samples were separated from whole blood with a minicentrifuge (ChemGlass) at 2,000 × g for 10 min before tests were performed. All human whole blood samples were obtained from a commercial provider (Research Blood Components, LLC), and thus no consent procedures are required for this project. Further details on Materials and Methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
D.E. and S.M. acknowledge primary funding support for this effort from Nutrition International, formerly the Micronutrient Initiative. Some elements of the reader device were developed under National Science Foundation Award 1343058 as well as National Institutes of Health Award 1R01EB021331.
Footnotes
Conflict of interest statement: The authors declare the following competing financial interests: D.E., S.M., and D.O. have an equity interest in VitaScan Inc., which is commercializing similar micronutrient diagnostic technology that is described herein.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711464114/-/DCSupplemental.
References
- 1.Zimmermann MB, Hurrell RF. Nutritional iron deficiency. Lancet. 2007;370:511–520. doi: 10.1016/S0140-6736(07)61235-5. [DOI] [PubMed] [Google Scholar]
- 2.Stevens GA, et al. Trends and mortality effects of vitamin A deficiency in children in 138 low-income and middle-income countries between 1991 and 2013: A pooled analysis of population-based surveys. Lancet Glob Health. 2015;3:e528–e536. doi: 10.1016/S2214-109X(15)00039-X. [DOI] [PubMed] [Google Scholar]
- 3.Underwood BA, Arthur P. The contribution of vitamin A to public health. FASEB J. 1996;10:1040–1048. [PubMed] [Google Scholar]
- 4.Semba R, Bloem M. The anemia of vitamin a deficiency: Epidemiology and pathogenesis. Eur J Clin Nutr. 2002;56:271–281. doi: 10.1038/sj.ejcn.1601320. [DOI] [PubMed] [Google Scholar]
- 5.Jang JT, Green JB, Beard JL, Green MH. Kinetic analysis shows that iron deficiency decreases liver vitamin A mobilization in rats. J Nutr. 2000;130:1291–1296. doi: 10.1093/jn/130.5.1291. [DOI] [PubMed] [Google Scholar]
- 6.Rosales FJ, et al. Iron deficiency in young rats alters the distribution of vitamin A between plasma and liver and between hepatic retinol and retinyl esters. J Nutr. 1999;129:1223–1228. doi: 10.1093/jn/129.6.1223. [DOI] [PubMed] [Google Scholar]
- 7.Sommer A, et al. Impact of vitamin A supplementation on childhood mortality: A randomised controlled community trial. Lancet. 1986;327:1169–1173. doi: 10.1016/s0140-6736(86)91157-8. [DOI] [PubMed] [Google Scholar]
- 8.Alleyne M, Horne MK, Miller JL. Individualized treatment for iron-deficiency anemia in adults. Am J Med. 2008;121:943–948. doi: 10.1016/j.amjmed.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sirisinha S, Edelman R, Suskind R, Charupatana C, Olson R. Complement and C3-proactivator levels in children with protein-calorie malnutrition and effect of dietary treatment. Lancet. 1973;301:1016–1020. doi: 10.1016/s0140-6736(73)90663-6. [DOI] [PubMed] [Google Scholar]
- 10.Suharno D, et al. Supplementation with vitamin A and iron for nutritional anaemia in pregnant women in West Java, Indonesia. Lancet. 1993;342:1325–1328. doi: 10.1016/0140-6736(93)92246-p. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization . Iron Deficiency Anaemia: Assessment, Prevention and Control: A Guide for Programme Managers. World Health Organization; Geneva: 2001. [Google Scholar]
- 12.Gamble MV, et al. Retinol binding protein as a surrogate measure for serum retinol: Studies in vitamin A–deficient children from the republic of the Marshall Islands. Am J Clin Nutr. 2001;73:594–601. doi: 10.1093/ajcn/73.3.594. [DOI] [PubMed] [Google Scholar]
- 13.Engle-Stone R, et al. Plasma retinol-binding protein predicts plasma retinol concentration in both infected and uninfected Cameroonian women and children. J Nutr. 2011;141:2233–2241. doi: 10.3945/jn.111.145805. [DOI] [PubMed] [Google Scholar]
- 14.Gorstein JL, Dary O, Shell-Duncan B, Quick T, Wasanwisut E. Feasibility of using retinol-binding protein from capillary blood specimens to estimate serum retinol concentrations and the prevalence of vitamin A deficiency in low-resource settings. Public Health Nutr. 2008;11:513–520. doi: 10.1017/S1368980007000821. [DOI] [PubMed] [Google Scholar]
- 15.Baeten JM, et al. Use of serum retinol-binding protein for prediction of vitamin a deficiency: Effects of HIV-1 infection, protein malnutrition, and the acute phase response. Am J Clin Nutr. 2004;79:218–225. doi: 10.1093/ajcn/79.2.218. [DOI] [PubMed] [Google Scholar]
- 16.Semba R, Yuniar Y, Gamble M, Natadisastra G. Assessment of vitamin A status of preschool children in Indonesia using plasma retinol-binding protein. J Trop Pediatr. 2002;48:84–87. doi: 10.1093/tropej/48.2.84. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization . Global Prevalence of Vitamin A Deficiency in Populations at Risk 1995–2005: WHO Global Database on Vitamin A Deficiency. World Health Organization; Geneva: 2009. [Google Scholar]
- 18.Erhardt JG, Estes JE, Pfeiffer CM, Biesalski HK, Craft NE. Combined measurement of ferritin, soluble transferrin receptor, retinol binding protein, and C-reactive protein by an inexpensive, sensitive, and simple sandwich enzyme-linked immunosorbent assay technique. J Nutr. 2004;134:3127–3132. doi: 10.1093/jn/134.11.3127. [DOI] [PubMed] [Google Scholar]
- 19.Christian P, Schulze K, Stoltzfus RJ, West KP. Hyporetinolemia, illness symptoms, and acute phase protein response in pregnant women with and without night blindness. Am J Clin Nutr. 1998;67:1237–1243. doi: 10.1093/ajcn/67.6.1237. [DOI] [PubMed] [Google Scholar]
- 20.Tomkins A. Assessing micronutrient status in the presence of inflammation. J Nutr. 2003;133:1649S–1655S. doi: 10.1093/jn/133.5.1649S. [DOI] [PubMed] [Google Scholar]
- 21.Thurnham D, McCabe G, Northrop-Clewes C, Nestel P. Effects of subclinical infection on plasma retinol concentrations and assessment of prevalence of vitamin A deficiency: Meta-analysis. Lancet. 2003;362:2052–2058. doi: 10.1016/s0140-6736(03)15099-4. [DOI] [PubMed] [Google Scholar]
- 22.Pearson TA, et al. Markers of inflammation and cardiovascular disease. Circulation. 2003;107:499–511. doi: 10.1161/01.cir.0000052939.59093.45. [DOI] [PubMed] [Google Scholar]
- 23.Zheng G, Lee SA, Antebi Y, Elowitz MB, Yang C. The ePetri dish, an on-chip cell imaging platform based on subpixel perspective sweeping microscopy (SPSM) Proc Natl Acad Sci USA. 2011;108:16889–16894. doi: 10.1073/pnas.1110681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L, et al. Solar thermal polymerase chain reaction for smartphone-assisted molecular diagnostics. Sci Rep. 2014;4:4137. doi: 10.1038/srep04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, Mehta S, Erickson D. Two-color lateral flow assay for multiplex detection of causative agents behind acute febrile illnesses. Anal Chem. 2016;88:8359–8363. doi: 10.1021/acs.analchem.6b01828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, et al. Nutriphone: A mobile platform for low-cost point-of-care quantification of vitamin B12 concentrations. Sci Rep. 2016;6:28237. doi: 10.1038/srep28237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemiroski A, et al. Universal mobile electrochemical detector designed for use in resource-limited applications. Proc Natl Acad Sci USA. 2014;111:11984–11989. doi: 10.1073/pnas.1405679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutledge RB, Skandali N, Dayan P, Dolan RJ. A computational and neural model of momentary subjective well-being. Proc Natl Acad Sci USA. 2014;111:12252–12257. doi: 10.1073/pnas.1407535111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Im H, et al. Digital diffraction analysis enables low-cost molecular diagnostics on a smartphone. Proc Natl Acad Sci USA. 2015;112:5613–5618. doi: 10.1073/pnas.1501815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roda A, et al. Smartphone-based biosensors: A critical review and perspectives. TrAC Trends Anal Chem. 2016;79:317–325. [Google Scholar]
- 31.Allen LH. Iron supplements: Scientific issues concerning efficacy and implications for research and programs. J Nutr. 2002;132:813S–819S. doi: 10.1093/jn/132.4.813S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.