Significance
Nanomaterials are developed for treating and diagnosing cancer, but only 0.7% (median) are delivered to a solid tumor. To address this delivery problem, we are examining each biological barrier to determine its impact on tumor delivery. Because the liver sequesters up to 70% of nanomaterials, in this study, we asked, if liver Kupffer cells were removed, what is the impact on tumor delivery? While we demonstrate that the tumor delivery increased up to 150 times, we achieved 2% for nanomaterials of different size, material, and tumor type. This suggests the need to focus on tumor pathophysiology to increase delivery efficiency, since this approach led to a greater availability of nanoparticles in the blood, but 98% did not accumulate in solid tumors.
Keywords: nanoparticle, cancer, liver, macrophage, tumor delivery
Abstract
A recent metaanalysis shows that 0.7% of nanoparticles are delivered to solid tumors. This low delivery efficiency has major implications in the translation of cancer nanomedicines, as most of the nanomedicines are sequestered by nontumor cells. To improve the delivery efficiency, there is a need to investigate the quantitative contribution of each organ in blocking the transport of nanoparticles to solid tumors. Here, we hypothesize that the removal of the liver macrophages, cells that have been reported to take up the largest amount of circulating nanoparticles, would lead to a significant increase in the nanoparticle delivery efficiency to solid tumors. We were surprised to discover that the maximum achievable delivery efficiency was only 2%. In our analysis, there was a clear correlation between particle design, chemical composition, macrophage depletion, tumor pathophysiology, and tumor delivery efficiency. In many cases, we observed an 18–150 times greater delivery efficiency, but we were not able to achieve a delivery efficiency higher than 2%. The results suggest the need to look deeper at other organs such as the spleen, lymph nodes, and tumor in mediating the delivery process. Systematically mapping the contribution of each organ quantitatively will allow us to pinpoint the cause of the low tumor delivery efficiency. This, in effect, enables the generation of a rational strategy to improve the delivery efficiency of nanoparticles to solid tumors either through the engineering of multifunctional nanosystems or through manipulation of biological barriers.
Nanomaterials are designed as drug carriers for the treatment of cancer, but few nanomedicines have advanced to clinical use. A recent metaanalysis showed that less than 0.7% (median) of administered nanoparticles are delivered into solid tumors (1). This low delivery efficiency can be attributed to phagocytic cells of the mononuclear phagocytic system sequestering nanomaterials due to a combination of cellular and physical properties. For example, Tsoi et al. (2) showed that, when nanomaterials enter the liver, the flow rate of the nanomaterials slows by 1,000 times in the liver sinusoid to increase interaction and uptake by hepatic Kupffer cells, B cells, and endothelial cells. The most common strategies to prevent phagocytic uptake by the liver is by coating the nanoparticle surface with neutrally charged antifouling polymers, such as polyethylene glycol (PEG), or by manipulating size and shape (3, 4). Even though this strategy has led to an increased blood half-life and tumor accumulation, these strategies do not allow us to determine the contribution of phagocytic cells in mediating the delivery process and have not effectively advanced nanomedicine formulations to the clinic. This begs us to ask the question, “What is the impact on nanoparticle delivery to tumors after removing some or all of the main liver cells that sequesters them?” Addressing this question allows us to determine the contribution of these cell types to the delivery process quantitatively. The results of these studies will enable us to systematically focus on the “culprit” biological system to increase delivery in future studies. Improving delivery efficiency to tumors is likely a balancing act between different organs. To figure this out, we must analyze one organ at a time.
Addressing this question requires a tool to deplete phagocytic liver cells. In our analysis of the literature, there have been a number of studies that have characterized different agents for such a purpose. Examples include gadolinium chloride (5, 6), methyl palmitate (7, 8), dextran sulfate (500 kDa) (9), carrageenan (10, 11), and clodronate liposomes (12). Ohara et al. (12) went a step further in their analysis and demonstrated the impact of depleting liver Kupffer cells in mediating the therapeutic effectiveness of Doxil by measuring tumor volume. It was difficult to determine the impact of the Kupffer cell depletion on nanoparticle delivery because Doxil can serve as a “chemotherapeutic agent as well as a macrophage inhibitor.” However, the results of these studies provided a chemical tool (e.g., clodronate liposome for Kupffer cell depletion) that allowed us to elucidate how liver macrophages impact the nanoparticle delivery process. This enabled the probing of the influence of nanoparticle formulations and tumor types in mediating the delivery process. Furthermore, by carefully selecting the proper nanoparticle type, we quantified the delivery process to allow our results to be fully compared in future studies. As the field of nanomedicine starts to database these quantitative results, analysis of a large dataset will lead to rational engineering strategies to overcome the delivery problem. In this study, we fully determined the impact of dose-dependent removal of Kupffer cells on nanoparticle tumor delivery and evaluated the toxicity to the animal after removal of these cells. We showed that removing all or a portion of the Kupffer cells can increase delivery of nanoparticles to the tumor up to 150 times. These enhancements were dependent on nanoparticle size, material composition, and tumor type.
Results
We designed a series of experiments to determine the impact of the liver Kupffer cells in mediating the tumor delivery process. These experiments involved the (i) dose-dependent removal of macrophages using dicholoromethylenediphosphonic acid liposomes (clodronate liposomes) (13) and then (ii) administration of nanoparticles (gold nanoparticles, silver, silica, and liposomes) in different xenograft models (ovarian, breast, skin, prostate, and lung cancer) (Fig. 1). At the beginning of this project, we focused on gold nanoparticles because of the versatility in engineering their sizes and surface chemistries and ease of quantification using both optical imaging (via fluorescent labeling) and elemental analysis with inductively coupled plasma–mass spectrometry (ICP-MS).
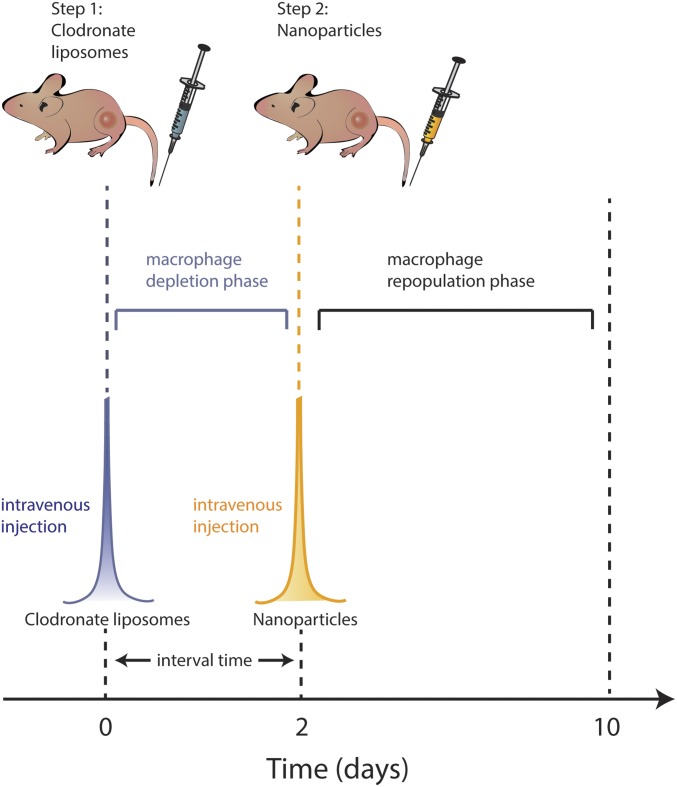
Fig. 1.
Two-step injection scheme to improve the delivery of nanoparticles to the tumor. Clodronate liposomes are i.v. injected into the mouse via the tail vein to remove a portion of Kupffer cells within 48 h. Gold nanoparticles are then i.v. injected. Removal of a portion of Kupffer cells ensure host immunity is not significantly suppressed and provides improved pharmacokinetics of gold nanoparticles by preventing off-target accumulation in the liver. Nanoparticle transport through the bloodstream is enhanced, and the subsequent delivery to the tumor is increased. Within 10 d from clodronate liposome administration, Kupffer cells are repopulated in the liver.
Gold Nanoparticle Functionalization.
We designed a library of gold nanoparticles (AuNPs) for assessment. AuNPs were synthesized, surface modified, and tagged with Alexa Fluor 750 (A750) as described previously by our group (14, 15). The surfaces of the nanoparticles were first functionalized with a mixed ligand film composed of 5-kDa methoxy-terminated PEG (5KmPEG) and 10-kDa amino-terminated PEG (10KamPEG) at a 4:1 ratio. The mixed PEG ligands were grafted to the nanoparticle surface at a density of 5 PEG·nm−2 to passivate the surface against the adsorption of serum proteins (4). The AuNPs were characterized using transmission electron microscopy, absorbance and fluorescence spectroscopy, and dynamic light scattering (SI Appendix, Figs. S1 and S2), and the results are summarized in Table 1.
Table 1.
Gold nanoparticle physicochemical properties
| Nanoparticle | Physicochemical properties | ||||
| Size | Surface ligands | Inorganic diameter,* nm | Hydrodynamic diameter, nm | ζ potential, mV | λLSPR, nm |
| 15 | Citrate | 15.4 ± 1.6 | 18.9 | −28.0 ± 0.8 | 519 |
| 50 | Citrate | 52.7 ± 6.9 | 57.9 | −36.7 ± 2.2 | 548 |
| Alexa 750–PEG | 94.7 | −9.9 ± 0.3 | |||
| 100 | Citrate | 109.1 ± 9.8 | 109.1 | −40.5 ± 1.1 | 582 |
| Alexa 750–PEG | 146.7 | −13.4 ± 0.6 | |||
| 200 | Citrate | 215.5 ± 15.7 | 190.9 | −48.2 ± 2.4 | 581, 823 |
| Alexa 750–PEG | 225.5 | −13.6 ± 0.7 | |||
Inorganic diameter was measured using transmission electron microscopy, and histogram distributions were compiled from the counting of ∼100 particles using ImageJ software. For citrate-stabilized gold nanoparticles, measurements were acquired in 0.02% (wt/vol) citrate solutions with the exception of 15-nm AuNPs; particles were measured as synthesized. For remaining AuNPs, hydrodynamic diameters were measured in PBS and ζ potentials were recorded in Hepes buffer. Precision reported is the SD of triplicate measurements; nanoparticle samples having a polydispersity index > 0.1 were discarded.
Characterization of Depletion of Phagocytes in the Liver and Spleen.
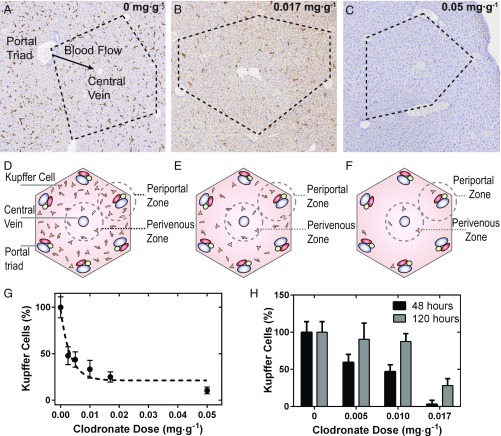
Clodronate liposomes were effective in killing phagocytic cells. Mice injected with clodronate liposomes (0–0.05 mg·g−1) were killed at 48 h postinjection. Clodronate liposomes depleted F4/80+ hepatic and splenic macrophages proportionally to dose (Fig. 2 A–C and SI Appendix, Fig. S3). The liver histological sections suggested that phagocytic Kupffer cells near the portal triads were depleted to a greater extent in comparison with the Kupffer cells near the central vein (16) (Fig. 2 A–C). It has been shown that 43% of Kupffer cells are located between adjacent portal triads. The number decreases successively to 25% toward the central vein (17). Blood flows from numerous portal triads into a single central vein (black arrow in Fig. 2A) (18), and therefore cells located near the portal triads are the first to interact with the nanoparticles. The relative number of Kupffer cells compared with controls decreased to 25% and to 10% when 0.017 and 0.050 mg·g−1 of clodronate liposomes were administered, respectively (Fig. 2). The annotations shown in Fig. 2 D–F are representative of the above histological sections. In Fig. 2 B and C, clear Kupffer cell depletion zones can be seen between adjacent portal triads. Furthermore, dosimetric depletion influenced the repopulation of Kupffer cells (Fig. 2H) in BALB/c mice. Our data suggest that, when only a portion of Kupffer cells are depleted, the livers of these animals repopulate to normal levels in 5 vs. 10 d. It is noteworthy that BALB/c mice were seen to have less Kupffer cells than CD1 nu/nu mice in comparison of hepatic histology (SI Appendix, Fig. S4).
Fig. 2.
Histological sections of mouse liver tissue immunohistochemically stained with anti–F4/80-HRP 48 h after administration of 0–0.05 mg·g−1 of clodronate liposomes (A–C). The histological sections are schematically represented in D–F. The liver contains a hexagonal microvascular repeating unit known as the hepatic lobule (black dashed lines). Blood flows from several portal triads (portal vein, hepatic artery, and bile ducts) and drains into a single central vein. At increasing doses of clodronate liposomes, clear depletion zones of Kupffer cells can be seen in periportal regions and between adjacent portal triads compared with perivenous regions (B and E) relative to controls (A). At a dose of 0.05 mg·g−1, near-complete depletion of Kupffer cells can be seen in the hepatic lobule shown in C. (G) Normalized percentage of Kupffer cells counted from fixed areas of CD1 nu/nu hepatic tissue sections from animals treated with clodronate liposomes (0.00–0.05 mg·g−1). (H) Depletion and repopulation of Kupffer cells in BALB/c mice. The number of Kupffer cells from fixed areas of tissue were quantified at 120 h and were normalized to the initial amounts at 48 h after administration of clodronate liposomes. It was found that 0.017 mg·g−1 was sufficient to completely deplete the livers in this strain of mice.
Clodronate Liposomes Improve the Pharmacokinetics and Delivery of Nanoparticles to Tumors.
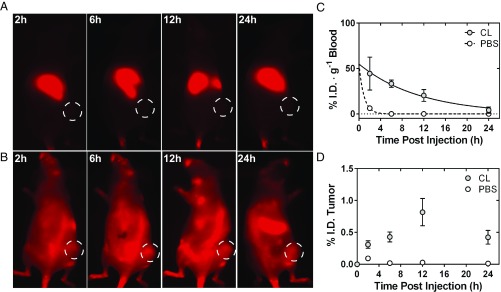
We first asked, if both the liver and spleen were completely depleted of phagocytic macrophages, what is the maximum increase in nanoparticle tumor delivery? Mice were injected with clodronate liposomes or PBS liposomes 48 h before administration of 100-nm AuNPs to deplete the liver and splenic macrophages (19). There was a redistribution of the AuNPs after administration of clodronate liposomes (Fig. 3 A and B). We also found that, after 48 h, gold nanoparticle uptake in the liver was minimized to less than 10% injected dose (I.D.) (SI Appendix, Fig. S5). Since fluorescence is a semiquantitative method, we further quantified the distribution of the AuNPs by using ICP-MS (Fig. 3 C and D). Clodronate liposomes increased the blood half-life of 100-nm AuNPs 13 times, from 0.64 to 8.00 h and increased their delivery to the tumor from 0.03 to 0.5% I.D. The full biodistribution analysis is shown in the SI Appendix, Fig. S6. The removal of phagocytic cells redistributed the administered nanoparticles and led to an increase in tumor accumulation of 13 times, 6 times higher than previously reported by Ohara et al. (12).
Fig. 3.
(A) In vivo fluorescence images of a single CD1 nu/nu SKOV3 xenograft at 2–24 h, after the i.v. injection of AuNPs that was treated with PBS liposomes. (B) Analogous images of a single animal after administration of AuNPs, albeit this animal was injected with clodronate liposomes 48 h prior. Improved circulation of the AuNPs can be visualized by a clearly resolved whole-body fluorescence image with fluorescence emerging from the position of the tumor xenograft (dashed white lines). (C) The normalized blood half-life of AuNPs as determined by ICP-MS in animals treated with clodronate liposomes (gray) and those with PBS liposomes (white). The curves were fit as single-phase exponential decays, and the administration of clodronate liposomes improved the half-life of the AuNPs to 8.00 h from 0.64 h. (D) The pharmacokinetic accumulation of AuNPs at the SKOV3 tumor from 2 to 24 h after administration of clodronate (gray) and PBS liposomes (white). The average tumor accumulation as determined by the integrated area under curve (AUC) as determined by ICP-MS was found to be 0.5% I.D. in animals treated with clodronate liposomes where control showed 0.03% I.D. CL, clodronate liposomes.
How Does Macrophage Depletion Relate to Nanoparticle Size?
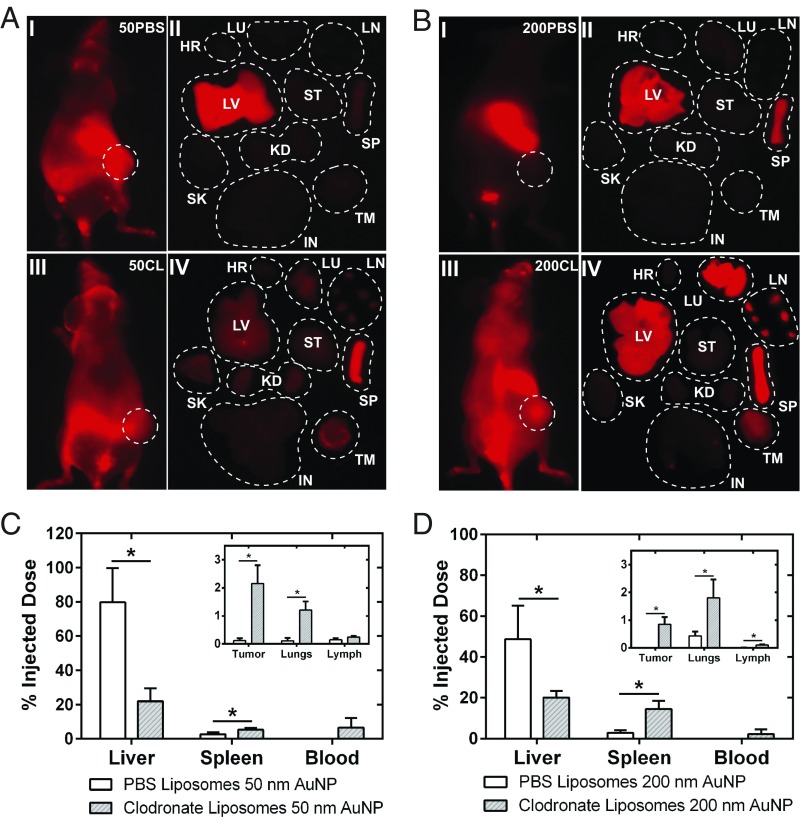
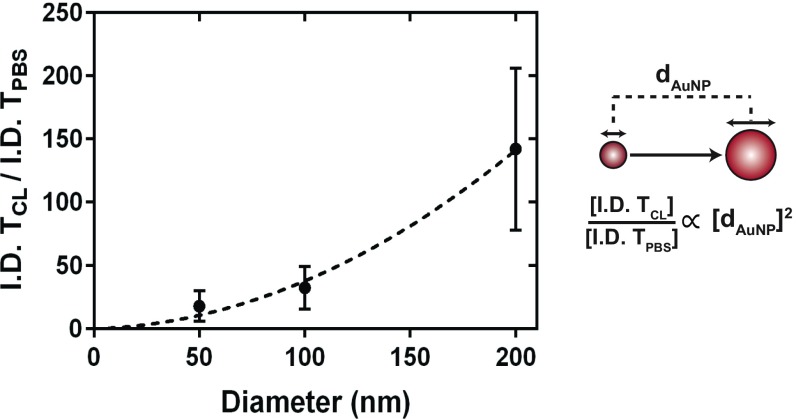
We next evaluated the impact of nanoparticle size in mediating the improvement in delivery efficiency. Macrophages are well-known to have a size-dependent interaction with nanoparticles (3). A range of AuNP size (50–200 nm) was selected. Fig. 4 A and B shows 24-h in vivo whole-animal fluorescence images and ex vivo fluorescence images of organs from animals that were administered 50- and 200-nm AuNPs, respectively. The whole-animal fluorescence images in Fig. 4A, I and III, appear similar, albeit the ex vivo organ fluorescence images in II and IV differ significantly. A similar redistributive trend was seen for 50-nm AuNPs that was displayed previously for 100-nm AuNPs in SI Appendix, Fig. S6. A four times reduction in nanoparticle accumulation in the liver liberated an excess of 250 times more nanoparticles in the blood after administration of clodronate liposomes (Fig. 4C). For these 50-nm AuNPs, we observed an increase in delivery efficiency to the tumor from 0.12 to 2.15% I.D. at 24 h. For 200-nm AuNPs, clear differences can be seen from the in vivo animal fluorescence images shown in Fig. 4B, I and III, resembling the biodistribution pattern for 100-nm AuNPs. The data in Fig. 4A show substantially improved pharmacokinetics for 200-nm AuNPs in comparison with the 50-nm AuNPs (Fig. 4B). Quantitative analysis of the 200-nm AuNP biodistribution using ICP-MS (Fig. 4D) highlighted an improvement of 150 times in the delivery efficiency of AuNPs to the tumor from <0.01 to 0.85% I.D. at 24 h. There was a direct relationship between nanoparticle size and delivery efficiency (Fig. 5), and the relationship is defined by Eq. 1:
| [1] |
Our data suggest that the relative increase in nanoparticle tumor delivery is proportional to the diameter of the nanoparticle when macrophages are depleted. The most substantial increase (150 times) was seen for 200-nm AuNPs. It is noteworthy that 50-nm AuNPs had the highest accumulation in the tumor at 24 h (2.15% I.D.) across all three nanoparticle sizes investigated and represents a 20-times increase relative to control animals (0.12% I.D.). Mathematical modeling of these data provides insight into the relative delivery efficiency that could be expected for a specific nanomaterial size. These results suggest that this two-step delivery strategy can improve the delivery of larger nanomaterials to tumors.
Fig. 4.
Biodistribution of 50- and 200-nm AuNPs 48 h after clodronate liposome administration. (A) In vivo animal (I + III) and ex vivo organ (II + IV) fluorescence images 24 h after tail vein injection of 50-nm AuNPs in control (Top)- and clodronate liposome (Bottom)-treated animals. Organ abbreviations: HR, heart; IN, intestines; KD, kidneys; LU, lungs; LV, liver; SK, skin; SP, spleen; ST, stomach; TM, tumor. The quantitative organ distribution as determined by ICP-MS is shown in C. A reduction in uptake of nanoparticles in the liver translated to a near 20-times increase in delivery of nanoparticles to the tumor. (B) In vivo animal (I + III) and ex vivo organ (II + IV) fluorescence images 24 h after administration of 200-nm AuNPs in animal treated with PBS liposomes (Top) and clodronate liposomes (Bottom). The quantitative organ distribution is shown in D. A twofold decrease in liver accumulation of 200-nm AuNPs translated to 142-times increase in the delivery of the nanoparticles to the tumor. Statistical significance was measured at *P < 0.05, **P < 0.01, and ***P < 0.001.
Fig. 5.
The delivery of AuNPs to SKOV3 tumors is improved as a function of nanoparticle diameter (dAuNP) using clodronate liposomes 48 h before AuNP administration. The relative increase in AuNP tumor delivery efficiency with clodronate liposomes (I.D. TCL) compared with controls (I.D. TPBS) was 18, 32, and 150 times for nanoparticle diameters of 50, 100, and 200 nm, respectively.
Gold Nanoparticle Tumor Delivery Efficiency Is Dependent on Clodronate Liposome Dose and Cancer Type.
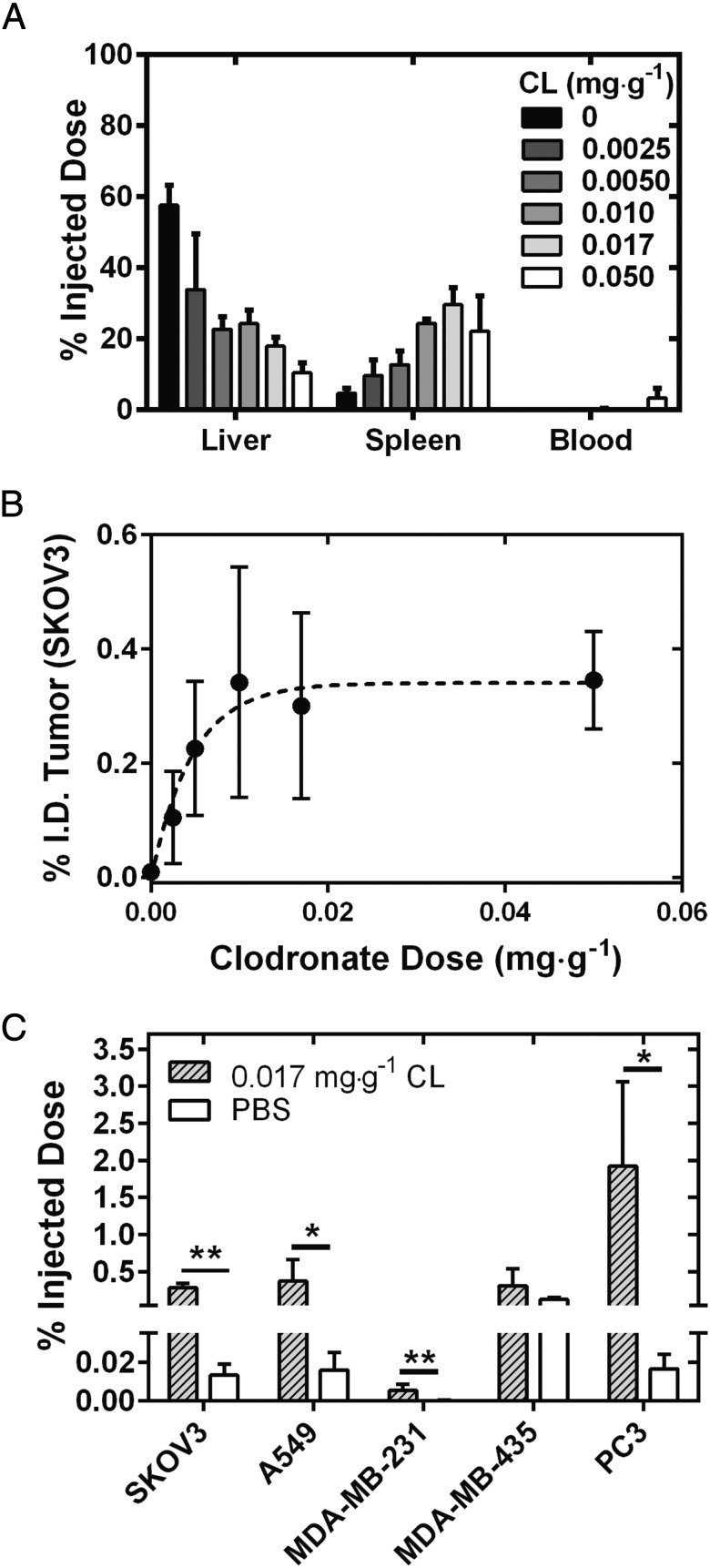
In the previous experiment, we used a dose of clodronate liposomes to completely remove all hepatic and splenic macrophages. However, if such a high dose (0.05 mg·g−1 of animal body weight) is required to improve AuNP delivery, this approach may not be viable for use in patients as Kupffer cells are an integral component of the innate immune system. Thus, we opted to evaluate whether improvements in nanoparticle delivery efficiency could be realized at lower doses of clodronate liposomes, leaving the host with some intact innate macrophages. Four additional doses of clodronate liposomes were investigated in the range of 0.0025 to 0.0170 mg·g−1 of animal body weight, and each dose was injected into the animal 48 h before AuNP administration. Analysis of the total gold content in the AuNP organ biodistribution also displayed a dose-dependent decrease in the amount of gold in the liver, with incremental increases in accumulation in the spleen as shown in Fig. 6A. The total amount of AuNPs in the liver decreased successively from 58 ± 6% I.D. (control) to 10 ± 3% I.D. Depleting 50% of Kupffer cells resulted in a 10-times increase in delivery of AuNPs to the tumor at 0.0025 mg·g−1 of clodronate liposomes (Fig. 6B). Interestingly, no statistically significant improvements were seen in AuNP tumor accumulation beyond 0.005 mg·g−1 dose of clodronate liposomes. These data suggest that complete hepatic and splenic depletion is not required to maximize the enhancement in tumor delivery of subsequently administered nanoparticles using this two-step delivery strategy. We next determined whether the improved AuNP delivery efficiency after clodronate liposome administration occurs in tumors beyond the SKOV3 ovarian cancer xenograft model. We investigated the delivery efficiency of AuNPs after administering a lower dose of clodronate liposomes (0.017 mg·g−1) in a s.c. xenograft of non–small-cell lung cancer (A549), and in orthotopic xenografts of breast cancer (MDA-MB-231), skin cancer (MDA-MB-435), and prostate cancer (PC3). We observed improvements of 20-fold relative to control animals for SKOV3, A549, and MDA-MB-231 tumors, while almost a 100-fold increase was seen in animals with PC3 orthotopic xenografts for the delivery of 100-nm AuNPs 48 h after administration of clodronate liposomes (Fig. 6C). Interestingly, no significant improvement in tumor delivery efficiency was seen for the MDA-MB-435 tumor model. This suggests that tumors present unique intrinsic barriers to nanoparticle accumulation. Of all of the tumor models investigated, PC3 tumors had some of the largest masses (0.7700 ± 0.4109 g), while MDA-MB-231 tumors grown on the mammary fat pad had the smallest masses (0.0078 ± 0.0065 g) (SI Appendix, Table S1). The results showed the highest and lowest AuNP accumulation of 1.92% I.D. and <0.01% I.D., respectively. These data are consistent with a previously reported study suggesting that tumor pathophysiology influences nanoparticle uptake (20). PC3 tumors also displayed the highest mass normalized AuNP accumulation of 2.09% I.D. per gram at 24 h (SI Appendix, Fig. S7). We saw no depletion of tumor-associated macrophages in SKOV3 tumors after administration of clodronate liposomes (SI Appendix, Fig. S8).
Fig. 6.
(A) Biodistribution at 24 h of 100-nm AuNPs as measured by ICP-MS in the liver, spleen, and blood at an increasing dosage of clodronate liposomes. A gradual decrease from 80% I.D. to less than 20% I.D. was seen for the liver, while the accumulation of AuNPs in the spleen increased from 10 to 40% I.D. It was found that less than 1% of AuNPs were seen circulating in blood at all other dosages of clodronate liposomes. (B) AuNP tumor accumulation as a percentage of the injected dose for increasing dosages of clodronate liposomes. (C) Administration of clodronate liposomes at a dose of 0.017 mg·g−1 improved the global delivery of nanoparticles to four of five xenograft tumor models investigated. The increased accumulation of A750-AuNPs in SKOV3 and A549 s.c. xenografts and MDA-MB-231 and PC3 orthotopic xenografts was 20–100 times. Statistical significance was measured at *P < 0.05, and **P < 0.01. CL, clodronate liposomes.
Macrophage Depletion Enhances the Tumor Delivery of Various Nanomaterials.
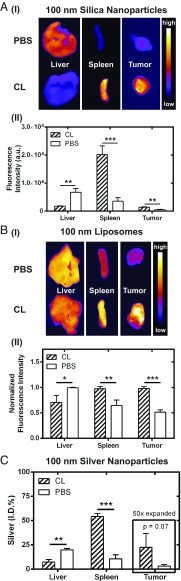
We evaluated whether macrophage depletion could improve the delivery of different nanoparticle types. To encompass a broad classification of materials, we selected silica nanoparticles (SiNPs), nanoliposomes, and silver nanoparticles (AgNPs) at a fixed diameter of 100 nm and surface modified with PEG ligands (SI Appendix, Figs. S9 and S10 and Table S2). Fig. 7 shows the hepatic, splenic, and tumoral tissue distribution of SiNPs, nanoliposomes, and AgNPs at 3 h postinjection, respectively. The clodronate liposome dose used was 0.017 mg·g−1, and animals were injected with clodronate liposomes or PBS liposomes 48 h before nanoparticle administration. Macrophage depletion increased the delivery of all particles to the tumor (Fig. 7). Enhancements in tumor delivery were two, five, and seven times for nanoliposomes, SiNPs, and AgNPs, respectively. Collectively, the data in Figs. 6 and 7 highlight the universality of this two-step delivery strategy to improve the tumor delivery of nanomedicines, both across various nanomaterials and types of cancer and using a dose that does not result in complete hepatic and splenic macrophage depletion.
Fig. 7.
(A, I) Ex vivo fluorescence images showing the distribution of 100-nm SiNPs in livers, spleens, and tumors 48 h after administration of clodronate liposomes (CL) or PBS liposomes (PBS). The average integrated fluorescence intensity of the corresponding tissues is shown in II. (B) The distribution of 100-nm nanoliposomes in the liver, spleen, and tumors is shown qualitatively by the ex vivo fluorescence images in I, and the average integrated fluorescence intensity normalized on an organ basis is given in II. (C) The organ distribution of 100-nm AgNPs was determined using ICP-MS. The silver signal from each tissue was normalized as a percentage of the injected dose. The Ag signal in the tumors was expanded by 50-fold for illustration on the same axis scale. Macrophage depletion reduced the liver accumulation of all three types of nanoparticles with resultant increases to splenic and tumor tissue. Nanoparticle delivery to the tumor increased between two and seven times and was dependent on the nanoparticle type. The dose of clodronate liposomes was 0.017 mg·g−1; mice were killed 3 h after injection of nanoparticles. Animals used in these experiments were SKOV3 tumor xenografts in CD1 nu/nu mice (n = 3–4). Statistical significance was measured at *P < 0.05, **P < 0.01, and ***P < 0.001.
Macrophage Depletion Causes No Systemic Toxicity but Increases Disease Onset in a Model of Polymicrobial Sepsis.
The data regarding enhancements in nanoparticle tumor delivery after macrophage depletion with clodronate liposomes are promising but arguably represents a double-edged approach in comparison with materials or drug design. Therefore, we asked, “What is the consequence of this immune suppression on the animal and could this impair their ability to combat infection?”
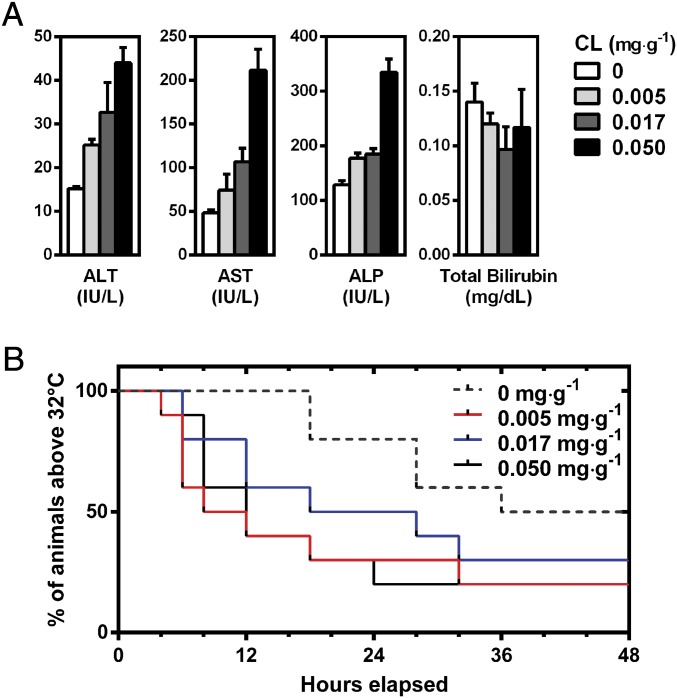
We first conducted a toxicology assessment using hematology and liver biochemistry assays on animals treated with a dosing scheme of clodronate liposomes from 0.00 to 0.05 mg·g−1 (Fig. 8A and SI Appendix, Figs. S11 and S12 and Tables S3–5). We saw an increased quantity of the parenchymal markers alanine aminotransferase (ALT), aspartate aminotransferase (AST), and alkaline phosphatase (ALP) after treatment with an increasing dose of clodronate liposomes, albeit the marker concentrations are still within the normal range for these animals (21). These observations have also been reported elsewhere when Kupffer cells were depleted with clodronate liposomes (22). Histological sections of CD1 nu/nu mice hepatic tissue showed no systemic or gross inflammation relative to controls (SI Appendix, Fig. S13). A similar dose-dependent depletion of Kupffer cells and splenic macrophages was seen in C57BL/6 mice (SI Appendix, Figs. S14 and S15). Furthermore, no statistically significant differences in hematological markers were seen across the doses of clodronate liposomes investigated against control animals (SI Appendix, Figs. S11 and S12).
Fig. 8.
(A) Hepatotoxicity analysis of serum markers alanine transaminase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and total bilirubin after i.v. administration of clodronate liposomes (0–0.05 mg·g−1). Data are represented as an average of n = 3, where error bars illustrate the SD of the measurement. (B) Survival curves of animals administered clodronate liposomes (0–0.05 mg·g−1) followed by induction of sepsis 48 h through CLP. Survival was defined by animals exhibiting hypothermia with a body temperature higher than 32 °C. Macrophage depletion with clodronate liposomes increased the onset of sepsis and reduced median survival time by threefold and overall survival by up to 30%. There was no statistical significance between animals receiving 0.017 mg·g−1 of clodronate liposomes and control animals (P = 0.19 by Mantel–Cox log-rank test). Each group consisted of n = 10 animals.
We sought to gain insight in the consequence of the host acquiring a severe microbial infection when macrophages were at their lowest levels. We selected an animal model of polymicrobial sepsis to answer this question as this is a severe complication that could arise from an infection. Furthermore, sepsis is a common problem for many patients receiving chemotherapy (23), and Kupffer cells are integral toward combating sepsis (24, 25). The data in Fig. 8B show total animal survival 48 h after induction of sepsis intraoperatively through a cecal ligation and puncture (CLP) procedure (26). We confirmed induction of the sepsis animal model by bacterial culture of blood collected from septic animals (SI Appendix, Fig. S16). Mice with body temperature below 32 °C were killed according to ethics guidelines of the animal use protocol where the endpoint was considered as a morbid state. Animals in groups treated with clodronate liposomes from 0.005 to 0.050 mg·g−1 are shown by the red, blue, and solid black lines in Fig. 8B. Clodronate liposomes increased the disease onset of sepsis where only 20% and 30% of animals exhibited mild hypothermia with body temperatures greater than 32 °C for all three treatment groups. Interestingly, there was no statistically significant difference between the group treated with 0.017 mg·g−1 clodronate liposomes and the control (P = 0.19, Mantel–Cox log rank test). However, the overall median survival for control animals was 38 h. Those animals receiving clodronate liposomes at 0.05, 0.017, and 0.005 mg·g−1 decreased to 12, 23, and 10 h, respectively. Our data show that survival was not dependent on clodronate liposome dose; rather, any degree of depletion increased the onset of sepsis. These data suggest that, if a patient were to acquire an acute infection that led to sepsis when their innate immunity was at peak suppression, their prognosis would be less favorable. Since depleting only a portion of Kupffer cells results in faster repopulation (Fig. 2H), this minimizes the duration of time between clodronate liposome and nanoparticle injection cycles that a patient is left with a weakened innate immune system. This reduces the risk of infection without sacrifice to enhancement in nanoparticle tumor delivery.
Discussion
The purpose of designing long-circulating nanoparticles is to enable a high concentration of nanoparticles to accumulate in the tumor. Here, we focused on the removal of different amounts of these nanoparticle sequestration cell types in the liver to determine their contribution to the delivery process. While our results show drastic improvements in the delivery efficiency of nanoparticles to tumors when these cells were removed, we were surprised that the increase was not more dramatic. By removing the contribution of these cells to the delivery process, we show that they are likely to contribute only 2% (maximum) to the delivery problem. This begs the issue of what are the contributions from the other organs, especially tumor (and how their pathophysiology mediates this process).
The calculation of tumor delivery efficiency assumes the nanoparticles are resident in the vessel and/or have entered into the tumor stroma over a period of time. In our strategy, we removed the liver cells that compete for the nanoparticles, which should theoretically allow a higher concentration to be available for tumor accumulation. However, if they do not accumulate in the target diseased tissue, they stay in circulation or be taken up by other cell types in the liver and potentially by cells in other organs. If they circulate long enough, they may be taken up by repopulated Kupffer cells. Hence, a high delivery efficiency to the tumor is a balancing act between the tissue kinetics of the different organs that are involved in the delivery process. This study shows the liver Kupffer cells may only contribute 2% to the delivery efficiency.
A next step to addressing the “delivery problem” would be to evaluate how each organ contributes to the delivery process. This may involve analysis of the spleen, lymph nodes, skin, and other organs that have been reported to sequester nanoparticles, as well as the tumor system. A full understanding of the contribution of each organ, from the organ, cellular, and molecular level, would help us to develop rational strategies to overcome the poor delivery efficiency. These results would allow us to consider how to design a multifunctional nanosystem that can avoid biological barriers, or how to use combinatory strategies to alter biological systems for effective nanoparticle delivery to the tumor. These basic studies are important to solving the “delivery problem.”
Materials and Methods
Mammalian Cell Culture.
Ovarian adenocarcinoma (SKOV3), breast adenocarcinoma (MDA-MB-231), melanoma (MDA-MB-435S), and nonsmall cell lungs carcinoma (A549) cell lines were cultured for inoculation of tumor xenografts. The SKOV3, MDA-MB-231, and A549 cell cultures were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin in T-175 tissue culture flasks (Nest Biotechnology Company) and kept at 37 °C in a 5% CO2 humid environment. Cells were passaged at 85% confluence and subdivided in 1/4 to 1/6 in fresh T-175 flasks. MDA-MB-435S cells were cultured identically except RPMI supplemented with 10% FBS, and 1% penicillin/streptomycin was used in place of DMEM.
Preparation of Human Tumor Xenograft Models.
All animal procedures and husbandry were conducted in accordance with the guidelines from the animal ethics committee from the Division of Comparative Medicine at the University of Toronto (animal protocol numbers 20010886, 20011813, and 20011962). Six-week-old female CD1 nu/nu mice were purchased from Charles River Laboratories. Animals were acclimatized for 1 wk following delivery. T-175 culture flasks at 85–90% confluence containing a given cell line were harvested using 0.25% (wt/vol) trypsin for 10 min at 37 °C in 5% CO2. The cell suspensions were centrifuged at 500 × g for 5 min, and the supernatant was discarded. The cell pellets were suspended in 1 mL of DMEM or RPMI, and the cell aliquot was counted with a hemacytometer. A 500-µL aliquot of the stock was mixed with 500 µL of Matrigel (Thermo Fisher Scientific) and kept on ice. To inoculate tumors, mice were subject to anesthesia initially at 5% (vol/vol) isoflurane in oxygen and later maintained using 3% (vol/vol) isoflurane in oxygen. Using a 1-mL syringe equipped with a 25-gauge needle, 200 µL of the stock cell–Matrigel mixture was injected s.c. in the animal’s right hind flank (SKOV3, A549, and MDA-MB-435S). For MDA-MB-231 orthotopic xenografts, the cell suspension was inoculated between the inguinal fourth and fifth mammary fat pad on either the right or left side (27). For SKOV3, A549, and MDA-MB-435S xenografts, between 4 and 6 × 106 cells were injected, while for MDA-MB-231, 6–8 × 106 cells were inoculated. Animals were then removed from anesthesia and allowed to recover under ambient conditions for 10 min before being placed back in their respective housing. Tumor growth was monitored by the Division of Comparative Medicine for ulceration. Mice were selected for experiments after tumor volume reached 1 cm3; MDA-MB-231 xenografts were selected when the tumor volume was 0.8 cm3. The PC3 orthotopic tumor xenografts were a gift from Gang Zheng, Toronto General Hospital, Toronto. Tumor volume was monitored using MRI, and animals were selected when the diameter reached 7 mm. Details on the inoculation of these xenografts can be found elsewhere (28).
Administration of Clodronate or PBS Liposomes, and Gold Nanoparticles.
Mice were tail vein injected with 200 µL of clodronate liposomes suspension (0.05 mg·mL−1) or 200 µL of saline (PBS) liposomes (control) using a 29-gauge insulin needle. For an average mouse of 20 g of body weight, this injected dose resulted in both hepatic and splenic macrophage depletion as stated by the manufacturer (13). For experiments investigating dose-dependent macrophage depletion, the stock clodronate liposomes were diluted to 0.017, 0.01, 0.005, and 0.0025 mg·mL−1 with sterile 1× PBS. After 48 h, A750-AuNPs were suspended in sterile PBS and injected into the animal via the tail vein. A fixed injection volume of 150 µL was used in the administration of all AuNPs. The injected dose was normalized to the total nanoparticle surface area across the different-sized AuNPs investigated. The surface area of a single AuNP with radius r was assumed as a sphere. For AuNPs with average diameters of 50, 100, and 200 nm, the surface areas were assumed to be 7,854, 31,416, and 125,664 nm2, respectively. For a standardized total injected surface area of 3.18 × 1015 nm2, the concentrations of 50-, 100-, and 200-nm AuNPs administered were 4.0, 1.12, and 0.28 nM, respectively.
In Vivo Fluorescence Imaging of Gold Nanoparticle Biodistribution.
Following administration of A750-AuNPs, animals were imaged using a Kodak in vivo multispectral imaging system (Bruker Corporation) at 2, 6, 12, and 24 h postinjection. Mice were initially placed under 4% (vol/vol) isoflurane in oxygen, and the anesthesia was maintained at 3% (vol/vol) during the course of the image. Excitation and emission bandpass filters at 750 and 830 nm (Carestream Health), respectively, were used in the acquisition of A750-AuNP fluorescence from whole-body animal images. The bandpass of each filter was ∼35 nm at FWHM. Images were acquired with an exposure and integration time of 10 min. Fluorescence images were modified using ImageJ software to enhance contrast and clarity; a fixed contrast setting was used to present fluorescence images from both experimental and control animals. To qualitatively assess AuNP biodistribution at the organ level, ex vivo images of extracted organs were also taken in Petri dishes. Identical organs from clodronate liposome-treated and PBS liposome-treated animals were also paired and imaged in groups for qualitative comparison.
Quantitative Blood and Organ Distribution of Gold Nanoparticles Using ICP-MS.
To quantitatively determine the AuNP blood and tumor pharmacokinetic profiles, mice were killed at 2, 6, 12, and 24 h after nanoparticle injection. Blood was collected via cardiac puncture using a 1-mL syringe equipped with a 25-gauge needle. The harvested organs included lungs, heart, liver, spleen, stomach, intestines, kidneys, axillary lymph nodes, brachial lymph nodes, inguinal lymph nodes, and the tumor. A skin sample was also taken from the animal’s posterior thoracic cavity. Average mass of the tumors resected from each mouse tumor model can be found in SI Appendix, Table S2. Organs and blood samples were weighed and placed into glass culture tubes. In addition to organ samples, a portion of or the total administered AuNP dose was also added to a separate tube. To the samples, 800 µL of concentrated nitric acid [69% (wt/vol)] was added. For regional lymph nodes, this volume was reduced to 200 µL of nitric acid. An acid control sample containing solely nitric acid was also prepared in triplicate. The samples were placed on a heat bath at 70 °C and allowed to digest overnight. The following day, 200 µL of concentrated hydrochloric acid [37% (wt/vol)] was added to each tube. To lymph node samples, the volume was reduced to 50 µL, as to maintain a 4:1 volume ratio between HNO3 and HCl. The organ digestate was maintained at 70 °C for an additional 3 h. The digested samples were transferred into 50-mL conical vials and diluted to a final volume of 40 mL with deionized water; samples containing lymph nodes were diluted to a total volume of 10 mL. The final acid concentration in each sample was 2% (vol/vol) HNO3 and 0.5% (vol/vol) HCl. Diluted samples were then passed through a 0.22-µm PES syringe filter (EMD Millipore–Merck). A calibration curve ranging from 1 × 10−3 to 1 × 102 µg·mL−1 of elemental gold (High-Purity Standards) prepared in 2% (vol/vol) HNO3 and 0.5% (vol/vol) HCl was used to determine the quantity of gold in each sample. Quantitative gold concentrations were determined using a NexION 350× ICP-MS (PerkinElmer). A 500-µL injection loop was used and sample solution was mixed with carrier [2% (vol/vol) HNO3 and 0.5% (vol/vol) HCl] and internal standard (Ir, 1,000 mg·L−1) before aspiration; several rinses were added between samples to minimize cross-contamination. The mass analyzer was set to Au 197 and Ir 192 for all measurements. Before analysis, the instrument was calibrated using a setup solution containing 1 µg·L−1 of Be, Ce, Fe, In, Li, Mg, Pb, and U in 1% HNO3 using the SmartTune method as indicated by the manufacturer. Quantitative analysis of total gold content in organs is presented as a normalized percentage of the injected dose. Since several AuNP ensembles were used over the course of this study, the data presented by Fig. 7 have been normalized to the average injected dose value of AuNPs determined from each measured replicate across the dosages of clodronate liposomes investigated. The blood half-life of AuNPs was determined as a percentage of the injected dose weighted against the amount of blood retrieved from the animal and normalized to 1.8 g; the average total blood content of a 20-g mouse. The decay curves were fit as single-exponential decays using GraphPad Prism 6 software. To determine the percentage of the injected dose of the AuNPs that were delivered to the tumor, the area under the curve (AUC) of the pharmacokinetic profile was determined using the trapezoidal approximation bounded by a time frame of 22 h.
Analysis of AgNP Biodistribution Using ICP-MS.
Quantitative determination of AgNP biodistribution using ICP-MS was done similarly as described for gold nanoparticles with the following modifications. SKOV3 xenograft mice were killed 3 h postinjection, and blood was collected via cardiac puncture. The organs analyzed included liver, spleen, blood, and the tumor. Organ samples were digested overnight using concentrated HNO3 and diluted to a final volume of 2% (vol/vol) before aspiration in the ICP-MS. The mass analyzer was set to Ag 107, where Ir 192 was used as an internal standard. The quantity of silver in each organ was determined using a silver standard curve and the quantitative organ biodistribution was normalized as a percentage of the injected dose to the quantity of silver measured in the injected aliquot.
Analysis of the Biodistribution of Liposomes and SiNPs.
The quantitative determination of liposome and SiNP organ biodistribution was done using fluorimetric analysis of organs imaged ex vivo. SKOV3 xenograft mice were injected with liposomes or SiNPs 48 h after initial administration of clodronate liposomes (0.017 mg·g−1) or PBS liposomes. Animals were then killed 3 h postinjection of liposomes or SiNPs, and blood was removed via cardiac puncture and extracted organs were washed with copious amounts of distilled water. Organs analyzed included livers, spleens, and tumors. Organs were grouped into clodronate liposomes-treated or control (PBS liposomes) and imaged adjacently in the same field using a Kodak in vivo multispectral imager. Organs were imaged using 650-nm excitation and fluorescence emission was collected using a 700-nm bandpass filter with FWHM ∼35 nm and exposure time of 1 min. Both liposomes and SiNPs contained dye analogs of Cy5 (SI Appendix, Fig. S9). Analysis of ex vivo fluorescence images of organs was done using ImageJ software (National Institutes of Health). The background signal of each image was determined by averaging three independent measurements of image area. Peripheries of organs were traced, and the average integrated fluorescence density was calculated for each organ. The integrated fluorescence intensities were averaged for each organ in the clodronate liposome- or control-treated animals and were used to quantitatively compare organ biodistribution of liposomes or SiNPs.
Immunohistochemistry and Histological Tissue Analysis.
Sections of liver, spleen, and tumor tissues were fixed in 10% formalin for 1 wk. The fixed tissues were then sent to the University Health Network Pathology Research Program at Toronto General Hospital or Toronto Centre for Phenogenomics for the remaining histological preparation. Tissue sections were stained with hematoxylin and eosin to observe any gross inflammation in hepatic and splenic tissue due to clodronate liposomes or AuNPs. Histology tissues were also treated with silver nitrate to observe AuNP distribution and immunohistochemically treated with F4/80 antibody and Ki67 to stain macrophages and cancer cells, respectively (29). The stained slices were imaged using an IX51 light microscope. For semiquantitative analysis of Kupffer cell populations in liver tissue, histological slides were sent to the Advanced Bioimaging Centre at Mount Sinai Hospital for full slide scans. The full images of the hepatic tissue sections were expanded to identical magnifications using Adobe Photoshop. A 20 × 20-cm frame was drawn centered on a hepatic central vein or portal triad from three independent positions in the tissue as shown by SI Appendix, Fig. S17. In these areas, Kupffer cells were identified using the brownish-red staining of horseradish peroxidase and the presence of a nucleus as the tissues were also counterstained with hematoxylin. From sequential counts in triplicate, an average Kupffer cell quantity was determined for each dose of clodronate liposomes investigated.
Hematology and Liver Biochemistry of Clodronate Liposomes-Treated C57BL/6 Mice.
C57BL/6 mice at 7–9 wk were injected with PBS or clodronate liposomes at dosages of 0.005, 0.017, and 0.05 mg·g−1 to create four groups of four animals per group (n = 4). At 48 h postinjection, animals were anesthetized with 4% (vol/vol) isoflurane in oxygen, and anesthesia was maintained at 3%. The thoracic cavity of the animal was exposed, and blood was collected through cardiac puncture using a 23-gauge needle. For hematology samples, blood was transferred to microfuge tubes containing dipotassium EDTA at a final concentration of 5–10 mM and kept on ice. For liver biochemistry analysis, serum was isolated from whole blood by centrifugation at 500 × g for 10 min. The serum was transferred to a fresh microfuge tube and kept on ice. Both blood and serum samples were sent for analysis at Toronto Centre for Phenogenomics immediately after collection and were analyzed within 1 d. Serum biochemistry markers of liver parenchyma included the following: ALT, ALP, aspartate transaminase, and bilirubin. Hematology markers included the following: red blood cell (RBC), hemoglobin (HGB), hematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), red blood cell distribution with (RDW), platelet (PLT), mean platelet volume (MPV), white blood cells (WBC), neutrophils (NE), lymphocytes (LY), monocytes (MO), basophils (BA), and eosinophils (EO).
CLP Sepsis Model in C57BL/6 Mice.
C57BL/6 mice at 7–9 wk were used in the induction of the CLP model of polymicrobial sepsis (26). Mice were injected 48 h before the CLP procedure with PBS liposomes or clodronate liposomes at dosages of, 0.005, 0.017, and 0.05 mg·g−1 to create four groups, each containing 10 animals (n = 10). It is noteworthy that animals in clodronate liposome groups were blinded to the surgeons, and all surgeries were conducted in a biosafety cabinet. Animals were anesthetized through inhalation of isoflurane at 4% (vol/vol) in oxygen; the maintenance concentration of isoflurane was 2.5% (vol/vol) throughout the duration of the procedure, and the animal was kept on a heat pad at 38 °C to maintain body temperature. The abdomen of the mouse was shaved and twice sterilized with 70% ethanol and betadine. Buprenorphine (0.15 mg·kg−1) was administered s.c. as preoperative analgesia. Mice were covered with slotted sterile drapes exposing only the sterile portion of the abdomen. A 1- to 1.5-cm skin incision was made along the midline exposing the abdominal muscle. A 1-cm incision was then made in the muscle layer to expose the peritoneum. The cecum was isolated, placed on gauze, and hydrated with sterile saline. A 6-0 absorbable silk suture was used to ligate the cecum 1 cm from the distal end. This accounted for a cecal ligation point that was ∼25% of the animal’s entire cecum. A single perforation was made midway in the ligated portion using a 21-gauge needle. A pinhead amount of fecal material was extruded using forceps, and the large intestine was returned to the peritoneal cavity. The incisions in the abdominal muscle and skin were closed using 3-0 to 5-0 absorbable silk suture. Animals were kept under a heat lamp for 15 min during recovery with access to food and water. Postoperative analgesia (buprenorphine, 0.15 mg·kg−1) was given every 10–12 h to ease pain and discomfort. Animals were monitored every 4–6 h for a total of 48 h following completion of the surgical procedure. Rectal body temperature was taken as hypothermia was used as an indirect indicator of sepsis. The endpoint in this study was defined by a rectal body temperature below 32 °C. Animals having body temperatures below this threshold were killed in accordance with ethics approval as established by the Animal Care Committee at the University of Toronto. To confirm induction of sepsis, blood was collected from the killed septic animals (n = 4) through cardiac puncture. Whole blood was diluted with sterile PBS, plated on LB agar plates (BD Biosciences), and incubated at 37 °C overnight. Agar plates were imaged using a standard smartphone camera (SI Appendix, Fig. S16). Control blood was collected through tail vein puncture of sterile animals (n = 4).
Statistical Analyses.
Data were analyzed by unpaired t tests or one-way or two-way ANOVA methods using GraphPad Prism 6 with Bonferroni correction. The Holm–Sidak approach was taken for multiple comparisons. Survival curves were analyzed by log-rank (Mantel–Cox) test. Data are presented as average values of three to four replicates, and precision reported is the sample SD, unless stated otherwise. Statistical significance between different experimental groups was measured at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
The authors thank Kim Tsoi and David Kongyu Zhang for fruitful discussions. This study was supported by Canadian Cancer Society Research Institute, Canadian Institutes of Health Research (CIHR) Grant MOP-130143, Natural Sciences and Engineering Research Council of Canada (NSERC) Grant 2015-06397, and a Collaborative Health Research Project (CHRP-493619-16). A.J.T. and W.P. acknowledge CIHR for provision of fellowships; W.P. also acknowledges Barbara and Frank Milligan and Cecil Yip graduate scholarships. Y.-N.Z. thanks NSERC, Wildcat Foundation, and Paul and Sally Wang for provision of fellowships. B.O. thanks Vanier Canada Graduate Scholarship, CIHR and the McLaughlin Centre for MD/PhD studentships, and the Donnelly Centre for graduate fellowships. D.D. acknowledges support from the China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713390114/-/DCSupplemental.
References
- 1.Wilhelm S, et al. Analysis of nanoparticle delivery to tumours. Nat Rev Mater. 2016;1:16014. [Google Scholar]
- 2.Tsoi KM, et al. Mechanism of hard-nanomaterial clearance by the liver. Nat Mater. 2016;15:1212–1221. doi: 10.1038/nmat4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J Am Chem Soc. 2012;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 4.Dai Q, Walkey C, Chan WCW. Polyethylene glycol backfilling mitigates the negative impact of the protein corona on nanoparticle cell targeting. Angew Chem Int Ed Engl. 2014;53:5093–5096. doi: 10.1002/anie.201309464. [DOI] [PubMed] [Google Scholar]
- 5.Diagaradjane P, Deorukhkar A, Gelovani JG, Maru DM, Krishnan S. Gadolinium chloride augments tumor-specific imaging of targeted quantum dots in vivo. ACS Nano. 2010;4:4131–4141. doi: 10.1021/nn901919w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renaud G, Hamilton RL, Havel RJ. Hepatic metabolism of colloidal gold-low-density lipoprotein complexes in the rat: Evidence for bulk excretion of lysosomal contents into bile. Hepatology. 1989;9:380–392. doi: 10.1002/hep.1840090307. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar S, Khan MF, Kaphalia BS, Ansari GAS. Methyl palmitate inhibits lipopolysaccharide-stimulated phagocytic activity of rat peritoneal macrophages. J Biochem Mol Toxicol. 2006;20:302–308. doi: 10.1002/jbt.20150. [DOI] [PubMed] [Google Scholar]
- 8.Cai P, Kaphalia BS, Ansari GAS. Methyl palmitate: Inhibitor of phagocytosis in primary rat Kupffer cells. Toxicology. 2005;210:197–204. doi: 10.1016/j.tox.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Gemsa D, Seitz M, Kramer W, Till G, Resch K. The effects of phagocytosis, dextran sulfate, and cell damage on PGE1 sensitivity and PGE1 production of macrophages. J Immunol. 1978;120:1187–1194. [PubMed] [Google Scholar]
- 10.Fowler EF, Thomson AW. Effect of carrageenan on activity of the mononuclear phagocyte system in the mouse. Br J Exp Pathol. 1978;59:213–219. [PMC free article] [PubMed] [Google Scholar]
- 11.Magaña IB, et al. Suppression of the reticuloendothelial system using λ-carrageenan to prolong the circulation of gold nanoparticles. Ther Deliv. 2015;6:777–783. doi: 10.4155/tde.15.33. [DOI] [PubMed] [Google Scholar]
- 12.Ohara Y, et al. Effective delivery of chemotherapeutic nanoparticles by depleting host Kupffer cells. Int J Cancer. 2012;131:2402–2410. doi: 10.1002/ijc.27502. [DOI] [PubMed] [Google Scholar]
- 13.van Rooijen N, van Kesteren-Hendrikx E. In: Macrophages: A Practical Approach. Paulnock DM, editor. Oxford Univ Press; New York: 2000. pp. 157–172. [Google Scholar]
- 14.Perrault SD, Chan WCW. Synthesis and surface modification of highly monodispersed, spherical gold nanoparticles of 50–200 nm. J Am Chem Soc. 2009;131:17042–17043. doi: 10.1021/ja907069u. [DOI] [PubMed] [Google Scholar]
- 15.Chou LYT, Chan WCW. Fluorescence-tagged gold nanoparticles for rapidly characterizing the size-dependent biodistribution in tumor models. Adv Healthc Mater. 2012;1:714–721. doi: 10.1002/adhm.201200084. [DOI] [PubMed] [Google Scholar]
- 16.Sleyster EC, Knook DL. Relation between localization and function of rat liver Kupffer cells. Lab Invest. 1982;47:484–490. [PubMed] [Google Scholar]
- 17.Lautt WW. Hepatic Circulation: Physiology and Pathophysiology. Morgan and Claypool Life Sciences; San Rafael, CA: 2009. [PubMed] [Google Scholar]
- 18.Ross MH, Kaye GI, Pawlina W. Histology: A Text and Atlas, With Cell and Molecular Biology. Lippincott Williams and Wilkins; Baltimore: 2003. [Google Scholar]
- 19.van Rooijen N, Hendrikx E. Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol Biol. 2010;605:189–203. doi: 10.1007/978-1-60327-360-2_13. [DOI] [PubMed] [Google Scholar]
- 20.Sykes EA, et al. Tailoring nanoparticle designs to target cancer based on tumor pathophysiology. Proc Natl Acad Sci USA. 2016;113:E1142–E1151. doi: 10.1073/pnas.1521265113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charles River 2011 Research models: C57BL/6 mice. Available at www.criver.com/files/pdfs/rms/c57bl6/rm_rm_d_c57bl6n_mouse.aspx. Accessed December 19, 2016.
- 22.Radi ZA, et al. Increased serum enzyme levels associated with Kupffer cell reduction with no signs of hepatic or skeletal muscle injury. Am J Pathol. 2011;179:240–247. doi: 10.1016/j.ajpath.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams MD, et al. Hospitalized cancer patients with severe sepsis: Analysis of incidence, mortality, and associated costs of care. Crit Care. 2004;8:R291–R298. doi: 10.1186/cc2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callery MP, Kamei T, Flye MW. Kupffer cell blockade increases mortality during intra-abdominal sepsis despite improving systemic immunity. Arch Surg. 1990;125:36–40, discussion 40–41. doi: 10.1001/archsurg.1990.01410130038005. [DOI] [PubMed] [Google Scholar]
- 25.Traeger T, et al. Kupffer cell depletion reduces hepatic inflammation and apoptosis but decreases survival in abdominal sepsis. Eur J Gastroenterol Hepatol. 2010;22:1039–1049. doi: 10.1097/MEG.0b013e32833847db. [DOI] [PubMed] [Google Scholar]
- 26.Toscano MG, Ganea D, Gamero AM. Cecal ligation puncture procedure. J Vis Exp. 2011;51:e2860. doi: 10.3791/2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain RK, Munn LL, Fukumura D. Mammary fat pad tumor preparation in mice. Cold Spring Harb Protoc. 2012;2012:1115–1116. doi: 10.1101/pdb.prot071514. [DOI] [PubMed] [Google Scholar]
- 28.Liu TW, et al. Inherently multimodal nanoparticle-driven tracking and real-time delineation of orthotopic prostate tumors and micrometastases. ACS Nano. 2013;7:4221–4232. doi: 10.1021/nn400669r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahmanzadeh R, et al. Ki-67 as a molecular target for therapy in an in vitro three-dimensional model for ovarian cancer. Cancer Res. 2010;70:9234–9242. doi: 10.1158/0008-5472.CAN-10-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.