Significance
Phosphorylation is a ubiquitous modification that has been implicated in signaling and other functions, but the atomic-level mechanisms are not completely understood. We identify a salt-bridge competition or “theft” mechanism wherein a phosphoserine, but not a phosphomimetic, breaks a pre-existing salt bridge, initiating a partial unfolding event and promoting new protein interactions. Structural elements underlying the theft occurred early in evolution and are found in 10% of homo-oligomers and 30% of hetero-oligomers. These findings identify a facile and evolutionarily accessible mechanism for reorganizing salt bridges and other electrostatic networks with only a single mutation to trigger a functional switch.
Keywords: phospho-swap, protein interaction, salt-bridge competition, Raf Kinase Inhibitory Protein, conformational change
Abstract
Phosphorylation is a major regulator of protein interactions; however, the mechanisms by which regulation occurs are not well understood. Here we identify a salt-bridge competition or “theft” mechanism that enables a phospho-triggered swap of protein partners by Raf Kinase Inhibitory Protein (RKIP). RKIP transitions from inhibiting Raf-1 to inhibiting G-protein–coupled receptor kinase 2 upon phosphorylation, thereby bridging MAP kinase and G-Protein–Coupled Receptor signaling. NMR and crystallography indicate that a phosphoserine, but not a phosphomimetic, competes for a lysine from a preexisting salt bridge, initiating a partial unfolding event and promoting new protein interactions. Structural elements underlying the theft occurred early in evolution and are found in 10% of homo-oligomers and 30% of hetero-oligomers including Bax, Troponin C, and Early Endosome Antigen 1. In contrast to a direct recognition of phosphorylated residues by binding partners, the salt-bridge theft mechanism represents a facile strategy for promoting or disrupting protein interactions using solvent-accessible residues, and it can provide additional specificity at protein interfaces through local unfolding or conformational change.
Phosphorylation is a ubiquitous posttranslational modification implicated in the regulation of innumerable processes (1). Phosphorylation often acts as a switch, controlling the formation of protein complexes that mediate function. However, beyond directly forming either favorable or unfavorable interactions at the binding interface, the possible modes of phospho-regulation are not clear (2, 3). Here, we investigate how phosphorylation of RKIP (PEBP1), a member of the phosphatidylethanolamine protein family, reorganizes a salt-bridge network to bring about a localized conformational change and an exchange of signaling partners. Bioinformatic analyses demonstrate the broader significance of this mechanism, which represents a general mechanism to regulate both homo-oligomeric and hetero-oligomeric protein interactions.
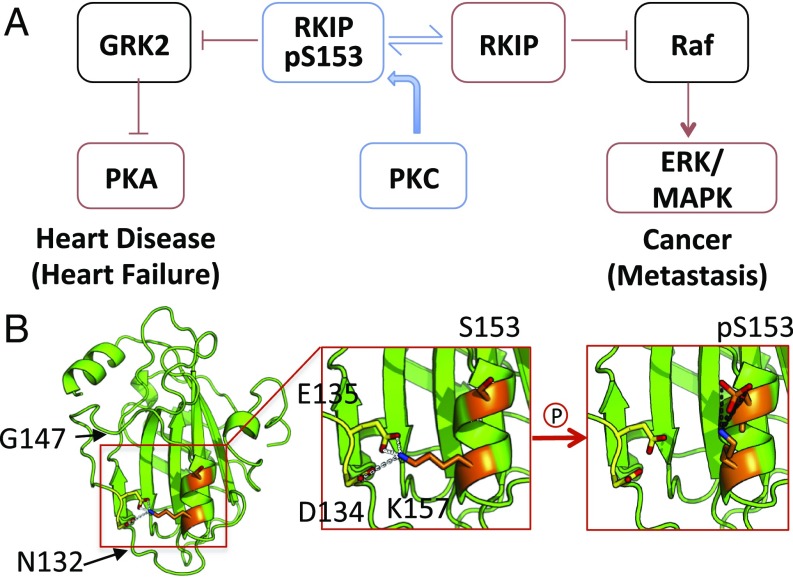
As a regulator of MAP kinase and G-Protein–Coupled Receptor (GPCR) signaling, RKIP prevents numerous pathological conditions including metastatic cancer (4–6) and heart disease (7, 8) (Fig. 1A). Well-characterized structurally by crystallography and NMR (9–11), RKIP assumes a highly conserved conformation with a pocket composed of a loop that interacts noncovalently with its C-terminal α-helix (Fig. 1B). Phosphorylation at S153 by protein kinase C (PKC) switches RKIP from binding Raf-1 to binding G-protein–coupled receptor kinase 2 (GRK2) (12–14), thus activating a new pathway (Fig. 1A).
Fig. 1.
RKIP function and structure. (A) Phosphorylation of S153 causes RKIP to inhibit GRK2 rather than Raf. RKIP acts to suppress heart disease and cancer. (B) Model of salt-bridge interactions and the proposed effects of S153 phosphorylation of RKIP. (B, Left) Key residues mapped onto the WT RKIP structure. (B, Middle) K157 interacts with D134-E135 in the WT RKIP crystal structure (yellow). (B, Right) A model of how pS153 could outcompete D134 and E135 for interaction with K157 (orange). Images were generated using PyMOL (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC).
To observe the effects of phosphorylating RKIP at S153 by NMR, we mixed the catalytic subunit of PKC with RKIP and analyzed the heteronuclear single quantum coherence (HSQC) spectra over 10 h. We also inserted a minimally perturbing P74L mutation shown previously to increase the phosphorylation rate of RKIP (10). Several lines of evidence indicate that phosphorylation, which had the proper mass shift (SI Appendix, Fig. S1), was nearly complete. These include the previously published result that only position S153 is phosphorylated by PKC (12–14) and a reduction in the S153 NMR peak height by greater than 80% (Fig. 2A and SI Appendix, Fig. S1).
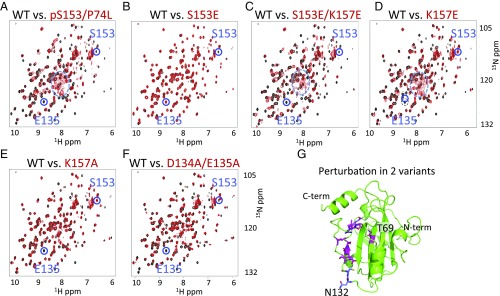
Fig. 2.
1H-15N HSQC NMR spectra. (A–F) Comparison of WT RKIP (black) to variants (red) indicates that RKIP adopts different conformational states upon S153 phosphorylation or mutation of residues involved in the salt bridge, whereas the phosphomimetic RKIPS153E is insufficient to induce the change. Peaks corresponding to S153 and E135 and the random coil region (dashed circle in A, C, and D) are highlighted. In the variants, the E135 peak is unperturbed only in S153E. Spectra were obtained at 25 °C. (G) Location of residues perturbed in the HSQC spectra of RKIPK157A (magenta) and RKIPD134A,E135A (magenta and blue).
Comparison of the NMR 1H-15N HSQC spectra of unphosphorylated and phosphorylated RKIPP74L revealed pronounced differences at more than one-third of the amide NH (NH) peaks (Fig. 2A). Changes included peak movement, line broadening, and disappearance of peaks for residues located near S153; however, these effects extended out to residues in the C-terminal helix located more than 30 Å away. These changes indicated that a subset of amide NHs experienced different chemical environments due to altered conformation(s), with some undergoing dynamical averaging between alternative conformations on the millisecond to microsecond timescale. An increase in the number of peaks in the random coil region (Fig. 2A, dashed circle) suggested a partial unfolding of residues 132–147 located in a well-folded loop region in the crystal structure. These findings indicated that the protein undergoes a large-scale perturbation upon PKC-induced phosphorylation of S153.
To probe the nature of the perturbation, we introduced mutations at or near S153. We first compared the HSQC spectrum of RKIPpS153,P74L to the phosphomimetic mutant RKIPS153E. We showed previously that the RKIPS153E variant bound Raf-1 instead of GRK2, and its HSQC spectrum was nearly identical to that of wild-type (WT) RKIP (10). Consistent with these findings, the singly charged RKIPS153E was insufficient to induce the large-scale perturbation of the HSQC spectrum observed with the authentic phosphorylation of S153 (compare Fig. 2 A and B).
To investigate why the phosphomimetic was insufficient, we examined the RKIP structure and noted that K157, located one helical turn away from S153, could be involved in the phosphorylation-induced structural changes. In the crystal structure, K157 forms a salt bridge with D134 and E135 on a nearby loop (Fig. 1B). We posited that the K157 side chain, upon phosphorylation of S153, could rotate and form a salt bridge with the double charged phosphate on pS153. Such a rotation of K157 would leave the negatively charged ends of D134 and E135 near each other without a compensating positive charge. As a result, the acidic groups should have a tendency to separate, and the loop containing the two residues would become partially disordered.
Support for the ability of pS153, but not the S153E phosphomimetic, to outcompete residues D134 and E135 for K157 comes from peptide studies showing that the salt bridge formed between a phosphoserine and a lysine located at positions i and i+4 along a helix is unusually stable (∼2 kcal⋅mol−1) relative to a standard K-E salt bridge (15). For RKIP, we propose that this enhanced stability enabled the pS153-K157 salt bridge to outcompete the D134/E135-K157 salt bridge and initiate the events leading to partial unfolding of the loop. Since typical single (+,−) salt bridges are generally weak (15), this result also provided a rationale for why some S-to-E phosphomimetics do not function as well as the phosphorylated versions.
In our “salt-bridge theft” model, the breakage of the D134/E135-K157 salt bridge is the critical event. To test the model, we disrupted the triad with an additional K157E substitution. As anticipated, this RKIPS153E,K157E variant had an HSQC spectrum matching that of RKIPpS153 (Fig. 2 C and D and SI Appendix, Fig. S2). Furthermore, few spectral differences were observed between the single RKIPK157E and the double RKIPS153E,K157E variants, indicating that the single K157E substitution was sufficient to trigger the conformational switch. We also examined less disruptive alanine substitutions on either the helix or the loop side, K157A and D134A/E135A, respectively. These substitutions caused structural changes approaching those seen in RKIPK157E, consistent with the loss of the salt bridge (Fig. 2 E and F and SI Appendix, Fig. S2). Substitution with larger tyrosine residues (D134Y/E135Y) also resulted in perturbations similar to those seen in K157E (SI Appendix, Fig. S2).
To clarify the nature of the differences between the K157 mutants substituted with charged versus uncharged residues, we analyzed an overlay of the K157E and K157A NMR spectra (SI Appendix, Fig. S2). This overlay highlights the differences observed between K157E and K157A, which are especially notable in the random coil region in the NMR spectrum of K157E and indicative of increased unfolding (Fig. 2D). This difference likely was a reflection of the more disruptive effects of three colocalized carboxylic acids in K157E versus two colocalized carboxylic acids in K157A.
Salt-bridge disruption in RKIPK157A caused a near-complete or total loss of NMR peak intensity for various residues including L68, L103, S104, V107, G108, E135, L138, R146, G147, and L184 (peaks for the intervening residues E105-Y106 and P136-N145 could not be resolved) (Fig. 2G). The perturbed residues in the RKIPD134A,E135A HSQC spectrum overlapped with those seen for RKIPK157A but also included N132 and C133 (Fig. 2G). Some peaks in the mutants were selectively lost while others had native-like intensities, suggesting that the region from L103 to G147 is partially unfolded upon disruption of the native salt-bridge triad.
The disruption of the D134/E135 loop upon the theft of K157 likely resulted from an electrostatic repulsion of the adjacent D134 and E135 residues. Studies of the pK shifts for a pair of neighboring glutamic acids by McIntosh and coworkers (16) noted a pKa increase for a glutamic acid of two units. This shift translates into an ∼3 kcal⋅mol−1 increase in proton affinity to the carboxylic acid, presumably resulting from the heightened negative potential due to the presence of the second glutamic acid. To create the heightened negative potential, additional energy is required to fold the protein with two nearby glutamic acids. There are also several other examples of single ionic locks that trigger dramatic structural reorganizations within a protein to facilitate protein activity-state transitions. For example, a similar E/DRY motif can be found in GPCRs with salt-bridge interactions between R and both E and D residues in consecutive positions (17). This salt bridge creates an ionic lock, maintaining the receptors in an inactive state. Mutagenesis of R is sufficient to transition the receptor to an active state. This motif is on the cytosolic face of the GPCR and therefore is not shielded from the influence of polar solvent or ionic interactions. Together, these studies suggest that RKIP has evolved to use the repulsion of two nearby glutamic acids to drive partial unfolding of a loop region.
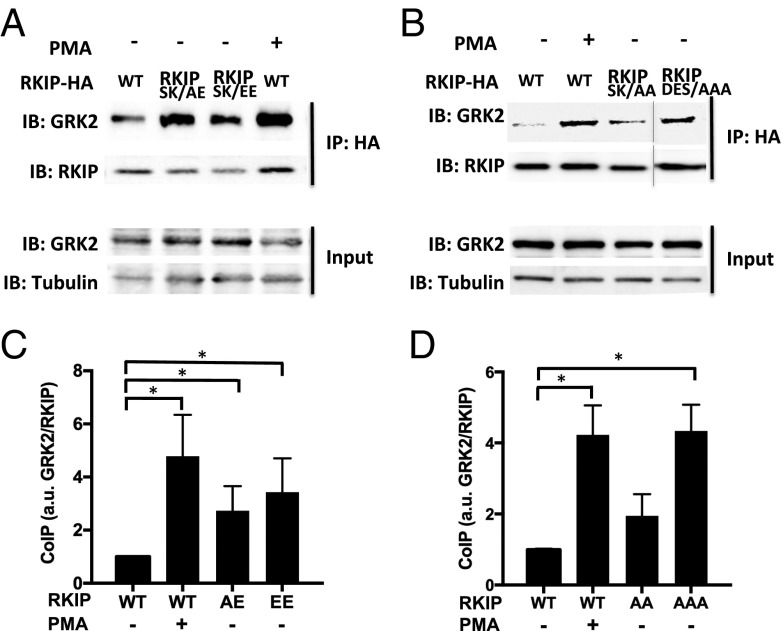
Analysis of GRK2 binding provided further evidence for the functional role of residues within the salt-bridge triad. To determine the degree to which RKIPK157E is a surrogate for RKIPpS153, we compared the ability of RKIPK157E to bind GRK2 with that of RKIPpS153 and the double variant RKIPS153E,K157E in 293T cells by coimmunoprecipitation (14). For all cell studies, we mutated S153 to either an alanine or a glutamic acid to prevent S153 phosphorylation. As previously observed (14), the RKIPS153E,K157E variant bound GRK2 instead of Raf-1 (Fig. 3). No significant difference in GRK2 binding was noted between RKIPpS153, generated by treating cells with phorbol-12-myristate-13-acetate (PMA), and the two variants RKIPS153A,K157E and RKIPS153E,K157E (Fig. 3 A and C). The triple alanine variant RKIPS153A,D134A,E135A was similarly able to bind to GRK2 (Fig. 3 B and D). By contrast, GRK2 binding to the double-alanine variant RKIPS153A,K157A was not statistically significant, possibly reflecting the more limited structural perturbation noted above. These findings provide additional support for both the disruption of the salt-bridge triad and the partial unfolding of the associated region as critical events leading to the GRK2-binding–competent state.
Fig. 3.
In vivo interactions between GRK2 and salt-bridge mutants of RKIP. (A and B) Cells expressing WT HA-RKIP or HA-RKIP variants were incubated with or without PMA (1 μM) for 10 min before precipitation with an anti-HA antibody and blotted for GRK2. Input represents 10% of total lysates used for immunoprecipitation assays: Representative coimmunoprecipitation assays using RKIP mutants (A) S153A/K157E (SK/AE) or S153E/K157E (SK/EE) or (B) S153A/K157A (SK/AA) or D134A/E135A/S153A (DES/AAA) are shown. (C and D) Plots of GRK2 bound to RKIP mutants: S153A/K157E (AE); S153E/K157E (EE); S153A/K157A (AA); D134A/E135A/S153A (AAA). Average of blot densities for GRK2 normalized to RKIP using (C) four or (D) three independent experiments including A is shown. Error bars indicate SEM. *P < 0.05 by a one-tailed Student’s t test.
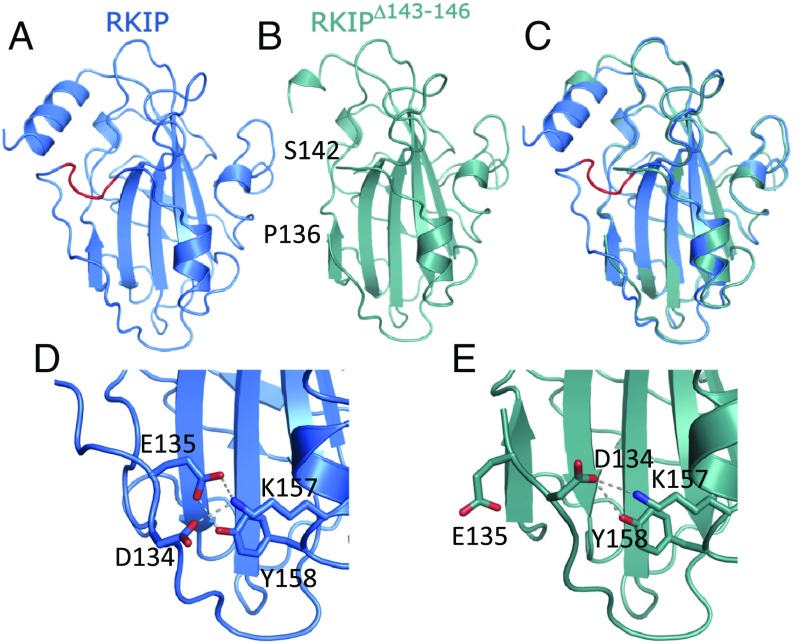
The crystal structure of an RKIP variant lacking residues 143–146 further validated our model. R146 is highly conserved, and this region is altered by the P74L mutation that increases S153 phosphorylation (10). The initial motivation for designing this deletion mutant was to mimic the conformational changes induced by S153 phosphorylation to further dissect the molecular mechanism of the Raf1-to-GRK2 switch of RKIP. The RKIPΔ143–146 deletion variant does indeed mimic the phosphorylated RKIPpS153 state (14). RKIPΔ143–146 binds GRK2 to a comparable level as RKIPpS153 and similarly to RKIPpS153, binds poorly to Raf1 (14). In addition, the deletion variant does not require S153 phosphorylation for these effects (14), further demonstrating that Δ143–146 largely simulates the structural change in RKIPpS153 induced by phosphorylation. We solved the crystal structure of RKIPΔ143–146 at a moderate resolution (2.7 Å) (Fig. 4 B and C and SI Appendix, Table S1). Notably, E135 now points away from K157 and no longer participates in the salt bridge (Fig. 4 D and E), leaving a salt bridge only between D134 and K157. Evidently, this interaction is too weak to maintain the original Raf-binding structure in solution (14). In addition, regions near R146 seem to be more flexible compared with other regions of the crystal structure indicated by larger B-factors relative to the average B-factor of the entire structure, suggesting that this part of the protein is perturbed. The RKIPΔ143–146 variant therefore serves as a useful tool to underscore the importance of local perturbation and partial unfolding of RKIP for GRK2 binding.
Fig. 4.
Crystal structures of WT RKIP (6ENS) and the RKIP deletion mutant (6ENT). (A) Ribbon presentation of the WT structure of RKIP. Highlighted in red are the residues deleted in the RKIPΔ143–146 variant. (B) Ribbon presentation of the RKIPΔ143–146 variant. The location of the residues flanking the deletion are noted. (C) Superposition of WT RKIP (blue) and the RKIPΔ143–146 variant (green). (D) Interactions of residue E135 with K157 and D134 in WT RKIP. Dashed lines indicate salt bridges and hydrogen bonds. (E) Interactions of residue D134 with K157 and Y158 in the RKIPΔ143–146 variant.
Since oligomerization could alter the interpretation of the NMR data, we examined whether RKIP forms a dimer by size-exclusion chromatography, multi-angle light scattering, and SDS/PAGE (SI Appendix, Figs. S3 and S4). Reduced RKIPK157E and RKIPS153E,K157E were nearly identical in size and molecular weight to monomeric WT RKIP (SI Appendix, Fig. S4). Furthermore, RKIPΔ143–146 crystallized as a monomer. Whereas RKIP may form oligomers in cells (14), phosphorylation at S153 transitions RKIP between monomeric states under reducing conditions.
The salt-bridge theft mechanism in RKIP involves breaking a salt bridge, which subsequently enables a switch in protein partners. We reasoned that a similar disruption of a salt bridge formed across a binding interface could be widespread in complexes controlled by phosphorylation. Consequently, we performed a bioinformatic search on ∼5,000 hetero-oligomers taken from a curated data set (18) to look for key features involving a phosphorylatable S/T pSite, a K or R Switch(+) and at least one D or E salt-bridge Partner(−) (Fig. 1B). The salt-bridge theft motif is defined as follows: (i) the pSite and Switch(+) are on the same chain at least two residues apart, while the Partner(−) is on another chain; (ii) the Switch(+) and Partner(−) are in contact with side chains within 3 Å (19); (iii) the Cα atoms of the pSite and the Switch(+) are within 8 Å (20, 21); (iv) the pSite is on the surface with a relative solvent accessible area above 25% (22, 23); (v) the pSite is a known or predicted phosphorylation site (24, 25); and (vi) the pSite is not at the binding interface, having no heavy atoms within 3 Å from any interfacial residue (19).
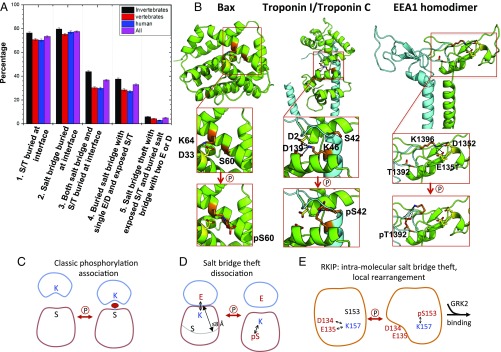
This analysis identified 33% (1,602/4,857) of total hetero-oligomers as having the necessary criteria for the salt-bridge theft mechanism with either known (5%) or predicted (28%) phosphorylation sites (Fig. 5A and SI Appendix, Tables S2 and S3). A similar analysis of homo-oligomers (18) revealed a 10% occurrence of the salt-bridge theft motif (2,048/20,685). From this set, along with additional criteria found in RKIP (namely, i and i+4 are in a helix and have two D/E in contact with K/R), we identified three candidates that had been previously studied by mutation or phosphorylation: Bax (1F16.pdb), Troponin I and C (1J1D, chains A and C), and Early Endosome Antigen 1 (EEA1) (1JOC, chains A and B). In RKIP, Bax, and Troponin I, the S/T-K pair was located at positions i and i+4 on an α-helix, whereas in EEA1, the i, i+4 pair was present in a β-turn.
Fig. 5.
Salt-bridge theft mechanism observed in RKIP and other proteins. (A) Bar plot of the frequencies of different interfacial properties among hetero-oligomers on a per-complex basis. Errors are the SD calculated assuming a binomial distribution. See SI Appendix, Table S2, for details. (B) Phosphorylation of Bax at S60 by PKA attracts K64, destabilizing the α-helix and activating translocation and cytochrome c release. Residues involved in the salt bridge (K64, D33) are indicated. Phosphorylation at S42 by PKC prevents the interaction of Troponin I (blue) with Troponin C (green) and inhibits Troponin I activity. Residues involved in the salt bridge are indicated (K46 on chain C; D2 and D139 on chain A). Phosphorylation of EEA1 at T1392 on chain B by the kinase p38 attracts K1396 on chain B, thus freeing D1352 on chain A to interact with phosphatidylinositol-3-phosphate within the endosomal membrane. Residues involved in the salt bridge are indicated (K1396 on chain B; D1352 and possibly E1351 on chain A). (C–E) Alternative models for phosphorylation-controlled protein association.
We postulate that the salt-bridge theft mechanism in Bax regulates local conformational changes along a single polypeptide chain that eventually leads to Bax oligomerization (26). Bax, an apoptotic protein that triggers release of cytochrome c from mitochondria (27), forms a salt bridge between K64 and D33 on two adjacent helices in its inactive “closed” conformation (Fig. 5B) (28). We expect that phosphorylation of S60 should appropriate K64, triggering the separation of the two helices to generate the active “open” conformation. This event facilitates oligomerization leading to cytochrome c release. In support of the theft mechanism, the loss of the interhelical salt bridge upon either a K64D or D33A mutation triggers cytochrome c release whereas the S60A mutation, which prevents phosphorylation, inhibits cytochrome c release (29). As in RKIP, a phosphomimetic substitution of the serine residue (S60D) was insufficient to fully activate Bax or trigger cytochrome c release, presumably because the singly charged residue, unlike the authentic phosphorylated serine, cannot outcompete the K64-D33 salt bridge (29).
The phosphorylation of Troponin I regulates heterodimer formation with Troponin C. Troponin C is a calcium-binding protein that interacts with Troponin I, eliciting a conformational change in Troponin I and muscle contraction (30). Phosphorylation of Troponin I at S42 on an α-helix in chain C disrupts the Troponin C/I interaction, releasing myofilament tension and decreasing sliding speed (31). K46 on Troponin I likely forms a salt bridge with D2 and D139 on Troponin C that is lost upon phosphorylation of S42 (Fig. 5B). Consistent with the theft mechanism, combining S42E and S44E substitutions decreases fiber tension and calcium sensitivity, whereas the S42A mutation enhances both parameters (32).
The phosphorylation of the homodimer EEA1 controls binding to phospholipids (33, 34), mediating endosomal trafficking by binding to phospholipid vesicles via phosphatidylinositol-3-phosphate (35). Phospholipid binding requires phosphorylation of T1392, and its mutation to alanine (T1392A) decreases this interaction (36, 37). The T1392 phosphorylation should attract K1396 (both on chain B), triggering a salt-bridge theft and freeing the salt-bridge partners (the adjacent D1352 and E1351 on chain A) to undergo local rearrangement and generate a new homodimer interface that interacts with endosomes (Fig. 5B) (33, 34).
The high frequency of the salt-bridge theft motif in hetero-oligomers suggests that it occurred early in evolution. Consistently, our motif search revealed a higher prevalence among invertebrates (38 ± 1%) relative to vertebrates (28 ± 1%) (Fig. 5A and SI Appendix, Table S2). However, RKIP (PEBP1) acquired S153 later in evolution whereas E135 and K157 are two of the most conserved residues within the PEBP family (SI Appendix, Fig. S5 and Table S4), indicating that this salt bridge antedates acquisition of the salt-bridge theft mechanism. Bax, EEA1, and Troponin I similarly acquired the salt-bridge theft motif at the vertebrate stage (SI Appendix, Fig. S5 and Tables S5–S7). Thus, the salt-bridge theft motif as a mediator of protein interactions is poised for regulation by the nascent kinome but may also be acquired later in evolution along with an expanded role for the kinome.
The salt-bridge theft mechanism that we describe here differs from the classic view of phosphorylation-controlled binding through protein domains, as exemplified by SH2 and 14–3-3 domains (38). In the latter case, the phosphorylation of a serine or threonine situated at the interface promotes binding (Fig. 5C). This mechanism involves an initially solvent-exposed serine or threonine. Possibly, the phosphorylation of residues at the interface could lead to dissociation; in this situation, however, the residues would not be as solvent-accessible. The high frequency (73%) of a hetero-oligomeric protein interface having either a serine or a threonine suggests that this is a viable mechanism for regulating oligomeric protein interactions (Fig. 5A and SI Appendix, Table S3), as supported by recent studies (3).
In contrast, our salt-bridge theft mechanism involves solvent-exposed serine or threonine residues that are not directly on the binding interface (Fig. 5 D and E). The mechanism builds on the high frequency of salt bridges (77%) at hetero-oligomeric protein interfaces (Fig. 5A and SI Appendix, Table S3). These charged residues are enriched two- to threefold at binding interfaces, as noted previously (39). By contrast, the frequency of S/T residues is the same throughout the protein whether on or near the interface or buried within the protein (SI Appendix, Table S3). If a solvent-exposed S/T is close enough to an interfacial salt bridge, then a pSer/pThr can compete for the bridge. Because only ∼30% of the protein interfaces feature the salt-bridge theft motif whereas salt bridges are present 77% of the time, the availability of S/T residues near the interface appears to be a limiting condition. When two acidic residues participate in the salt bridge, more extensive conformational changes can occur. This option is present at lower levels than the salt-bridge dyads for both hetero-oligomers (5%) and homo-oligomers (1%) (SI Appendix, Table S2).
Whereas the traditional model posits that phosphorylation modulates protein interactions by directly altering the binding interface, the salt-bridge theft mechanism has several advantages as an additional route for regulation by the kinome. Due to solvent accessibility, the theft allows for facile removal or addition of the phosphate whereas direct binding across the interface allows only for facile phosphorylation. Therefore, phosphorylation leading to interface disruption would be more likely to occur with the salt-bridge theft mechanism. In support of this hypothesis, the examples that we have highlighted here for the salt-bridge theft all involve phosphorylation-induced disruption of protein interactions. Troponin I phosphorylation dissociates a heterodimer, and EEA1 disrupts a homodimer (Fig. 5D), whereas RKIP and Bax phosphorylation disrupt interacting polypeptide chains within a single protein, enabling local unfolding that facilitates binding to GRK2 (Fig. 5E) or Bax, respectively.
Finally, disruption of the salt bridge, especially one that involved two negative charges, may induce protein remodeling through partial unfolding due to the colocalization of two negatively charged side chains. Similar reorganizations of electrostatic networks and allosteric effects resulting from disrupted charge clusters have also been observed in other systems, including PKA and VraR (40, 41). These examples as well as others (42) suggest that neither mono- nor divalent cation binding would completely recover the energy invested in the colocalization of D134 and E135 in RKIP, and, thermodynamically, the partial unfolding of the loop is favored.
In this article, we have demonstrated the presence of a phosphorylation-triggered salt-bridge competition or “theft” mechanism for regulating RKIP/GRK2 association. The motif exists in one-third of hetero-oligomers and is enriched in invertebrates. The mechanism could be an early mode of introducing phosphorylation-controlled binding, in part as it uses an existing salt bridge, an interaction known to be enhanced at protein–protein interfaces (39). The original salt bridge is left intact, in contrast to serine phosphorylation that directly participates in the salt bridge across an interface (43). Thus, the regulation using the theft mechanism is controlled through a nearby solvent-accessible residue that would be easier to substitute or phosphorylate. Our computational analysis suggests that this mechanism may be broadly operative in controlling protein oligomerization.
Materials and Methods
NMR Experiments.
All HSQC spectra were collected on a 500-MHz magnet with a Bruker AVANCE III console at 25 °C with a typical protein concentration of 0.5 mM. Spectra were processed using NMR Pipe and CARA software packages.
Phosphorylation for NMR Studies.
A 400 μM 15N RKIPP74L solution was prepared in 20 mM Hepes, pH 7.5, with 0.5 mM EGTA, 2 mM DTT, and 200 nM PKC. MgCl2 and ATP were then added to final concentrations of 5 mM each. Phosphorylation was carried out at 37 °C for 10 h before being reduced to 25 °C for NMR measurements.
Further descriptions of the methods are listed in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Helmholtz-Zentrum Berlin for the allocation of synchrotron radiation beamtime and the staff of beamline MX 14.1 for technical assistance and Drs. Gianluigi Veglia and Jonggul Kim for valuable discussions. This work was supported by Grants GM087630 (to M.R.R.), GM55694 (to T.R.S.), Deutsche Forschungsgemeinschaft FZ82 (to K.L., C.K., and H.S.) and SFB688 and TPA17 (to K.L.), the German Ministry of Research and Education and the Ministry for Innovation, Science and Research of the Federal State of North Rhine-Westphalia (K.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.J. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank, www.rcsb.org [PDB ID codes 6ENS (RKIP) and 6ENT (Δ143-146 variant)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711543114/-/DCSupplemental.
References
- 1.Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2012;367:2513–2516. doi: 10.1098/rstb.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis FP. Phosphorylation at the interface. Structure. 2011;19:1726–1727. doi: 10.1016/j.str.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Nishi H, Hashimoto K, Panchenko AR. Phosphorylation in protein-protein binding: Effect on stability and function. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Z, et al. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate. 2006;66:248–256. doi: 10.1002/pros.20319. [DOI] [PubMed] [Google Scholar]
- 5.Dangi-Garimella S, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yun J, et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30:4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid E, et al. Cardiac RKIP induces a beneficial β-adrenoceptor-dependent positive inotropy. Nat Med. 2015;21:1298–1306. doi: 10.1038/nm.3972. [DOI] [PubMed] [Google Scholar]
- 8.Lorenz K, Rosner MR, Brand T, Schmitt JP. Raf kinase inhibitor protein: Lessons of a better way for β-adrenergic receptor activation in the heart. J Physiol. 2017;595:4073–4087. doi: 10.1113/JP274064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banfield MJ, Barker JJ, Perry AC, Brady RL. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- 10.Granovsky AE, et al. Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol Cell Biol. 2009;29:1306–1320. doi: 10.1128/MCB.01271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark MC, et al. NMR assignment of rat Raf kinase inhibitor protein. J Biomol NMR. 2006;36:4. doi: 10.1007/s10858-005-4424-y. [DOI] [PubMed] [Google Scholar]
- 12.Corbit KC, et al. Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem. 2003;278:13061–13068. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz K, Lohse MJ, Quitterer U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature. 2003;426:574–579. doi: 10.1038/nature02158. [DOI] [PubMed] [Google Scholar]
- 14.Deiss K, Kisker C, Lohse MJ, Lorenz K. Raf kinase inhibitor protein (RKIP) dimer formation controls its target switch from Raf1 to G protein-coupled receptor kinase (GRK) 2. J Biol Chem. 2012;287:23407–23417. doi: 10.1074/jbc.M112.363812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Errington N, Doig AJ. A phosphoserine-lysine salt bridge within an alpha-helical peptide, the strongest alpha-helix side-chain interaction measured to date. Biochemistry. 2005;44:7553–7558. doi: 10.1021/bi050297j. [DOI] [PubMed] [Google Scholar]
- 16.Platzer G, Okon M, McIntosh LP. pH-dependent random coil (1)H, (13)C, and (15)N chemical shifts of the ionizable amino acids: A guide for protein pK a measurements. J Biomol NMR. 2014;60:109–129. doi: 10.1007/s10858-014-9862-y. [DOI] [PubMed] [Google Scholar]
- 17.Rovati GE, et al. The DRY motif and the four corners of the cubic ternary complex model. Cell Signal. 2017;35:16–23. doi: 10.1016/j.cellsig.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA. 3D complex: A structural classification of protein complexes. PLoS Comput Biol. 2006;2:e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue LC, Dobbs D, Bonvin AM, Honavar V. Computational prediction of protein interfaces: A review of data driven methods. FEBS Lett. 2015;589:3516–3526. doi: 10.1016/j.febslet.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput Biol. 2017;13:e1005324. doi: 10.1371/journal.pcbi.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact prediction in CASP10. Proteins. 2014;82:138–153. doi: 10.1002/prot.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J, Wang S. AcconPred: Predicting solvent accessibility and contact number simultaneously by a multitask learning framework under the conditional neural fields model. Biomed Res Int. 2015;2015:678764. doi: 10.1155/2015/678764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan C, Wu F, Jernigan RL, Dobbs D, Honavar V. Characterization of protein-protein interfaces. Protein J. 2008;27:59–70. doi: 10.1007/s10930-007-9108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue Y, et al. GPS: A comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–W187. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullah S, et al. dbPAF: An integrative database of protein phosphorylation in animals and fungi. Sci Rep. 2016;6:23534. doi: 10.1038/srep23534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung TC, et al. Solution structure of apoptotic BAX oligomer: Oligomerization likely precedes nembrane insertion. Structure. 2015;23:1878–1888. doi: 10.1016/j.str.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Zheng JH, Viacava Follis A, Kriwacki RW, Moldoveanu T. Discoveries and controversies in BCL-2 protein-mediated apoptosis. FEBS J. 2016;283:2690–2700. doi: 10.1111/febs.13527. [DOI] [PubMed] [Google Scholar]
- 28.Cartron PF, et al. Involvement of the N-terminus of Bax in its intracellular localization and function. FEBS Lett. 2002;512:95–100. doi: 10.1016/s0014-5793(02)02227-5. [DOI] [PubMed] [Google Scholar]
- 29.Arokium H, et al. Substitutions of potentially phosphorylatable serine residues of Bax reveal how they may regulate its interaction with mitochondria. J Biol Chem. 2007;282:35104–35112. doi: 10.1074/jbc.M704891200. [DOI] [PubMed] [Google Scholar]
- 30.Burkart EM, et al. Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J Biol Chem. 2003;278:11265–11272. doi: 10.1074/jbc.M210712200. [DOI] [PubMed] [Google Scholar]
- 31.Kooij V, et al. PKCα-specific phosphorylation of the troponin complex in human myocardium: A functional and proteomics analysis. PLoS One. 2013;8:e74847. doi: 10.1371/journal.pone.0074847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyle WG, Sumandea MP, Solaro RJ, De Tombe PP. Troponin I serines 43/45 and regulation of cardiac myofilament function. Am J Physiol Heart Circ Physiol. 2002;283:H1215–H1224. doi: 10.1152/ajpheart.00128.2002. [DOI] [PubMed] [Google Scholar]
- 33.Callaghan J, Simonsen A, Gaullier JM, Toh BH, Stenmark H. The endosome fusion regulator early-endosomal autoantigen 1 (EEA1) is a dimer. Biochem J. 1999;338:539–543. [PMC free article] [PubMed] [Google Scholar]
- 34.Macé G, Miaczynska M, Zerial M, Nebreda AR. Phosphorylation of EEA1 by p38 MAP kinase regulates mu opioid receptor endocytosis. EMBO J. 2005;24:3235–3246. doi: 10.1038/sj.emboj.7600799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumas JJ, et al. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 36.Ramanathan HN, Zhang G, Ye Y. Monoubiquitination of EEA1 regulates endosome fusion and trafficking. Cell Biosci. 2013;3:24. doi: 10.1186/2045-3701-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 38.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 39.Xu D, Tsai CJ, Nussinov R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 1997;10:999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]
- 40.P Barros E, et al. Electrostatic interactions as mediators in the allosteric activation of protein kinase a RIα. Biochemistry. 2017;56:1536–1545. doi: 10.1021/acs.biochem.6b01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leonard PG, Golemi-Kotra D, Stock AM. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc Natl Acad Sci USA. 2013;110:8525–8530. doi: 10.1073/pnas.1302819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearlman SM, Serber Z, Ferrell JE., Jr A mechanism for the evolution of phosphorylation sites. Cell. 2011;147:934–946. doi: 10.1016/j.cell.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.