Significance
We show that the temporal order of replication (replication timing, RT), normally an extremely stable cell type-specific chromosomal property, is altered in cells from two different premature aging (progeroid) diseases. By converting patient cells to stem cells and redifferentiating them as a model of disease progression, we identified the TP63 gene as one of the earliest RT alterations and altered RT was associated with abnormal TP63 gene expression. TP63 mutations have been linked to other diseases that share clinical features of progeroid syndromes. These findings introduce an approach for disease marker discovery, identify molecular abnormalities distinguishing progeroid diseases from natural aging, and point to TP63 as a molecular link to the pathophysiological manifestations of progeroid diseases.
Keywords: DNA replication timing, progeroid diseases, TP63, RT signatures
Abstract
Progeroid syndromes are rare genetic disorders that phenotypically resemble natural aging. Different causal mutations have been identified, but no molecular alterations have been identified that are in common to these diseases. DNA replication timing (RT) is a robust cell type-specific epigenetic feature highly conserved in the same cell types from different individuals but altered in disease. Here, we characterized DNA RT program alterations in Hutchinson–Gilford progeria syndrome (HGPS) and Rothmund–Thomson syndrome (RTS) patients compared with natural aging and cellular senescence. Our results identified a progeroid-specific RT signature that is common to cells from three HGPS and three RTS patients and distinguishes them from healthy individuals across a wide range of ages. Among the RT abnormalities, we identified the tumor protein p63 gene (TP63) as a gene marker for progeroid syndromes. By using the redifferentiation of four patient-derived induced pluripotent stem cells as a model for the onset of progeroid syndromes, we tracked the progression of RT abnormalities during development, revealing altered RT of the TP63 gene as an early event in disease progression of both HGPS and RTS. Moreover, the RT abnormalities in progeroid patients were associated with altered isoform expression of TP63. Our findings demonstrate the value of RT studies to identify biomarkers not detected by other methods, reveal abnormal TP63 RT as an early event in progeroid disease progression, and suggest TP63 gene regulation as a potential therapeutic target.
Progeroid syndromes arise from mutations that affect the nuclear lamina or DNA repair and share phenotypic characteristics with natural aging (1). One of the most studied is the Hutchinson–Gilford progeria syndrome (HGPS) caused by a point mutation in the LMNA gene that encodes two of the major components of the nuclear lamina: lamin A and C. The mutation activates an alternative splicing site, resulting in a truncated protein referred to as “progerin” (2, 3). HGPS patients display multiple anomalies including alopecia, loss of body fat, limited growth, scleroderma, and cardiovascular complications that eventually lead to their premature death (4). At the cellular level, expression of progerin leads to its accumulation in the nuclear envelope (5), which is linked to multiple nuclear defects such as abnormal morphology, altered chromatin organization, loss of heterochromatin, deficiencies in DNA-damage response, and impaired antioxidative pathways (6, 7). Intriguingly, HGPS is one of several disorders known as “progeroid syndromes” that, despite their pathophysiological similarities, arise from mutations in genes with distinct functions and have different cellular alterations (1). For example, Rothmund–Thomson syndrome (RTS) results from a mutation in the DNA helicase Q4 (RECQL4) and does not show the characteristic nuclear defects of HGPS, but patients present similar clinical symptoms (8, 9). Thus, despite recent progress in the characterization of cellular phenotypes associated with HGPS (6, 7, 10), little is known about the mechanisms linking the cellular defects to pathophysiological manifestations of the disease.
Previously, we demonstrated that the temporal order of genome duplication (replication timing, RT) is linked to chromatin organization and is regulated during development coordinated with gene activation (11, 12). Hence, RT constitutes a readily measurable functional readout of large-scale chromatin organization and transcriptional potential that can be exploited to detect alterations in disease. Here, we analyzed cells from progeroid HGPS and RTS patients to determine if the RT program is altered and the extent to which RT alterations in progeria are associated with the RT changes observed during normal aging. We identified a progeroid-specific RT signature containing genomic regions that replicate early in progeroid cells but late in cells from healthy individuals across a wide range of ages. Among the RT abnormalities, we identified TP63 as a gene marker for progeroid syndromes. TP63 alterations have not been observed previously in progeroid patients but have been associated with other diseases that share clinical manifestations. Additionally, when cells derived from HGPS and RTS patients were reprogrammed to induced pluripotent stem cells (iPSCs), all RT differences with normal cells were erased, but when these iPSCs were redifferentiated back to fibroblast cells, the abnormal RT of TP63 reappeared, suggesting that this change is an early epigenetic event in progeroid disease progression. Moreover, the TP63 RT abnormality was associated with an altered ratio of TP63 isoform expression, which previously has been linked to cellular senescence defects and multiple developmental alterations. These results implicate TP63 in the progression of progeroid disease, suggest a provocative link between abnormal RT and altered gene-variant expression, and demonstrate the utility of RT profiling to identify novel avenues in disease research.
Results
RT Abnormalities in HGPS.
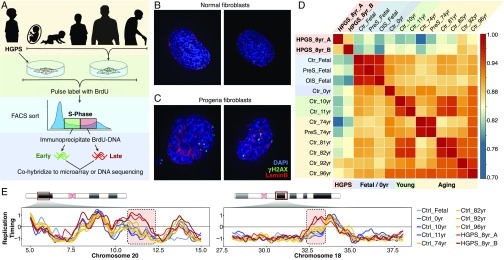
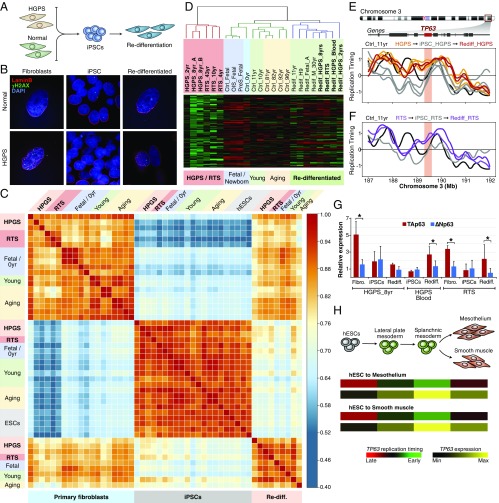
We measured the RT programs of progeroid and normal fibroblasts and characterized changes in RT upon reprogramming to iPSCs and redifferentiation back to fibroblasts. Overall, we generated 61 genome-wide RT datasets of fibroblasts, iPSCs, and redifferentiated cells derived from progeroid patients and healthy donors (Fig. 1A and Table S1). We first confirmed the known HGPS cellular abnormalities (13, 14), such as altered nuclear morphology and increased number and size of γH2AX foci associated with DNA damage (Fig. 1 B and C). Since earlier studies observed similar alterations in cells undergoing in vitro-induced senescence (15, 16), we also analyzed cells entering replicative or oncogene-induced senescence (OIS). Consistent with previous findings demonstrating that RT is a highly stable epigenetic property (17), we found strong genome-wide correlation of RT among all samples (Fig. 1D). Nonetheless, progeria cells were separated from normal fibroblasts, albeit more correlated with cells from the oldest healthy donors (Fig. 1D). Overall, RT differences across all samples comprise 7% of the autosomal genome, with 25% of those RT changes specific for HGPS. Exemplary RT profiles showing alterations specific for progeria patient cells are shown in Fig. 1E.
Fig. 1.
RT abnormalities in HGPS. (A) Depiction of cell samples analyzed by genome-wide RT profiling. Primary fibroblasts derived from progeroid syndrome patients (HGPS, far left) and from healthy fetal (IMR90), neonatal (BJ), young (10- and 11-y-old), and aged (74- to 96-y-old) donors were pulse labeled with BrdU and sorted into early and late S-phase, and RT programs were obtained by either array hybridization or next-generation sequencing (Table S1). (B and C) Representative immunofluorescence staining of primary fibroblasts from healthy persons (B) or HGPS patients (C). (Magnification: 60×.) (D) Correlation matrix of genome-wide RT programs of HGPS fibroblasts and cells from distinct healthy donors representing natural aging and cellular senescence. OIS, oncogene-induced senescence; PreS, cellular senescence induced by serial passaging. (E) RT profiles of exemplary chromosome segments showing RT alterations in progeria (HGPS). The RT profiles are displayed as log2 ratios of signals from early and late S-phase fractions. Positive values correspond to early replication, and negative values correspond to late replication.
A Specific RT Signature Distinguishes HGPS Cells.
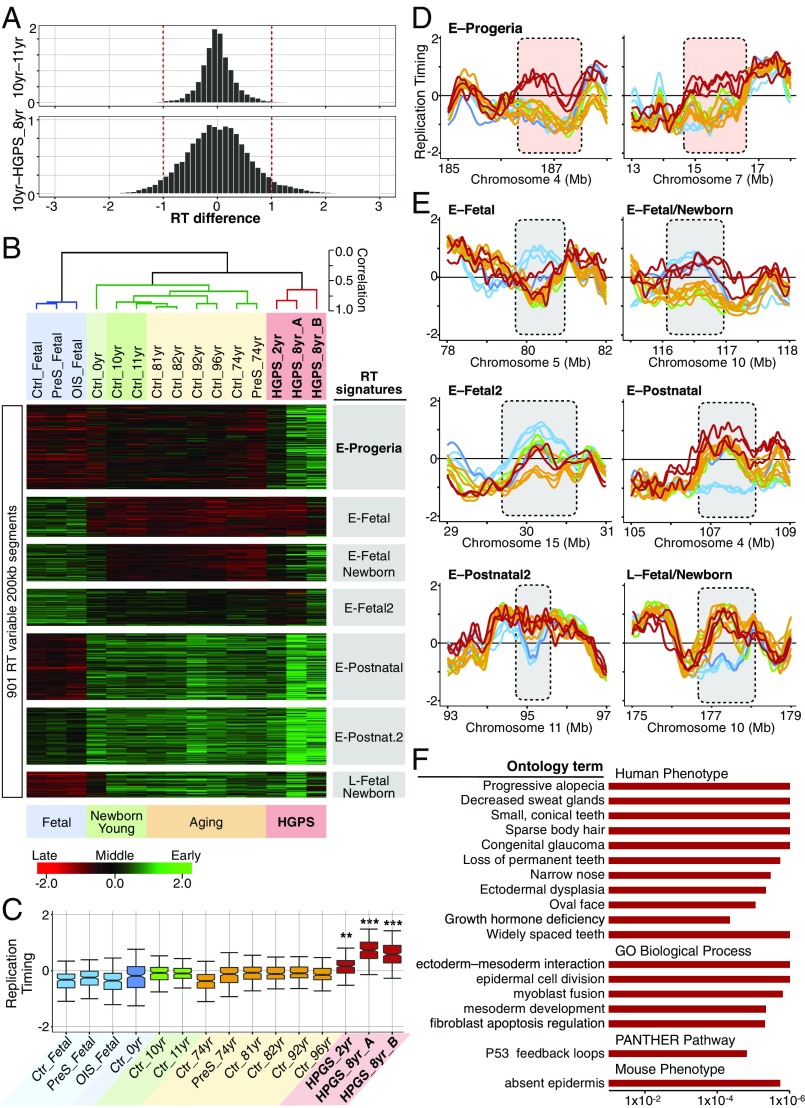
Previously, we characterized the RT changes during human development and identified RT signatures that distinguish each cell lineage (11). Here, we identified RT signatures between samples of different age or disease groups. Briefly, RT-variable regions were identified and clustered by unsupervised analysis (Fig. 2A and see Methods) into three major groups: fetal fibroblasts, healthy postnatal (0–96 y) cells, and HGPS cells (Fig. 2B). Moreover, we identified a specific RT signature containing regions that replicate early only in progeria (E-progeria regions) but replicate later in cells from all healthy donors, including cells from naturally aging donors, and in cells entering cellular senescence (Fig. 2 B–D). Although several genomic regions replicate later during early S phase in progeria than in the age-matched controls (Fig. 2A), we did not identify an RT signature of chromosomal segments that replicates late during S phase in progeria but early in all normal cells (Fig. 2B). The progeria-specific RT signature includes only 1.6% of the genome (Dataset S1). Additional RT signatures identified differences between fetal cells and all postnatal fibroblasts (Fig. 2 B and E and Dataset S1); however, all the fetal datasets were derived from a single cell line (IMR90), so their significance is uncertain. To determine the biological significance of the RT signatures and their relationship to disease pathogenesis, we performed gene ontology (GO) analysis on each of them (Fig. S1). Our results revealed that the E-progeria regions are strongly associated with phenotypic characteristics of the disease (Fig. 2F). Intriguingly, none of the ontology terms has been previously associated with progeroid syndromes, but the terms have been associated with other genetic diseases.
Fig. 2.
Identification of an HGPS-specific RT signature. (A) RT differences between progeroid cells and normal fibroblasts are bigger than the differences between age-matched controls. RT-variable regions were identified as those that display differences ≥1 in pairwise comparisons between all samples analyzed. (B) A progeria-specific RT signature distinguishes HGPS fibroblasts from normal cells. Unsupervised clustering analysis of RT-variable regions identified specific RT signatures (labeled in gray boxes). The heat map shows the RT ratios [= log2(early/late)]. A correlation threshold of 0.6 was used to define three main sample clusters. k-means analysis of variable segments defined RT signatures. (C) Genomic regions from the E-progeria RT signature replicate significantly differently in HGPS cells (**P < 1 × 10−5, ***P < 2 × 10−16 based on pairwise t-test with Bonferroni adjustment). (D and E) RT profiles of exemplary genomic regions from each RT signature. Samples are color-coded as in C. (F) Ontology analysis revealed that RT abnormalities in progeria are strongly associated with the phenotypic characteristics of HGPS disease. Statistical significance was measured by binomial test using the GREAT tool (48, 49).
TP63 Is a Marker of HGPS.
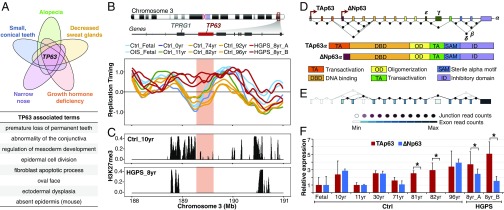
To identify candidate markers of HGPS, we examined the genes within each of the GO terms. Surprisingly, from the 200 genes within the genomic regions that replicate early only in progeria cells we found only a single gene common in all the GO terms: TP63, which encodes the tumor-suppressor protein p63 (Fig. 3A). Remarkably, multiple GO terms associated with TP63 match the progeroid pathophysiological symptoms, suggesting that TP63 is associated with the disease phenotype (Fig. 3A). TP63 alterations have not been previously observed in progeroid patients but have been observed in other disorders characterized by developmental abnormalities. TP63 replicates early only in progeria cells but replicates late in fibroblasts from all healthy donors (Fig. 3B); since RT is often correlated with transcriptional activity (11, 12), we reasoned that abnormal TP63 RT might be associated with altered gene regulation. Consistently, analysis of datasets obtained from a previous study (18) showed depletion of H3K27me3 throughout the TP63 locus relative to healthy donors (Fig. 3C). Importantly, these datasets were derived from one of the patient cell lines analyzed here for RT and gene expression, cell line HGADFN167.
Fig. 3.
The TP63 gene is a marker of HGPS. (A) Overlap analysis of the genes within GO terms reveals TP63 as unique gene linked with the progeroid phenotypic characteristics. Additional GO terms annotated for the E-progeria RT signature and linked to TP63 abnormalities are shown in the table. (B) TP63 replicates early in progeria cells but late in all fibroblasts derived from healthy donors as well as in cells entering senescence. (C) Loss of H3K27me3 peaks at the TP63 gene in HGPS cells (cell line HGADFN167 = HGPS_8yr). A healthy donor of similar age is shown. (H3K27me3 datasets were obtained from ref. 18.) (D) TP63 exon organization and functional domains. (E) Skin tissue from healthy donors predominantly expresses ∆Np63 isoforms. Exon and junction RNA-seq expression levels are color-coded according to read numbers. (F) Isoform-specific expression analysis by qRT-PCR demonstrates an altered ratio of TP63 variants expression in HGPS. Relative expression was normalized against HPRT1 gene and fetal fibroblasts. Significant differences are shown (*P < 0.05).
TP63 is a member of the p53 family and encodes multiple variant protein isoforms with distinct functions (19, 20). Alternative promoters, located >150 kb apart from each other, drive the expression of the main isoforms: TAp63 (containing the 5′ transactivation domain) and ∆Np63 (N-terminally truncated). Additionally, alternative splicing sites produce distinct C-terminal variants (Fig. 3D). To determine the patterns of TP63 isoform expression in healthy individuals, we analyzed RNA-sequencing (RNA-seq) data from the Genotype-Tissue Expression (GTEx) Project (21). The highest expression was detected in skin, and ∆Np63 was the predominant family of isoforms (Fig. 3E). Since GTex data derive from heterogeneous cell populations within each tissue, we analyzed transcriptome data from earlier studies (7, 18) by extracting specific microarray probes that hybridize to each isoform. We found a slight but significant unbalanced expression of TP63 in HGPS (not previously reported), with TAp63 expressed at higher levels than ∆Np63 isoforms (Fig. S2). To validate these findings in the cells from this study, we analyzed isoform-specific expression by qRT-PCR. We detected similar expression levels of both TP63 isoforms in cells derived from young healthy donors but higher levels of TAp63 isoforms in cells derived from HGPS patients (Fig. 3F). Our results demonstrate that abnormal RT of TP63 is correlated with an altered ratio of TP63 variants in the progeroid diseases.
Abnormal TP63 RT Is Not Dependent on Progerin Expression.
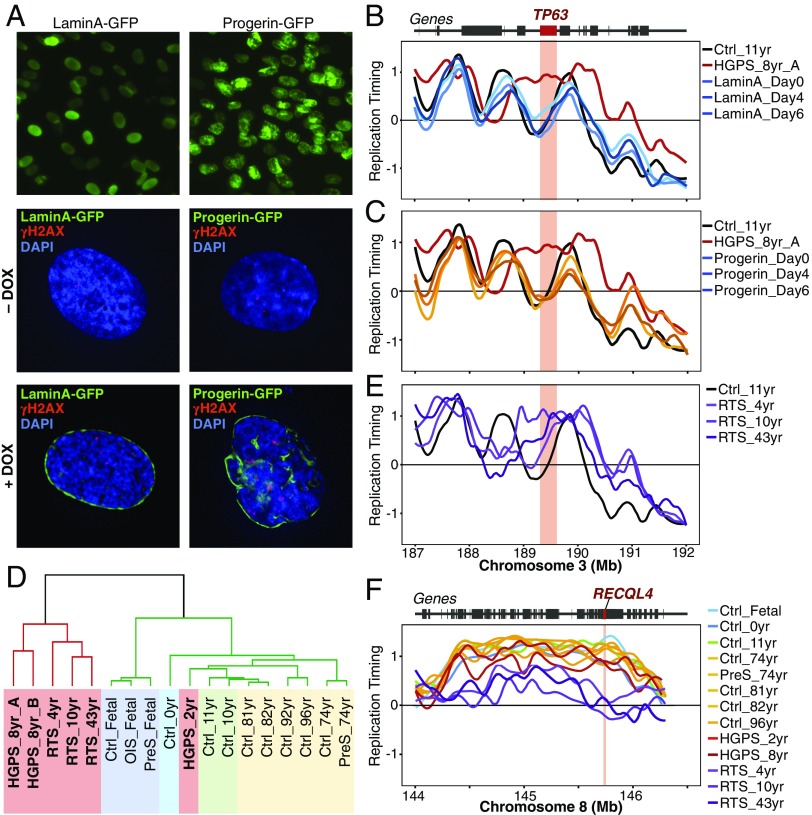
Ectopic expression of progerin in WT cells recapitulates the HGPS cellular defects (7). Hence, we analyzed whether TP63 alterations are dependent on progerin. First, we confirmed that progerin overexpression in WT cells induced the HGPS nuclear alterations vs. a control cell line overexpressing lamin A (Fig. 4A and Fig. S3). Nuclear alterations matching the HGPS cellular phenotype were detected as early as day 2 after induction (7). We analyzed RT up to 6 d after progerin overexpression and found that, despite nuclear alterations, no significant RT changes were detected genome wide, including at the TP63 locus. Consistently, we did not find unbalanced expression of TP63 isoforms after progerin overexpression (Fig. S3). These results demonstrate that the expression of progerin is not sufficient to alter the RT or isoform expression of TP63 (Fig. 4 B and C and Fig. S3). These results are, however, consistent with our previous findings showing that RT is robust to depletion of many factors that disrupt chromatin architecture (17) and underscore the importance of the RT alterations that we do detect in progeroid disease.
Fig. 4.
TP63 abnormal RT is not dependent on progerin expression and is observed in other progeroid syndromes. (A) Progerin overexpression in WT cells recapitulates nuclear alterations of HGPS. Normal fibroblasts with doxycycline (DOX)-inducible GFP-tagged lamin A or progerin were analyzed by immunofluorescence. (Magnification: 60×.) (B and C) RT remains unaltered at the TP63 locus upon overexpression of lamin A or progerin. RT of normal fibroblasts is shown in black, and RT of HGPS cells is shown in red; RT profiles after overexpression of progerin-GFP and lamin A-GFP for 0–6 d are shown. (D) Unsupervised clustering analysis of fibroblasts derived from HGPS and RTS patients and healthy donors of distinct ages. RT-variable chromosomal segments were identified (differences ≥1 in pairwise comparisons between all samples analyzed). Branches of the dendrogram were constructed based on the correlation values (distance = 1 − correlation value), and a correlation threshold of 0.4 was used to define the two main sample clusters: progeroid samples and normal fibroblasts. (E) Altered replication of the TP63 locus is also observed in fibroblasts from RTS patients. (F) Abnormal RT at the RECQL4 locus is specific for RTS cells. RT profiles of RTS fibroblasts are shown in comparison with cells derived from healthy donors of distinct ages and from HGPS patients.
Altered RT in RTS.
To determine whether altered TP63 RT is also detected in progeroid diseases that are not associated with progerin expression, we analyzed fibroblasts from RTS patients. RTS is a progeroid syndrome characterized by poikiloderma, sparse hair, skeletal and dental abnormalities, and a high risk of cancer and is linked to inactivating mutations of the RECQL4 gene (22). Notably, we found that 75% of the genomic regions that replicate early in progeria also replicate early in RTS fibroblasts. In fact, RTS and HGPS cells clustered together, demonstrating that the two progeroid syndromes share RT abnormalities (Fig. 4D). Among the RT alterations shared by HGPS and RTS was TP63 (Fig. 4E). Importantly, these results confirm that abnormal TP63 RT is independent of progerin expression. Moreover, we also detected the same unbalanced TP63 isoform expression in cells from RTS patients and from HGPS patients (Fig. S2), demonstrating that altered regulation of TP63 expression is common to these two progeroid diseases. Interestingly, additional RT abnormalities in RTS cells, which are not seen in HGPS cells, include late replication of the RECQL4 gene (Fig. 4F).
TP63 Recapitulates Its Abnormal RT upon Differentiation of iPSCs.
We analyzed whether the abnormal progeroid RT program can be reset by reprogramming primary cells to iPSCs (Fig. 5A). HGPS-iPSCs showed normal morphology and decreased signal for γH2AX (Fig. 5B). Moreover, the RT program was completely reprogrammed in all iPSCs, regardless of the age of the donors or progeroid disease (Fig. 5C and Fig. S4). Next, we redifferentiated the iPSCs and confirmed that the fibroblast RT program can be reestablished in the redifferentiated cells (Fig. 5C and Fig. S4). However, in contrast with previous studies showing reestablishment of altered nuclear morphology, loss of heterochromatin, and increases in DNA damage and mitochondrial reactive oxygen species after redifferentiation (23, 24), we found that redifferentiated cells from both progeroid syndromes retained normal RT globally, including most of the progeroid-specific RT signature regions (Fig. 5D, red branch). In fact, 95% of the genomic regions with altered RT in progeroid cells replicate normally in the redifferentiated cells (Fig. S4). Nonetheless, 10 specific genomic regions reacquired the progeroid-specific abnormal RT pattern upon redifferentiation (Fig. 5D and Fig. S4). Among these few loci the TP63 locus reacquired abnormal RT very early during redifferentiation of iPSCs from patients with either of the progeroid diseases (passages 3–4), despite being fully reset to “healthy” late RT at the pluripotent stage (Fig. 5 E and F). Moreover, increased expression of TAp63 isoforms is also observed after redifferentiation of progeroid, but not healthy, iPSCs (Fig. 5G).
Fig. 5.
Altered RT of TP63 is an early marker of progeroid diseases. (A) Reprogramming of primary cells to iPSCs and redifferentiation back to fibroblasts as a model of progeroid disease progression. (B) Representative immunofluorescence images of normal and HGPS cells confirm that reprogrammed iPSCs revert to the HGPS nuclear abnormalities which are spontaneously reestablished upon differentiation. (Magnification: 60×.) (C) Genome-wide RT correlation of primary cells, iPSCs, and redifferentiated cells from normal, HGPS, and RTS donors. (D) Clustering analysis of fibroblasts and redifferentiated cells from iPSCs. Only regions from the E-progeria RT signature are shown. (E and F) Altered TP63 RT is reestablished in redifferentiated HGPS and RTS cells. (G) TP63 isoforms expression by qRT-PCR in primary cells, iPSCs, and redifferentiated cells. Relative expression was normalized against the HPRT1 gene. Significant differences are shown (*P < 0.05). (H) TP63 changes to early replication during the first stages of mesoderm differentiation and switches back to late replication in later stages and is highly induced only when the gene is replicated early during S-phase. Re-diff, redifferentiated.
RT Changes in TP63 During Normal Human Development.
To understand the biological significance of TP63 alterations, we analyzed gene expression and RT during human development. We found that the TP63 locus is late replicating in human ES cells (hESCs), changes transiently to early replication during cell-fate commitment, and changes back to late replication in the differentiated cell types of all differentiation pathways analyzed (Fig. 5H and Fig. S5). Moreover, TP63 transcriptional induction is restricted to early stages of mesodermal differentiation correlated with early replication (Fig. 5H and Fig. S5). Finally, analysis of RNA-seq data from ENCODE show that TP63 expression is higher in hESCs than in differentiated fetal and newborn fibroblasts, correlated the with enrichment of H3K27me3 (Fig. S5). These results show that early TP63 replication is a normal epigenetic state restricted to the early stages of cell differentiation when the gene is highly expressed. The locus then switches back to late replication in coordination with epigenetic down-regulation of the gene.
Discussion
Despite recent advances in the characterization of progeroid diseases and the dissection of molecular mechanisms associated with its cellular defects (7, 10), the link between cellular alterations and the physiological abnormalities observed at the organismal level remain unknown. Moreover, progeroid diseases arise from mutations in functionally different genes, but no common defects have been identified among cells from patients with these different mutations. We performed a comprehensive genome-wide characterization of RT abnormalities in progeroid diseases compared with natural aging, revealing a molecular link between progeroid diseases of different origin. In contrast to numerous studies linking abnormalities of these congenital disorders to natural age-related defects (1), we identified a specific RT signature that distinguishes progeroid syndromes from natural aging. More importantly, GO analysis revealed an intriguing association of TP63 RT alterations with many of the phenotypic defects characteristic of this family of disorders. Although TP63 alterations have not been previously linked to progeroid syndromes, alterations in this gene are linked to developmental defects. In fact, genotyping studies have identified TP63 mutations associated with ectrodactyly-ectodermal dysplasia and cleft-lip/palate (EEC3), Rapp–Hodgkin ectodermal, acro-dermato-ungual-lacrimal-tooth (ADULT), and ankyloblepharon–ectodermal defects–clefting (AEC) syndromes (20). All these disorders show alterations of the skin, hair, teeth, nails, and sweat glands, and all these alterations are also observed in progeroid syndromes. Consistently, p63−/− mice have epidermal developmental defects and a shorter life span (25–28), and TP63 is required for proper skin stratification (29, 30). Moreover, TP63 deficiency induces cellular senescence and causes accelerated aging phenotypes in mice (31). Until this study, why TP63 alterations were not identified in progeroid disease remained a mystery.
A long-standing correlation between early replication and gene expression has been observed in all eukaryotes (12, 32). Hence, we explored whether altered RT in progeroid syndromes is associated with abnormal TP63 regulation. A previous study showed a genome-wide depletion of H3K27me3 in HGPS patients (18). Our analysis of data derived from that study also detected loss of H3K27me3 at the progeroid-specific RT signature regions (Fig. S6) including the TP63 locus (Fig. 3C). Additionally, the RT abnormalities in progeroid syndromes at the TP63 locus include the TP63-regulated 1 gene (TPRG1), which is also depleted of H3K27me3 in the disease (Fig. 3 B and C). TPRG1 encodes a cytoplasmic protein of unknown function and is highly expressed in epithelial cells under the control of TP63 (33). Although no alterations in TP63 expression were detected in previous HGPS studies (7, 18, 23, 34), none of the prior studies distinguished isoform expression. In fact, closer inspection of microarray data derived from two of these studies (7, 18) revealed a slight but significantly altered ratio of the expression of TP63 variants in HGPS. Moreover, we confirmed by qPCR that the TAp63 isoform is expressed at higher levels than the ∆Np63 isoform in HGPS and RTS. Interestingly, fibroblasts derived from two aged (81-y-old and 82-y-old) donors expressed only TAp63 isoforms, suggesting that alterations in TP63 regulation may also occur during natural aging through mechanisms not associated with early RT. While we cannot explain why the sample from the 96-y-old patient did not show the skew, this could be due to a difference between chronological and biological aging. Many of the HGPS cellular alterations are thought to be dependent on the accumulation of progerin (23, 24, 35–37). However, alterations in RT and TP63 regulation are independent of the LMNA mutation, as overexpression of progerin does not alter the RT of TP63 (Fig. 4 B and C), and cells from patients with RTS, which do not express progerin, also display the TP63 RT alteration and preferential expression of the TAp63 isoform (Figs. 4E and 5G). Hence, our study has identified progeroid-specific alterations in TP63 RT that are independent of progerin expression, are shared in HGPS and RTS patients, and are associated with a depletion of H3K27me3 and skewed variant expression but we did not find increased overall steady-state levels of TP63 transcript.
We also employed ESC differentiation to characterize normal development and iPSC reprogramming and redifferentiation as a model of disease progression. Distinct progeria-specific hallmarks (nuclear envelope alterations, γH2AX foci, and loss of heterochromatin) are reset in reprogrammed iPSCs; however, these hallmarks are reestablished spontaneously upon redifferentiation (23, 24, 37). Consistently, RT abnormalities in HGPS cells revert to normal in progeroid iPSCs (Fig. 5C). It has been speculated that the resetting of HGPS alterations in iPSCs is due to the lack of progerin expression in pluripotent cells (23, 24). However, the fact that RT abnormalities in cells derived from RTS patients also revert to normal in iPSCs suggests that the establishment of pluripotency, rather than down-regulation of progerin, is responsible for the RT resetting in iPSCs. Moreover, in contrast to previous studies showing a resetting of cellular defects in iPSCs but spontaneous reestablishment of these alterations after redifferentiation (23, 24), we found that fibroblasts derived from progeroid iPSCs retained normal RT, except for few regions, including the TP63 locus. Since alterations in RT occur early during differentiation, even before changes in nuclear morphology are evident, early RT and skewed isoform usage of TP63 constitutes a potential early marker of the progeroid phenotype. Consistently, fibroblasts from the youngest HGPS patient (HGPS_2 y) showed fewer RT differences with normal fibroblasts (Figs. 2B and 4D) than did fibroblasts from older patients (HGPS_8yr_A and HGPS_8yr_B) with the same mutation (1824C > T) but still included the TP63 RT difference, suggesting that an increase in the number of RT changes is associated with progeroid disease progression and that RT alterations at the TP63 locus are one of the earliest progeroid defects.
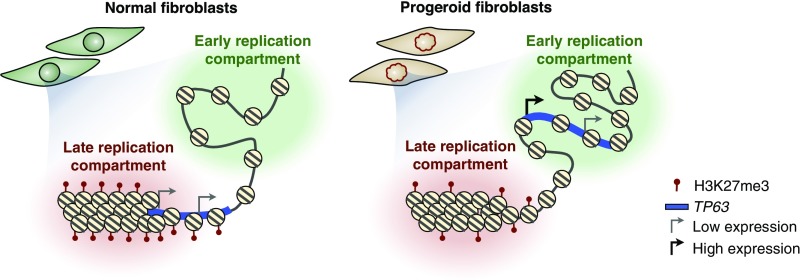
Together, our results support a model in which TP63 is embedded in a late-replicating compartment of the nucleus where its transcription is restricted to low basal levels. During differentiation, transcriptional activation of TP63 is coordinated with a switch to early replication, while in fully differentiated cells the gene is lowly expressed and switches back to the late-replicating compartment (Fig. 6). Hence, alterations in TP63 in progeroid syndromes might be due to failure of RT to switch back to late replication, potentially leading to an altered isoform expression pattern (Fig. 6). An altered increase in the expression of TAp63 isoforms could then lead to abnormal control of cell differentiation of epithelial cells, resulting in the characteristic phenotype of progeroid patients (Fig. S7). Although we can only speculate about the mechanisms linking RT alterations to isoform expression, the correlation observed in six progeroid patients from two distinct genetic origins and reappearing again in four redifferentiated cell samples constitutes compelling evidence that altered RT is linked, directly or indirectly, to the altered expression of TP63 isoforms. One formal possibility is that, since the TAp63 promoter is normally later replicating than ΔNp63 when they show balanced expression, but both promoters are equally early replicating when showing imbalanced expression, the Tap63 promoter could be more affected by its larger RT shift. Unraveling the mechanisms linking the abnormalities of TP63 regulation to the clinical symptoms of progeroid patients will be challenging due to the multiple TP63 isoforms with distinct functions and the very limited lifespan of the primary cells from progeroid patients and because, so far, no cellular abnormalities have been directly linked to the pathophysiological abnormalities of the disease. In fact, the alterations described here identify cellular abnormalities that are common to both HGPS and RTS. However, evidence suggests that proper modulation of expression ratios between TP63 isoforms is essential for epithelial development (20); abnormal TP63 regulation affects maintenance and homeostasis of the epidermis (19, 38, 39). In fact, mice deficient in TP63 have multiple developmental defects including truncated limbs, craniofacial anomalies, lack of epidermis, hair follicles, teeth, and mammary, lachrymal, and salivary glands (25, 28, 40, 41). Moreover, a mouse knockin of the TP63 mutation observed in EEC3 syndrome patients develops multiple ectodermal defects and craniofacial abnormalities (42). The roles of each TP63 isoform and their downstream targets are also the subject of active investigation, and it has been suggested that TP63 prevents aging and cellular senescence by controlling the proliferation and maintenance of stem cell precursors in mouse models (26). Overexpression of the TAp63 group of isoforms induces cellular senescence in mice fibroblasts (43), while ∆Np63 isoforms are necessary for proper differentiation of epithelial cells (44) and are down-regulated in a mouse model of progeria (45). TP63 alterations also have been associated with cardiac defects; TAp63 is required for cardiac differentiation of ESCs (46, 47), and p63−/− mice have severe cardiomyopathy (19). Although further experimentation is needed to elucidate the mechanisms linking TP63 alterations to the progeroid phenotype, abnormal RT associated with the skewed expression of gene variants opens new avenues of investigation into progeroid diseases. Specifically, abnormal RT may be associated with altered proximity to transcription machinery or altered regulation of the alternative promoters of TP63.
Fig. 6.
Model of TP63 alterations in progeroid diseases. Schematic depiction of TP63 locus organization and gene regulation in normal and progeroid fibroblasts. In normal cells, TP63 is localized to late replication compartments where it is expressed at low levels (consistent with an increased enrichment of H3K27me3). In contrast, in cells from patients with progeroid diseases the TP63 gene replicates early and is depleted of H3K27me3, which is associated with unbalanced isoform expression.
Materials and Methods
Primary fibroblasts obtained from Coriell Institute for Medical Research), the Progeria Research Foundation, and from donors from the Centre Hospitalier Régional Universitaire (CHRU) Montpellier cohort (Table S1) were reprogrammed to iPSCs and redifferentiated back to fibroblasts. All human cell lines were managed according to the guidelines in the Declaration of Helsinki, and donor privacy and confidentiality were ensured by the cell banks’ blinding of samples with a numerical identification code. Experimental details are described in SI Materials and Methods.
All datasets generated in this study are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; GEO series GSE98475) and on The Florida State University ReplicationDomain database at www.replicationdomain.org. H3K27me3 data were obtained from GSE41764, gene expression data from HGPS, and normal fibroblast data were extracted from GSE41751 and GSE69391, and RT and transcription datasets during human ESCs differentiation toward endoderm, mesoderm, ectoderm, and neural crest were extracted from GSE63428 and GSE63592.
Supplementary Material
Acknowledgments
We thank Ruth A. Didier for assistance with flow cytometry, Jared Zimmerman for data deposition in public repositories, Nard Kubben and Tom Misteli for sharing the progerin-GFP and lamin A-GFP cell lines, Armando Aranda-Anzaldo for critical reading of the manuscript, and la Région Languedoc Roussillon/Occitanie for a Ph.D. Student Fellowship. This work was supported by NIH Grants GM083337 and GM085354 (to D.M.G.), la Ligue Nationale Contre le Cancer Grant “Programme Labellisation Equipe 2015” (EL2015.LNCC/JML to J.-M.L.), INGESTEM National Infrastructure in Biology and Health, and le Centres Hospitaliers Universitaires de Montpellier.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98475) and on The Florida State University ReplicationDomain database at www.replicationdomain.org.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711613114/-/DCSupplemental.
References
- 1.Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- 2.De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055–2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vidak S, Foisner R. Molecular insights into the premature aging disease progeria. Histochem Cell Biol. 2016;145:401–417. doi: 10.1007/s00418-016-1411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalo S, Kreienkamp R. DNA repair defects and genome instability in Hutchinson-Gilford progeria syndrome. Curr Opin Cell Biol. 2015;34:75–83. doi: 10.1016/j.ceb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubben N, et al. Repression of the antioxidant NRF2 pathway in premature aging. Cell. 2016;165:1361–1374. doi: 10.1016/j.cell.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu L, Jin W, Wang LL. Aging in Rothmund-Thomson syndrome and related RECQL4 genetic disorders. Ageing Res Rev. 2017;33:30–35. doi: 10.1016/j.arr.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Fu W, Ligabue A, Rogers KJ, Akey JM, Monnat RJ., Jr Human RECQ helicase pathogenic variants, population variation and “missing” diseases. Hum Mutat. 2017;38:193–203. doi: 10.1002/humu.23148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, et al. Loss of H3K9me3 correlates with ATM activation and histone H2AX phosphorylation deficiencies in Hutchinson-Gilford progeria syndrome. PLoS One. 2016;11:e0167454. doi: 10.1371/journal.pone.0167454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera-Mulia JC, et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 2015;25:1091–1103. doi: 10.1101/gr.187989.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera-Mulia JC, Gilbert DM. Replication timing and transcriptional control: Beyond cause and effect-part III. Curr Opin Cell Biol. 2016;40:168–178. doi: 10.1016/j.ceb.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson EC, Manning B, Zhang H, Lawrence JB. Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J Cell Biol. 2013;203:929–942. doi: 10.1083/jcb.201306073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandra T, et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015;10:471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dileep V, Rivera-Mulia JC, Sima J, Gilbert DM. Large-scale chromatin structure-function relationships during the cell cycle and development: Insights from replication timing. Cold Spring Harb Symp Quant Biol. 2015;80:53–63. doi: 10.1101/sqb.2015.80.027284. [DOI] [PubMed] [Google Scholar]
- 18.McCord RP, et al. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23:260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paris M, Rouleau M, Pucéat M, Aberdam D. Regulation of skin aging and heart development by TAp63. Cell Death Differ. 2012;19:186–193. doi: 10.1038/cdd.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanbokhoven H, Melino G, Candi E, Declercq W. p63, a story of mice and men. J Invest Dermatol. 2011;131:1196–1207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- 21.GTEx Consortium Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suter AA, et al. Rothmund-Thomson syndrome: Novel pathogenic mutations and frequencies of variants in the RECQL4 and USB1 (C16orf57) gene. Mol Genet Genomic Med. 2016;4:359–366. doi: 10.1002/mgg3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu G-H, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller JD, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 26.Su X, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki D, Sahu R, Leu NA, Senoo M. The carboxy-terminus of p63 links cell cycle control and the proliferative potential of epidermal progenitor cells. Development. 2015;142:282–290. doi: 10.1242/dev.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 29.Borrelli S, et al. The p63 target HBP1 is required for skin differentiation and stratification. Cell Death Differ. 2010;17:1896–1907. doi: 10.1038/cdd.2010.59. [DOI] [PubMed] [Google Scholar]
- 30.Keyes WM, et al. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8:164–176. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keyes WM, et al. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera-Mulia JC, Gilbert DM. Replicating large genomes: Divide and conquer. Mol Cell. 2016;62:756–765. doi: 10.1016/j.molcel.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonini D, et al. Tprg, a gene predominantly expressed in skin, is a direct target of the transcription factor p63. J Invest Dermatol. 2008;128:1676–1685. doi: 10.1038/jid.2008.12. [DOI] [PubMed] [Google Scholar]
- 34.Csoka AB, et al. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004;3:235–243. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez S, Coppedè F, Sagelius H, Eriksson M. Increased expression of the Hutchinson-Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur J Hum Genet. 2009;17:928–937. doi: 10.1038/ejhg.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Melino G, Memmi EM, Pelicci PG, Bernassola F. Maintaining epithelial stemness with p63. Sci Signal. 2015;8:re9. doi: 10.1126/scisignal.aaa1033. [DOI] [PubMed] [Google Scholar]
- 39.Su X, Chakravarti D, Flores ER. p63 steps into the limelight: Crucial roles in the suppression of tumorigenesis and metastasis. 2013;13:136–143. doi: 10.1038/nrc3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolff S, et al. The alpha/beta carboxy-terminal domains of p63 are required for skin and limb development. New insights from the Brdm2 mouse which is not a complete p63 knockout but expresses p63 gamma-like proteins. Cell Death Differ. 2009;16:1108–1117. doi: 10.1038/cdd.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keyes WM, Mills AA. p63: A new link between senescence and aging. Cell Cycle. 2006;5:260–265. doi: 10.4161/cc.5.3.2415. [DOI] [PubMed] [Google Scholar]
- 42.Moretti F, et al. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120:1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano R-A, et al. ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosengardten Y, McKenna T, Grochová D, Eriksson M. Stem cell depletion in Hutchinson-Gilford progeria syndrome. Aging Cell. 2011;10:1011–1020. doi: 10.1111/j.1474-9726.2011.00743.x. [DOI] [PubMed] [Google Scholar]
- 46.Rouleau M, et al. TAp63 is important for cardiac differentiation of embryonic stem cells and heart development. Stem Cells. 2011;29:1672–1683. doi: 10.1002/stem.723. [DOI] [PubMed] [Google Scholar]
- 47.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 48.Saldanha AJ. Java Treeview–Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 49.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.