Significance
With the advent of advanced proteomic technologies, a unique generation of plasma biomarkers is likely to arise in the foreseeable future. One of the fundamental practical problems in developing such biomarkers for clinical use is the lack of a high-throughput, robust, and reproducible system for validating candidate biomarkers. Here, we report the development of a system that is suitable for validating a large number of candidate biomarkers in a quantitative and massively parallel manner. In addition to describing this system [called sequential analysis of fractionated eluates by selected reaction monitoring (SAFE-SRM)], we have used it to discover a peptide biomarker for ovarian cancer that may prove to have clinical value.
Keywords: clinical proteomics, selected reaction monitoring, plasma biomarker, peptide biomarker, cancer diagnostics
Abstract
We here describe a selected reaction monitoring (SRM)-based approach for the discovery and validation of peptide biomarkers for cancer. The first stage of this approach is the direct identification of candidate peptides through comparison of proteolytic peptides derived from the plasma of cancer patients or healthy individuals. Several hundred candidate peptides were identified through this method, providing challenges for choosing and validating the small number of peptides that might prove diagnostically useful. To accomplish this validation, we used 2D chromatography coupled with SRM of candidate peptides. We applied this approach, called sequential analysis of fractionated eluates by SRM (SAFE-SRM), to plasma from cancer patients and discovered two peptides encoded by the peptidyl-prolyl cis–trans isomerase A (PPIA) gene whose abundance was increased in the plasma of ovarian cancer patients. At optimal thresholds, elevated levels of at least one of these two peptides was detected in 43 (68.3%) of 63 women with ovarian cancer but in none of 50 healthy controls. In addition to providing a potential biomarker for ovarian cancer, this approach is generally applicable to the discovery of peptides characteristic of various disease states.
Nearly a quarter of a million women will be diagnosed with ovarian cancer this year, and more than 140,000 women will die from their disease (1). If ovarian cancer is diagnosed and treated at early stages, before the cancer has spread outside the ovary, the 5-y relative survival rate is over 90% (1). However, only 15% of all ovarian cancers are found at such early stages and the prognosis for patients whose cancers are discovered at late stages is dismal (1). There is thus a widely recognized need for the development of biomarkers that could potentially detect ovarian cancers earlier. There have been numerous attempts to use conventional biomarkers, such as CA-125 or HE-4, or to use ultrasound, for such detection (2–5). Although some show promise, none of them is recommended for screening by the US Preventive Services Task Force because they too frequently lead to “important harms, including major surgical interventions in women who do not have cancer” (6).
Proteins have historically been the most widely used and most successful type of biomarkers for use in cancer patients, although they are generally applied in diagnostic rather than screening settings (7, 8). Major advances in proteomics have inspired renewed efforts to develop improved biomarkers for ovarian and other cancers (9–12). Some of the most sophisticated of these use unbiased approaches wherein proteins from cancer patients and normal individuals are proteolytically digested and the resultant peptides are assessed via MS technologies. A variety of candidate peptides are often discovered through such approaches (13). The next step in such biomarker discovery is often rate-limiting for biomarker discovery: how does one narrow down the large list of candidate peptides to a more manageable list that does not compromise quantification, sensitivity, or specificity? We here describe a peptide-centric platform for developing biomarkers that specifically addresses this issue. Moreover, we show that peptides isolated directly from plasma, rather than from cancer tissues, can be used for the discovery of cancer biomarkers.
Results
Study Design.
This study was designed to identify and validate proteomic biomarkers for cancers using a combination of qualitative and quantitative MS techniques. Most previous studies in this area have begun with the analysis of cancer tissues, and then attempted to determine whether cancer-specific proteins or peptides could be identified in the plasma. In the current study, we attempted to identify candidate peptides directly from the plasma. The study was executed in three discrete phases: phase 1, global plasma proteomic profiling of samples from cancer patients and healthy individuals, yielding 641 candidate peptide markers from 188 genes; phase 2, implementation of a selected reaction monitoring (SRM)-based assay, called sequential analysis of fractionated eluates by SRM (SAFE-SRM), to evaluate each of the 641 candidate peptide markers in additional plasma samples, yielding two peptides from peptidyl-prolyl cis–trans isomerase A (PPIA) as promising biomarkers; and phase 3, evaluation of the performance of these two peptides in an independent set of cancer patients and controls using SAFE-SRM. Phase 1 was performed on an Orbitrap mass spectrometer, which is most suitable for qualitative analysis of large numbers of proteins, while phases 2 and 3 were conducted on a triple-quadrupole mass spectrometer, most suitable for quantitative analyses of selected analytes. A total of 266 plasma samples from different donor sources was evaluated during the three phases of this study (Table S1).
Phase 1: Identification of Candidate Biomarkers from Cancer Patients.
To identify potential protein biomarkers for cancers, we first created four pooled human plasma samples composed of equal volumes of plasma from 50 normal healthy individuals, 18 patients with ovarian cancer, 13 patients with pancreatic cancer, and 18 patients with colorectal cancer (Dataset S1). All patients with cancer had advanced disease so as to maximize the likelihood that high concentrations of putative biomarkers would be found in the plasma. An antibody-based plasma depletion was performed to remove 14 highly abundant proteins, such as albumin and immunoglobulins, from each of the four pools. Each pool was then digested with trypsin and the resultant peptides were differentially labeled with iTRAQ. iTRAQ labeling allows the four pools to be mixed and analyzed in a single MS experiment. The pools were then analyzed to assess whole proteomes (Fig. 1A and Fig. S1A). In a separate experiment, the pooled plasma samples were enriched for glycoproteins before trypsin digestion and iTRAQ labeling to reveal potential differences in the peptides derived from glycosylated proteins (Fig. S1B).
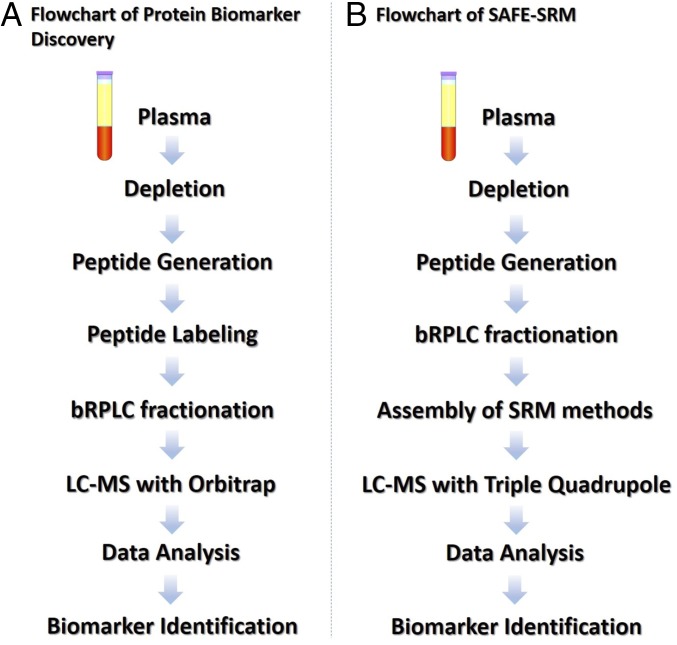
Fig. 1.
Workflow of plasma biomarker identification and validation. Plasma biomarker discovery and identification were conducted through labeling-dependent quantitative proteomics, such as iTRAQ or TMT assays (A); plasma biomarker validation was conducted through SAFE-SRM (B).
Problems with the reproducibility of large-scale proteomics experiments such as those we carried out are well known (14), so we performed replicates of the entire workflow outlined in Fig. S1. In total, 223,602 peptides were identified through these analyses, representing 10,789 unique peptides from 1,249 unique proteins (Datasets S2 and S4). The relative abundances of each of these peptides in the plasma samples from cancer patients and normal individuals were then calculated using an empirical-Bayes modified t test (Materials and Methods). A total of 8,069 unique peptides was quantified in at least two replicates, and the correlation for the abundances of these peptides between the replicates was 0.74 (95% CI, 0.73–0.75). As described in detail in Materials and Methods, our analyses eventually yielded 641 peptides derived from 188 proteins with significantly increased abundance in the pooled cancer plasma samples compared with the pooled normal controls (Dataset S4).
Phase 2a: Development of SAFE-SRM.
The validation of hundreds of potential peptide biomarkers is a daunting task. The difficulty is exacerbated by the fact that the abundances of peptides from plasma proteins are generally low and the abundances of different peptides vary considerably within this low range. We developed an approach to tackle these challenges, with five major components. First, the 641 peptides of interest were individually synthesized, but not highly purified, so as to keep costs manageable. Second, an SRM method was created for each of these peptides. Each of the 641 methods was optimized for the collision energies and dwell times of the precursor ions that yielded the highest intensities of the postcollision peptide-specific transitions of major interest. The dwell time given to each peptide was inversely proportional to the peptide’s intensity measured from a human plasma peptide sample spiked with equal amounts of synthetic peptides. This feature permitted the instrument to spend more time on detecting the peptides with lower signal intensities, thereby improving the overall ion statistics for the detection of low-abundance peptides. This protocol led to the identification of 4,384 transitions (approximately seven transitions per peptide; Dataset S5).
Third, the peptides were fractionated using basic pH reversed-phase liquid chromatography (bRPLC), yielding 96 fractions organized into 32 “fraction groups” each containing three sequential fractions; 20 fraction groups were selected for further analysis. Fourth, the peptides in each fraction group were separated by an orthogonal high-performance liquid chromatography (HPLC) method based on hydrophobic interactions (C18-RPLC). Finally, continuous elutes from the second HPLC column were analyzed using an SRM method composed of the collision energies, dwell times, and transitions that had been preoptimized using the synthetic peptides noted above. We termed this approach SAFE-SRM (Fig. S2).
One advantage of SAFE-SRM is that it employs a two-dimensional chromatographic fractionation. The individual fractions contain much less peptide than the total, thereby reducing ion suppression from unwanted peptides and increasing the signal-to-noise ratio. A second advantage of SAFE-SRM is that it converts the qualitative approach used for peptide discovery to a quantitative approach during the validation phases. Finally, the method is highly tolerant to fluctuations in elution times that are commonly observed in bRPLC chromatography because sequential fractions are redundantly tested for peptide abundances (Materials and Methods).
To assess the performance of SAFE-SRM, we chose six peptides with different hydrophobicity characteristics in HPLC and synthesized them as heavy isotope-labeled forms (SI Materials and Methods). We then mixed these peptides and performed a standard SRM analysis using the optimized collision energies and dwell times described above. All six peptides were detected at high confidence, as expected. However, when we spiked these peptides into trypsin-digested samples generated from normal plasma as described above, their average intensities were only around 5% of that obtained with the pure peptides, and three of the six peptides were not detectable at all. When this spiked sample was analyzed with SAFE-SRM, all six peptides could be detected, with an intensity that averaged 70% of that obtained with the pure peptides (Fig. 2).
Fig. 2.
Peptide detectability by SAFE-SRM in complex samples. Six heavy-isotope–labeled peptides (peptide 1: IQLVEEELDR*; peptide 2: VILHLK*; peptide 3: IILLFDAHK*; peptide 4: TLAESALQLLYTAK*; peptide 5: LLGHLVK*; peptide 6: GLVGEIIK*, where * indicates C13 and N15 heavy-isotope–labeled amino acids) were synthesized and used to evaluate the sensitivity of SAFE-SRM in detecting low amount of peptides in complex samples. One femtomole of each peptide was detected by conventional SRM (A). However, when 1 fmol of these peptides was added to trypsin-digested plasma samples, they were much more difficult to detect (B). bRPLC fractionation was able to increase the sensitivity of standard SRM, but with a large variance between runs (C). SAFE-SRM with optimized dwell and cycling time allowed detection of all six peptides, at intensities averaging 70% of the intensities of the free peptides (D).
Phase 2b: Testing of Candidate Peptides by SAFE-SRM.
We began by using SAFE-SRM to evaluate the four plasma pools used for the initial iTRAQ-based discovery phase of the study. We expected that the peptides detectable in these pooled samples would be those least likely to be affected by ion suppression, the coelution of unwanted peptides in the same chromatographic fractions, or other technical issues. After careful examination, 318 out of the 641 tested peptides proved to be reproducibly detectable in the pooled samples through 1,990 transitions (6.3 transitions per peptide; Dataset S5). These 318 peptides were mapped to 121 proteins.
We then used SAFE-SRM to evaluate 94 individual plasma samples, none of which was used in the discovery phase. Forty-eight of these samples were from normal individuals and 14, 14, and 18 were from patients with colorectal cancers, ovarian cancers, and pancreatic cancers, respectively (Dataset S1). SAFE-SRM abundance scores were calculated for the 318 peptides in each of the 94 individual and 4 pooled plasma samples (Dataset S6). We used statistical methods to determine whether any peptide or combination of peptides was able to accurately classify the origin of a sample from the peptide signatures. For this purpose, we randomly selected approximately one-half of the samples for training (27 from healthy donors and 7, 7, and 9 samples from patients with colorectal cancers, ovarian cancers, or pancreatic cancers, respectively). The remaining half of the samples were used to test the performance of the classifiers derived from the training samples.
A recursive, leave-one-out cross-validation strategy was used to estimate the predictive performance of the classification model as it evolved. The peptides yielding the highest cross-validated classification scores on the training set were first selected. Data on other peptides were then searched to determine whether any second peptide could increase the classification score. This process of selecting a peptide biomarker to be added was repeated until no further increases in the classification score could be achieved by addition of other peptides. Using this approach, several combinations of peptides with excellent classification potential were identified (Fig. 3 A and B).
Fig. 3.
Ovarian cancer prediction by peptide biomarkers. (A) Mean square errors (MSEs) of ovarian cancer prediction of all 318 peptides are plotted with the peptides ranked by MSE from the best predictors to the worst predictors. (B) The 10 best peptide biomarkers are shown; the peptide VSFELFADK from peptidyl-prolyl cis–trans isomerase A was the best predictor. (C) The ovarian cancer prediction performance of PPIA peptide VSFELFADK was further improved by combining with another peptide, FEDENFILK, from the same protein.
The best performance was observed for the classification of ovarian cancers with a combination of several markers. The top single peptide marker for ovarian cancers was VSFELFADK from PPIA (also known as Cyclophilin-A). We then determined whether any of the other peptides from PPIA among those in the 318-peptide set could be added to the classifier without decreasing specificity and found that a second peptide from PP1A (FEDENFILK) could be added in this way (Fig. 3C). Using peptide abundance levels resulting in 100% specificity among 36 normal samples, we found that VSFELFADK and FEDENFILK yielded 75.0% and 78.6% sensitivities, respectively. The Pearson correlation coefficient for the two PPIA peptides was 0.83 (95% CI, 0.78–0.87). At least one of the two peptides was elevated in 23 (82.1%) of the 28 samples.
Phase 3: Validation.
The dataset used to form the classifier was large: 1,990 transitions from 318 peptides tested in each of 98 samples. It is well known that overfitting is possible in such experiments and that independent validations of any classifier are mandatory. We therefore evaluated a separate cohort of 73 cases, consisting of plasma from 35 ovarian cancer cases and 38 samples from healthy individuals or patients with other cancer types (Dataset S7). In these 73 cases, SAFE-SRM was performed, but the only transitions analyzed were those corresponding to the two peptides from PPIA plus a peptide from Fibronectin, which we found to be expressed at similar levels in all samples and was thereby used for normalization. The relative abundances required for a positive score were predetermined from the results in phase 2b described above. Examples of the SAFE-SRM profiles for these peptides in ovarian cancer patients and normal individuals are shown in Fig. S3. Twenty (57.1%; 95% CI, 40–73%) of the 35 plasma samples from ovarian cancer cases scored positive for VSFELFADK from PPIA, while none of the 14 samples from normal individuals scored positive (specificity of 100%; 95% CI, 89–100%). For the second peptide FEDENFILK from PPIA, 14 (40.0%; 95% CI, 24–58%) of the 35 plasma samples from ovarian cancer cases were scored as positive, and, as for the first PPIA peptide, none of the 14 samples from healthy individuals scored positive. All of the plasma samples scoring positive for the FEDENFILK peptide also scored positive for the VSFELFADK from the same protein. Twenty-four patients with pancreatic cancer were tested in this assay, and only one of them (4.2%; 95% CI, 0.2–23.1%) scored positive for peptide VSFELFADK, and none for peptide FEDENFILK (Dataset S7).
It was notable that 11 of 17 (64.7%) of the plasmas from patients with early-stage ovarian cancers scored positive for PPIA peptides, while 32 of 46 (69.6%) of the plasmas from patients with more advanced cancers scored positive (combining phase 2b and phase 3; Dataset S7). For comparison, CA125 levels were measured in a subset of the same cohort. CA125 was elevated in 20 of 63 ovarian cancer patients and in none of 50 healthy controls. The elevations in CA125 and PPIA did not completely overlap, so that the sensitivity for detection of either CA125 or PPIA levels was 74.6% (95% CI, 62.1–84.7%), higher than either alone (see Venn diagram in Fig. S4).
Discussion
There are some important differences between the approach described here and most of those used in the past to identify protein biomarkers for cancer. Most studies start with the analysis of tumors, searching for proteins that are expressed at higher levels than in the corresponding normal tissues. It is then determined whether the identified proteins are elevated in the circulation of cancer patients. Although there are advantages to this approach, proteins that are expressed at high levels in a tumor are not necessarily released into the plasma. Moreover, such proteins can sometimes be expressed in normal tissues other than those initially used for comparison. In contrast, we initiated our efforts with a search for peptides that were found at higher levels in the plasma of cancer patients than in the plasma of normal individuals. The advantage of this approach is that, should such peptides be identified, they immediately become candidate biomarkers. This approach eliminates the step at which so many other biomarkers fail, that is, in the experiments necessary to show that proteins expressed at higher levels in tumors can actually be found in the plasma at high levels.
On the other hand, our approach is limited by the low abundance of tumor-specific peptides compared with other peptides found in the plasma. Although we depleted a variety of abundant proteins at the start of our discovery process (Materials and Methods), these proteins cannot be totally removed and peptides derived from them are still at much higher levels than any tumor-specific peptide and can confound analysis. It is possible that many tumor-specific peptides in the plasma escape detection, either because they are masked by more abundant proteins as a result of ion suppression or because they are not at high enough levels to be detected by the Orbitrap mass spectrometer used in the discovery phase of our process.
A second distinguishing feature of our approach is that the analytes are peptides rather than proteins. Several studies have used state-of-the-art proteomics methods to discover new proteins, and then validated them by SRM (15, 16). However, these studies were focusing on developing protein biomarkers through peptides, rather than directly using peptides as biomarkers. One advantage of using peptides is that they are relatively resistant to degradation by proteases; even if the parent protein is degraded by proteases in the extracellular space surrounding the tumor or in the circulation, small peptides may survive. This may be responsible for the observations on α1-antitrypsin that we made during the current study. Fourteen peptides from this protein were confirmed to be detectable in phase 2 of our study (Dataset S6). They did not pass the requirements for a potentially useful biomarker in the subsequent validation phases, but we were able to compare their relative abundances in plasma to those of circulating α1-antitrypsin protein levels previously reported by others. Tountas et al. (17) reported an average increase in α1-antitrypsin protein levels of 1.12-fold (486 ± 18 mg/100 mL vs. 434 ± 13 mg/100 mL) in pancreatic cancer patients over that observed in normal individuals. Pérez-Holanda et al. (18) reported an average increase in α1-antitrypsin protein levels of 1.4-fold in colorectal cancer patients over that observed in normal individuals. In contrast, we found a much larger increase in peptides from the α1-antitrypsin protein: averages of 20-fold, 36-fold, and 59-fold increases in patients with pancreatic cancer, colorectal cancer, and ovarian cancer, respectively, over that observed in normal individuals (Dataset S6). Similarly, we found a 13.3-fold increase of the peptides from another protein, DJ-1, in pancreatic cancer patients, while an ELISA revealed only a 2.9-fold increase in the protein level in a previous study (19). The reason for these dramatic differences are not clear, although we speculate that it could be related to the following factors: binding of the target protein to other proteins or macromolecules in the circulation, thereby masking the antibody-binding site in antibody-based assays; cancer-specific posttranslational modifications of the target proteins, similarly masking its binding to antibodies; or degradation of the released protein in the tumor cells or their environment, destroying the antibody binding site. The differences we noted in the apparent abundances of proteins and their derived peptides in the circulation are not unprecedented. Yassine et al. (20) also reported a discrepancy between antibody-based tests and SRM-based tests for α1-antitrypsin. An ELISA for this protein showed a 1.5-fold increase in α1-antitrypsin proteins in plasma samples from patients with diabetes over that in normal individuals, while an SRM assay on the same samples revealed a 10-fold increase.
PPIA catalyzes the cis–trans isomerization of peptide bonds preceding proline (21). The protein is predominantly located in the cytosol but also can be secreted extracellularly, perhaps accounting for our ability to detect peptides derived from it in plasma. Although there are no prior reports of the use of PPIA as a biomarker, there are many published connections between this protein and cancer. PPIA has been reported to regulate cell proliferation, prevent apoptosis, and defend against oxidative stress (22–24). Microarray analysis showed down-regulation of focal adhesion signaling in response to PPIA knockdown in human endometrial cancer cells (25) and in cholangiocarcinoma cell lines (26). Whether PPIA expression is causally related to the neoplastic process is not essential to its potential use as a biomarker. For example, widely used cancer biomarkers, such as CA19-9 and CA125, are not known to play an etiologic role in the cancer types in which they are used.
In sum, we present a generalizable method for discovering disease-specific peptides in the circulation and present data suggesting that peptides from PPIA may prove to be useful diagnostic markers for ovarian cancer. The next stage of this work will involve development of high-throughput methods to measure PPIA peptides in a large cohort of individuals with ovarian cancers. For this purpose, we are currently attempting to develop antibodies reactive with the two PPIA peptides that might be used for ELISA.
Materials and Methods
Plasma Samples.
Plasma samples from a total of 266 individuals were obtained, comprising 96 healthy individuals, 81 patients with ovarian cancer, 51 with pancreatic cancer, and 38 with colorectal cancer. The plasma samples and clinical data were obtained from The Ontario Tumor Bank, Indivumed, Innovative Research, and The Johns Hopkins Hospital. This study was approved by the Institutional Review Boards for Human Research at each participating institution, and complied with Health Insurance Portability and Accountability Act. Informed consent was obtained from all patients. Selected clinical features of the 266 patients and histopathologic characteristics of their tumors are listed in Dataset S1.
Quantitative Proteomics Assays for Normal and Cancer Plasma Samples.
Plasma samples were prepared for iTRAQ analysis as described in SI Materials and Methods. iTRAQ labeling-dependent quantitative proteomics assays were performed to evaluate the proteomic difference between normal plasma and cancer plasma samples. The pipeline included plasma depletion, denaturation, reduction, alkylation, enrichment for glycoproteins, trypsin digestion, desalting, iTRAQ labeling, strong cation exchange (SCX) cleaning, and bRPLC fractionation followed by Orbitrap MS analysis and quantitative proteomics data analysis using in-house–developed R scripts. Detailed procedures are provided in SI Materials and Methods.
Selection of 641 Peptides as Potential Cancer Biomarkers for Further Validation.
A total of 204 proteins was shared by at least two out of three whole-plasma iTRAQ proteomics datasets. Eighty-seven of these proteins were selected as potential cancer biomarkers for further SRM-based validation based on their abundance test score in the empirical modified eBayes t test (see details in SI Materials and Methods). A total of 461 proteotypic peptides from these proteins was selected as SRM quantifying targets (approximately five target peptides per protein). Of these 461 peptides, 208 were directly observed in our experiments and an additional 253 peptides were added from querying several databases, including PeptideAtlas, PRIDE, etc. (27–29). We also identified 180 peptides in our iTRAQ datasets that did not meet our rigorous criteria for initial selection but which we considered reasonable candidate biomarkers on the basis of their biologic properties. Altogether, we selected 641 SRM target peptides from phase 1 of our study that were carried forward to the validation phase (Dataset S4).
Development of SAFE-SRM Assays.
A total of 4,384 transitions targeting the 641 target peptides in our study was optimized by using synthetic peptides. For each synthetic peptide, a set of optimized collision energies and dwell times was obtained (Dataset S5). An HPLC fractionation was performed to separate the 641 synthetic peptides into 96 fractions based on each peptide’s hydrophobicity in a weak basic environment (pH 8.2). A total of 96 peptide fractions was then organized into 32 groups comprising three sequential fractions each, according to the scheme shown in Fig. S2. Each of these groups was subjected to fractionation through a C18-based HPLC coupled to the Agilent 6490 triple-quadrupole mass spectrometer. SRM assays covering all 4,384 transitions were performed in each of the groups to determine the optimum parameters for detecting each peptide. After identifying the SAFE-SRM fraction group ID for each peptide, a unique SAFE-SRM method was constructed for each fraction group, and the SRM transitions in sequential groups that eluted just before or just after the target group were also incorporated into the method (Fig. S2). The SAFE-SRM group ID for each peptide is listed in Dataset S5, where each ID refers to the bRPLC fractionation plate shown on Fig. S2. After initial method-building steps using standard peptides, we were able to pare the number of groups that needed to be analyzed in the final HPLC-MS step from 32 to 20. A total of 318 of the 641 peptides was reproducibly observed in at least one of these 20 groups, yielding 1,990 detectable transitions (average of 6.3 transitions per peptide).
SAFE-SRM Assays.
The 200-μL plasma samples from each individual were processed using the procedures described in SI Materials and Methods. Lyophilized plasma peptide samples were reconstituted in 2 mL of 10 mM triethylammonium bicarbonate (pH 8.2) with 3% acetonitrile. Peptide fractionation was performed on an Agilent 1260 HPLC system with a C18 column at pH 8.2. The two HPLC mobile phase solvents were 10 mM triethylammonium bicarbonate (solvent A), and 10 mM triethylammonium bicarbonate with 90% acetonitrile (solvent B). A 120-min HPLC gradient method was applied with a flushing step for the first 20 min to remove salt, and this was followed by a 96-min gradient with solvent B increasing from 0 to 100%. The 96 fractions from a plasma peptide sample were collected in a Protein LoBind plate (Eppendorf), and the peptides eluted during each 1-min window were collected in each well. Peptide fractions were combined according to the scheme shown in Fig. S2A and vacuum dried. Dried peptides were then reconstituted using 40 μL of SRM solvent A and spiked with 3 fmol of heavy isotope-labeled K-Ras wild-type (WT) peptides (LVVVGAGGVGK*) before another online fractionation on an Agilent 1290 UHPLC system at pH 3. Fractionated samples were continuously injected into the Jet Stream ESI source of an Agilent 6490 triple-quadrupole mass spectrometer operated in SRM positive-ion mode.
Analysis of SAFE-SRM Assays.
A set of assays composed of 20 different SRM methods for all groups were performed to quantify the abundance of each of the 318 peptides. Twenty datasets were generated by the mass spectrometer using the 20 SAFE-SRM methods for each plasma sample and were imported into Skyline 3.6 for data analysis (30). We improved the labeled reference peptide (LRP) method (31) through a dual-control approach to adjust for the variance of sample preparation efficiency and fluctuations of mass spectrometer sensitivity. The first control was a heavy-isotope–labeled mutant KRAS protein spiked into the plasma sample before sample preparation. The second control was a heavy-isotope–labeled WT KRAS peptide spiked into each group before running on the final HPLC-MS (28). The abundance of a target peptide was represented by the total area under the curve (AUC) of all its transitions normalized to the total AUC of all transitions from the 3-fmol heavy-isotope (heavy-lysine residue)–labeled K-Ras WT peptides (LVVVGAGGVGK). Variations in sample preparation were adjusted by normalizing the abundance of each peptide from a given sample to the abundance of the peptides derived from the heavy-isotope–labeled K-Ras mutant (G12D) protein purchased from Origene. We selected six peptides derived from this heavy-isotope amino acid (heavy-lysine and heavy-arginine)–labeled protein for this adjustment. Peptide sequences and optimized transition parameters are listed in Dataset S5.
A SAFE-SRM abundance score (S) was calculated for each of the 318 peptides in every sample. Assume that Pi,j,k is the integrated intensity of a peptide i in sample j fraction k, Nj,k is the integrated intensity of the K-Ras WT heavy control peptide in sample j, fraction k, and Mj is the integrated intensity of the median abundance K-RAS protein peptide in sample j. Let Si,j be the abundance score of peptide i in sample j; therefore, Si,j can be calculated as follows:
where for Mj:
In this study, 71 out of 318 peptides were repeatedly detected across two adjacent SAFE-SRM groups. The abundance of such peptides in each sample was calculated by summing the normalized abundance scores in adjacent SAFE-SRM runs where the peptides were detected.
Reproducibility of the SAFE-SRM pipeline was measured by calculating the reproducibility ratio (RR) for sample j as follows:
RR value for each sample processed through SAFE-SRM pipeline was listed in Dataset S7.
Cancer Proteomic Biomarker Identification.
To identify the best peptide classifiers, stepwise forward selection logistic regression was employed in MATLAB. First, a logistic regression model was fit to the training set of 50 samples, including 27 known healthy samples and 7, 7, and 9 known colorectal, ovarian, and pancreatic cancer plasma samples using the 318 peptide abundance scores. Leave-one-out cross-validation was used to estimate predictive performance of each model. The peptide yielding the lowest cross-validated misclassification rate on the training set was selected for inclusion in the model. If more than one peptide achieved the lowest misclassification rate, ties were broken by selecting the peptide that produced the greatest model likelihood. This process of selecting a peptide biomarker to be added to the model was repeated until no further decrease in cross-validated misclassification rate could be achieved by addition of a peptide. To find a subset of peptides from the same protein that could achieve perfect classification, the same stepwise forward selection procedure was applied for each potential biomarker protein. After identifying the best classifiers, predictive performance of models fit to different combinations of the peptide biomarkers was compared on an additional 48 samples in a blind manner. The predictive models constructed by combinations of best peptide classifiers and by each individual best peptide classifier were evaluated on an additional cohort of 73 samples in a blind manner.
Supplementary Material
Acknowledgments
This work was supported by The Virginia and D. K. Ludwig Fund for Cancer Research, The Lustgarten Foundation for Pancreatic Cancer Research, The Conrad N. Hilton Foundation, and National Institutes of Health Grants P50-CA62924, CA06973, and GM07309.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited via ProteomeXchange (identifier PXD008094).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712731114/-/DCSupplemental.
References
- 1.Howlader N, et al. SEER Cancer Statistics Review, 1975–2011. National Cancer Institute; Bethesda: 2014. [Google Scholar]
- 2.Fishman DA, et al. The role of ultrasound evaluation in the detection of early-stage epithelial ovarian cancer. Am J Obstet Gynecol. 2005;192:1214–1221; discussion 1221–1222. doi: 10.1016/j.ajog.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 3.Li J, et al. HE4 as a biomarker for ovarian and endometrial cancer management. Expert Rev Mol Diagn. 2009;9:555–566. doi: 10.1586/erm.09.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholler N, Urban N. CA125 in ovarian cancer. Biomarkers Med. 2007;1:513–523. doi: 10.2217/17520363.1.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Gorp T, et al. HE4 and CA125 as a diagnostic test in ovarian cancer: Prospective validation of the Risk of Ovarian Malignancy Algorithm. Br J Cancer. 2011;104:863–870. doi: 10.1038/sj.bjc.6606092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyer VA. U.S. Preventive Services Task Force Screening for ovarian cancer: U.S. Preventive Services Task Force reaffirmation recommendation statement. Ann Intern Med. 2012;157:900–904. doi: 10.7326/0003-4819-157-11-201212040-00539. [DOI] [PubMed] [Google Scholar]
- 7.Clarke-Pearson DL. Clinical practice. Screening for ovarian cancer. N Engl J Med. 2009;361:170–177. doi: 10.1056/NEJMcp0901926. [DOI] [PubMed] [Google Scholar]
- 8.Locker GY, et al. ASCO ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 9.Sallam RM. Proteomics in cancer biomarkers discovery: Challenges and applications. Dis Markers. 2015;2015:321370. doi: 10.1155/2015/321370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crutchfield CA, Thomas SN, Sokoll LJ, Chan DW. Advances in mass spectrometry-based clinical biomarker discovery. Clin Proteomics. 2016;13:1. doi: 10.1186/s12014-015-9102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrads TP, Petricoin EF., 3rd The Obama Administration’s Cancer Moonshot: A call for proteomics. Clin Cancer Res. 2016;22:4556–4558. doi: 10.1158/1078-0432.CCR-16-0688. [DOI] [PubMed] [Google Scholar]
- 12.Kim HJ, Lin D, Lee HJ, Li M, Liebler DC. Quantitative profiling of protein tyrosine kinases in human cancer cell lines by multiplexed parallel reaction monitoring assays. Mol Cell Proteomics. 2016;15:682–691. doi: 10.1074/mcp.O115.056713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harsha HC, et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabb DL, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbatiello SE, et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol Cell Proteomics. 2015;14:2357–2374. doi: 10.1074/mcp.M114.047050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteaker JR, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011;29:625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tountas Y, Sparos L, Theodoropoulos C, Trichopoulos D. Alpha 1-antitrypsin and cancer of the pancreas. Digestion. 1985;31:37–40. doi: 10.1159/000199175. [DOI] [PubMed] [Google Scholar]
- 18.Pérez-Holanda S, Blanco I, Menéndez M, Rodrigo L. Serum concentration of alpha-1 antitrypsin is significantly higher in colorectal cancer patients than in healthy controls. BMC Cancer. 2014;14:355. doi: 10.1186/1471-2407-14-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, et al. DJ-1, a novel biomarker and a selected target gene for overcoming chemoresistance in pancreatic cancer. J Cancer Res Clin Oncol. 2012;138:1463–1474. doi: 10.1007/s00432-012-1205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yassine HN, et al. The application of multiple reaction monitoring and multi-analyte profiling to HDL proteins. Lipids Health Dis. 2014;13:8. doi: 10.1186/1476-511X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, et al. Cyclophilin A promotes human hepatocellular carcinoma cell metastasis via regulation of MMP3 and MMP9. Mol Cell Biochem. 2011;357:387–395. doi: 10.1007/s11010-011-0909-z. [DOI] [PubMed] [Google Scholar]
- 22.Choi KJ, et al. Overexpressed cyclophilin A in cancer cells renders resistance to hypoxia- and cisplatin-induced cell death. Cancer Res. 2007;67:3654–3662. doi: 10.1158/0008-5472.CAN-06-1759. [DOI] [PubMed] [Google Scholar]
- 23.Al-Ghoul M, et al. Comparative proteomic analysis of matched primary and metastatic melanoma cell lines. J Proteome Res. 2008;7:4107–4118. doi: 10.1021/pr800174k. [DOI] [PubMed] [Google Scholar]
- 24.Qi YJ, et al. Proteomic identification of malignant transformation-related proteins in esophageal squamous cell carcinoma. J Cell Biochem. 2008;104:1625–1635. doi: 10.1002/jcb.21727. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Gou J, Xu J. Down-regulation of focal adhesion signaling in response to cyclophilin A knockdown in human endometrial cancer cells, implicated by cDNA microarray analysis. Gynecol Oncol. 2013;131:191–197. doi: 10.1016/j.ygyno.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 26.Obchoei S, et al. Cyclophilin A enhances cell proliferation and tumor growth of liver fluke-associated cholangiocarcinoma. Mol Cancer. 2011;10:102. doi: 10.1186/1476-4598-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desiere F, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, et al. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci USA. 2011;108:2444–2449. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLean B, et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, et al. Methods for peptide and protein quantitation by liquid chromatography-multiple reaction monitoring mass spectrometry. Mol Cell Proteomics. 2011;10:M110.006593. doi: 10.1074/mcp.M110.006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.