Significance
Standard in vitro assays to assess genotoxicity frequently generate positive results that are subsequently found to be irrelevant for in vivo carcinogenesis and human cancer risk assessment. Currently used follow-up methods, such as animal testing, are expensive and time-consuming, and the development of approaches enabling more accurate mechanism-based risk assessment is essential. We developed an in vitro transcriptomic biomarker-based approach that provides a robust biomarker reflecting stress-signaling responses. The biomarker correctly identifies the vast majority of irrelevant genotoxicity results from in vitro chromosome damage assays. TGx-DDI, a multigene biomarker for DNA damage-inducing agents, is the first biomarker that not only shows convincing interlaboratory and intralaboratory reproducibility, but also performs accurately in a system suitable for high-throughput screening.
Keywords: TGx-DDI, transcriptomic biomarker, high-throughput screening, genotoxicity, DNA damage response
Abstract
Interpretation of positive genotoxicity findings using the current in vitro testing battery is a major challenge to industry and regulatory agencies. These tests, especially mammalian cell assays, have high sensitivity but suffer from low specificity, leading to high rates of irrelevant positive findings (i.e., positive results in vitro that are not relevant to human cancer hazard). We developed an in vitro transcriptomic biomarker-based approach that provides biological relevance to positive genotoxicity assay data, particularly for in vitro chromosome damage assays, and propose its application for assessing the relevance of the in vitro positive results to carcinogenic hazard. The transcriptomic biomarker TGx-DDI (previously known as TGx-28.65) readily distinguishes DNA damage-inducing (DDI) agents from non-DDI agents. In this study, we demonstrated the ability of the biomarker to classify 45 test agents across a broad set of chemical classes as DDI or non-DDI. Furthermore, we assessed the biomarker’s utility in derisking known irrelevant positive agents and evaluated its performance across analytical platforms. We correctly classified 90% (9 of 10) of chemicals with irrelevant positive findings in in vitro chromosome damage assays as negative. We developed a standardized experimental and analytical protocol for our transcriptomics biomarker, as well as an enhanced application of TGx-DDI for high-throughput cell-based genotoxicity testing using nCounter technology. This biomarker can be integrated in genetic hazard assessment as a follow-up to positive chromosome damage findings. In addition, we propose how it might be used in chemical screening and assessment. This approach offers an opportunity to significantly improve risk assessment and reduce cost.
There is a critical need for improved, accurate, and reliable toxicity assays to expedite the health risk assessment of chemical agents. Genotoxicity leads to genetic changes, such as mutations, chromosome damage, and consequent genomic instability, progressing to cancer. Thus, genotoxicity testing is a crucial component of safety evaluation for drugs and chemicals (1, 2). The genotoxicity testing battery includes standard in vitro mutation and in vitro chromosomal damage (CD) assays. The latter include assessments of various cytogenetic abnormalities and/or micronucleus formation. A high incidence of chemicals with positive findings on CD assays that are not reproducible in vivo is of considerable concern to industry and regulatory agencies (3). Many of these “positive” findings are not caused by initial DNA damage, but rather arise as a result of cytotoxicity or other nongenotoxic mechanisms (2–4). Despite the progress in refinement of testing protocols for standard genotoxicity in vitro chromosome damage assays, irrelevant positives remain major challenges to industry and regulatory agencies (3, 5); additional examples are listed in Table S1.
The differentiation of relevant from irrelevant in vitro results is crucial for the interpretation of positive findings in the context of risk to human health. Such irrelevant positive results typically require expensive and time-consuming follow-up tests involving animal testing. When cost is a consideration, or when animal testing is not feasible (6, 7), potentially useful chemicals may be excluded from further commercial development. As conceptually depicted in the current Food and Drug Administration guidance for industry for drug development, investigating genotoxic mechanisms during lead optimization, candidate selection, and/or Investigational New Drug (IND) application requires effective experimental follow-up strategies. Since broad mechanism-based assays are not available and currently used follow-up methods are laborious and time-consuming, experimental approaches enabling mechanism-based risk assessment are needed.
We previously developed an in vitro transcriptomic biomarker-based approach that provides biological relevance based on stress signaling responses for assessment of chemicals showing positive results in the standard genotoxicity testing battery (8). Taking advantage of a modern toxicogenomic approach, we constructed a reference database containing global gene expression profiles of 28 model agents with a broad range of known toxic mechanisms. A transcriptomic biomarker, TGx-DDI (designated TGx-28.65 in ref. 8), which discriminates DNA damage-inducing (DDI) agents from other agents, was derived from this initial reference dataset. DDI agents include DNA-reactive agents that are known to be directly genotoxic, along with indirect-acting agents causing DNA damage either by inhibition of topoisomerase action or blockage of DNA synthesis. Three test agents consisting of a known genotoxicant, a nongenotoxic stress agent, and a nongenotoxic agent with irrelevant positive CD results were successfully categorized in that study as a proof of application; however, before this method is added to the standard in vitro genotoxicity battery, comprehensive performance validation is required to assess its robustness and feasibility.
In the present study, a performance evaluation and validation exercise was undertaken to thoroughly evaluate the ability of this biomarker approach to identify agents that cause DNA damage, and to clearly identify any potential limitations of its use in genetic safety risk assessment. In the first phase, intralaboratory performance was assessed using a variety of DDI and non-DDI agents. The ability of the biomarker to predict DDI/non-DDI agents was also considered using published work from another laboratory and publicly available data. Next, a panel of 45 chemicals with known mechanisms of action were analyzed to explore the context of use, including 10 agents with irrelevant findings in CD assays. This project included support by and advice from a large consortium of scientists from industry, government agencies, and academia organized by the Health and Environmental Science Institute (HESI). Our experimental design for this phase consists of a concentration setting experiment followed by a microarray analysis of global gene expression alterations. As explained previously (8), concentration-ranging is essential to identify effective doses for triggering stress signaling, and timing is critical to avoid later nonspecific effects. However, we emphasize that application of this biomarker in compound testing requires concentration–response experiments. In this phase of the study, a standardized workflow for experiment and data analysis is proposed for the TGx-DDI application in both pharmaceutical and chemical testing.
The inventory of chemicals mandated by the Toxic Substances Control Act contains 73,757 chemicals that have been reported by manufacturers as being in commercial use as of February 2001, and this number is continually increasing. Thus, a thorough assessment of the health effects of chemicals present in the environment and marketplace poses a serious challenge for regulatory agencies worldwide. In vitro high-throughput screening (HTS) has been proposed as a first-tier screen in chemical assessments (9). To adapt our transcriptomic biomarker (identified and validated using microarray technology), we developed a TGx-DDI high-throughput cell-based assay using the nCounter system. Here we present our evaluation of the robustness of the TGx-DDI nCounter assay in identifying DDI agents and its concordance with the output of the microarray approach.
Results
Technical Performance Evaluation.
To demonstrate the technical robustness and reproducibility of the cell culture and exposure conditions, the microarray method, and overall comparability with the learning set data used for TGx-DDI identification, we conducted four independent replicate transcriptomic experiments in which TK6 cells were exposed to 80 μg/mL cisplatin alongside concurrent 0.9% NaCl (vehicle) controls. As shown in Table S2, the correlation coefficients across the replicates were >0.95, indicating that this technical system is highly reproducible. Four additional agents were selected from the original training set to confirm the reproducibility of DDI prediction using the TGx-DDI biomarker: a DNA alkylating agent [methyl methanesulfonate (MMS)], a topoisomerase inhibitor (etoposide), an HDAC inhibitor (oxamflatin), and ionizing radiation (IR) (4 Gy). Dose–response studies were conducted using a qRT-PCR indicator gene panel comprising ATF3, CDKNIA, and GADD45A to determine the specific concentration that triggered a robust response (i.e., greatest overall increases in mRNA levels for these test transcripts) for each chemical agent (as described in ref. 8), and the selected concentrations for these agents were identical to the previously determined ones (8). At the selected concentrations, the microarray results of the three agents and IR were used to classify these agents with the TGx-DDI biomarker, and the expression profiles for each agent were compared with the previously published dataset (Fig. S1A). The microarray results derived for the TGx-DDI biomarker genes compared favorably with our previous work. As anticipated, the treatments clustered with their expected categories by two-dimensional clustering (2DC) using the TGx-DDI biomarker (Fig. S1A). Taken together, these experiments demonstrate that this model system and technology generate robust and comparable data in our laboratory that are highly reproducible.
Extensive interlaboratory validation of the biomarker is beyond the scope of the present study; however, the performance of the biomarker was explored at Health Canada. This analysis confirmed the ability of the biomarker to correctly classify nine genotoxic and four nongenotoxic chemicals in TK6 cells (10). Moreover, the biomarker correctly classified five genotoxic and 10 nongenotoxic agents using Affymetrix DNA microarrays from HepaRG cells (publicly available data from another laboratory; ref. 11).
Selection of Validation Compounds.
Based on feedback for our toxicogenomic approach proposed in our earlier Voluntary Exploratory Data Submissions (VXDS) (4) and ongoing interactions with the Food and Drug Administration, a strategy was developed to evaluate the performance of the TGx-DDI biomarker with a set of chemicals that covered five mechanistic classes spanning DDI and non-DDI mechanisms:
-
•
Class 1: DDI agents that interact directly with DNA that should be detected as positive in the in vitro CD assays. This group of agents includes alkylating and cross-linking agents, and serves as a positive control for detection of direct DNA-reactive mechanisms.
-
•
Class 2: DDI agents that interact indirectly with DNA. Topoisomerase inhibitors and intercalators are highly potent indirect genotoxicants. Antimetabolites, such as nucleoside analogs, cause CD in vitro. We note that some antimetabolites may show effects only after longer exposures than provided in our 4-h assay, and inclusion of these agents tests the limits of the experimental design.
-
•
Class 3: agents that interact indirectly with DNA via effects on the cell cycle, regulation of apoptosis, and interaction with the mitotic apparatus. This class includes aneugens that are microtubule inhibitors, which are non-DDI because they cause aneugenicity through spindle interference, and in vitro CD-positive kinase inhibitors that are not relevant genotoxicants in vivo because they are typically positive only at doses that are not physiologically relevant.
-
•
Class 4: non-DDI compounds with a “clean” genotoxicity profile, including negativity in vitro CD assays. This class serves as negative controls for testing the transcriptomic biomarker.
-
•
Class 5: compounds known to have irrelevant positive results in in vitro genotoxicity assays. This class includes such agents as caffeine, nongenotoxic carcinogens, apoptosis inducers, and other chemicals that have been reported as positive in in vitro CD assays but for which the genotoxicity findings are understood as irrelevant.
Based on the foregoing, we selected 45 chemicals (Table 1 and Table S3) from the literature and used expert knowledge to populate each class (5).
Table 1.
Classes of test compounds
| Class | Definition | CD | Validation set | Previously tested* |
| 1 | Genotoxins that interact directly with DNA | Positive | 8 | 3 |
| 2 | Genotoxins that interact indirectly with DNA | Positive | ||
| Topo inhibitors, including DNA intercalators | 5 | 2 | ||
| Antimetabolites | 5 | 3 | ||
| 3 | Genotoxins that interact indirectly with DNA | Positive | ||
| Effect on cell cycle and mitotic apparatus | ||||
| Antimitotic agents | 3 | 4 | ||
| Kinase inhibitors (in vitro positive) | 3 | None | ||
| Heavy metals | None | 3 | ||
| 4 | Non–DNA-reactive chemicals, in vitro negative | Negative | ||
| Kinase inhibitors (in vitro negative) | 2 | None | ||
| Nongenotoxic carcinogens | 3 | None | ||
| General pathways | 2 | None | ||
| Others | 3 | None | ||
| 5 | Irrelevant positives | Positive | 11 | 1 |
The number of compounds in the previous study (8).
Concentration Optimization.
As discussed in more detail previously (8, 10–14), a sufficient concentration of the test agent is required to trigger a measurable transcriptional response; such concentrations may differ from other toxicologic endpoints. Therefore, to determine an appropriate concentration for transcriptomic profiling, we performed a dose-range finder experiment for all test compounds as described by Li et al. (8). Six concentrations of each agent were used to assess mRNA changes in three indicator genes (ATF3, CDKNIA, and GADD45A) by qRT-PCR. The concentration for each agent showing the strongest induction of the indicator genes was then selected. In addition, concordance of responses in the indicator genes was confirmed before samples were pooled for microarray analysis. If none of the indicator genes was induced in concentration setting experiments, then the IC50 value was selected for the microarray analysis. The IC50 value was determined through a standard MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay at 24 h using 10 concentrations and three replicates. Based on this cytotoxicity assay, the selected concentrations for microarray analysis were not overtly cytotoxic for any test agent (Fig. S2). If there was neither cytotoxicity nor induction of expression changes in the gene panel, a concentration of 1 mM was used for microarray analysis, in accordance with the revised International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use guidance on genotoxicity testing of pharmaceuticals (15).
We note that selection of a single concentration and pooling of replicate samples for microarray analysis (described below) are specific to biomarker development and validation, where multiple compounds were used in each class. Future application in substance testing should be undertaken using a dose–response design with samples in triplicate, as has been described in a case study on the TGx-DDI biomarker in chemical testing (16).
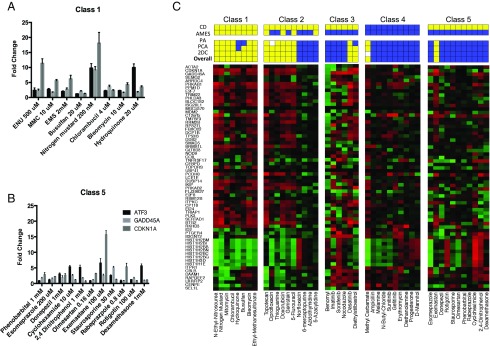
The concentration determination results of all five classes are presented in Fig. S3. As an example, responses for the indicator genes at the selected concentrations for chemicals in classes 1 and 5 are shown in Fig. 1 A and B, respectively. All chemicals in class 1 except busulfan induced robust responses in at least one gene at the selected concentration. Only GADD45A was induced by at least twofold in cells treated with busulfan at the selected concentration. Higher concentrations of busulfan did not cause greater induction of these indicator genes (Fig. S3F), suggesting transcriptional inhibition at high concentrations. The treatment of all but one compound, bleomycin, at the selected concentrations resulted in a minimum 30% reduction in cell viability at 24 h (Fig. S2); however, cells treated with bleomycin showed an 80% decline in viability at 24 h. The concentration setting was based on qRT-PCR results for all class 5 compounds except rotigotin, which did not induce any of the indicator genes at the concentrations tested, including cytotoxic doses. Therefore, the IC50 for cell viability was selected for the microarray experiment. In contrast, the other class 5 compounds induced at least one of the three indicator genes at the selected concentrations. Aside from exemastan, rabeprazpole, and rotigotin, these class 5 compounds were not cytotoxic, with ≥80% viability (Fig. S2).
Fig. 1.
Prediction of the probability that the test agents are DDI or non-DDI using the TGx-DDI transcriptomic biomarker. (A and B) Representative transcriptional responses for concentration-optimization indicator genes ATF3, CDKN1A, and GADD45A, measured by qRT-PCR. The ratio designates the relative change in gene expression compared with vehicle-treated control cells. Results are shown for the concentrations selected for subsequent microarray experiments. (C) Forty-five chemicals were grouped based on mechanistic properties (Table 1); 2DC heatmaps are shown for each class of agents, and prediction results are listed above. Three methods were used to predict DDI-positive (yellow), and the overall prediction (Bottom) is based on positive results with any of these three methods. (Top and Middle) Published results from the CD and Ames assays. Yellow and blue indicate positive and negative findings, respectively; white boxes indicate indeterminate classification.
TGx-DDI Transcriptomic Biomarker Evaluation.
Following the concentration determination, a microarray analysis was performed for each test compound. RNAs from three replicates in the concentration setting experiment were pooled together and used for microarray analysis. Cisplatin or IR was used in parallel during each batch of experiments as a positive control and to assess batch variation (Fig. S1B). The TGx-DDI transcriptomic biomarker panel was used to classify each chemical as DDI or non-DDI using 2DC, principal component analysis (PCA), and probability analysis (PA). Hierarchical clustering and PCA were used as an initial unsupervised method to explore the data. As shown in Fig. S4, category assignment was first determined by the position of the test chemical in the tree structure of the dendrogram generated by 2DC, or in the PCA plot. Finally, TGx-DDI–based prediction was conducted by applying the shrunken centroids approach for posterior probability analysis (17). This was done by determining the extent of gene expression changes for each of the biomarker genes from the DDI and non-DDI centroids. A DDI call was based on P > 0.9 of the compound being in that class, and vice versa for a non-DDI call. A chemical was considered unclassified if it did not meet these criteria.
Fig. 1C shows the TGx-DDI heatmap for chemicals in all five classes. PA, PCA, and 2DC results using the TGx-DDI biomarker for validation chemicals are shown by colored boxes above each heatmap, along with the published CD and Ames assay results. Yellow and blue represent positive and negative results, respectively. To decrease the probability of false negatives, we used a three-pronged approach for overall final classification. A chemical was classified as DDI if it gave a positive call in any one of the TGx-DDI biomarker analyses described above (2DC, PCA, or PA prediction), and was classified as non-DDI if it did not meet any of these criteria.
Overall, application of our three-pronged analytical approach yielded the expected classifications, with a few exceptions. Specifically, all agents in class 1 were classified as DDI, all but one agent (methyl carbamate) in class 4 were classified as non-DDI, and all but one agent (exemastan) in class 5 were classified as non-DDI. Class 3 agents were classified as non-DDI with two exceptions; both dasatanib and diethylstilbestrol gave DDI calls. More than one-half of the class 2 agents gave DDI calls.
Development of the TGx-DDI nCounter Assay.
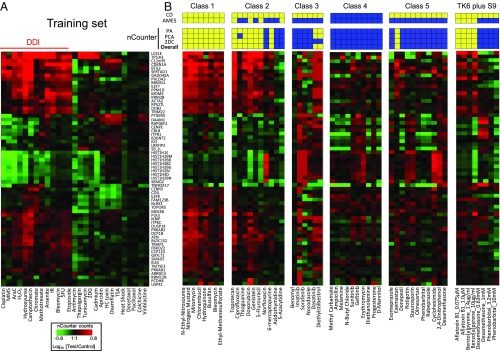
To meet the need for a multiplex detection system suitable for HTS, we developed a TGx-DDI assay applying nCounter, a direct digital counting technology. First, we assessed the robustness of TGx-DDI nCounter assay by comparing the results of the training set agents in TK6 cells to those using microarrays (Fig. S5A). The TGx-DDI code set includes an optimized TGx-DDI gene set and eight housekeeping genes. The housekeeping genes were selected based on stability and detectable expression levels. A high correlation was observed between nCounter assay and microarray results for TGx-DDI (Fig. S5B).
To validate the sensitivity and specificity of DDI prediction of the TGx-DDI nCounter assay, 45 test compounds were evaluated using nCounter technology. nCounter assays were performed on 100 ng of total RNA using the same RNA samples from the microarray analysis. Applying our three-pronged analytical approach for classification, we classified compounds as DDI or non-DDI based on the nCounter assay data (Fig. 2). In addition to the 45 compounds in five classes, we validated four additional chemicals requiring metabolic activation that were used in a previous study (11) in which this approach was adapted to DDI agents requiring metabolic activation. Fig. 2A shows the heatmap of the training set agents using the TGx-DDI nCounter assay, and Fig. 2B shows the heatmap for compounds in different classes and compounds requiring metabolic activation. The overall classification results for all test compounds are shown as colored boxes immediately above the heatmap labeled “overall.” Classification of the majority of the compounds was consistent with the microarray results (Fig. 1); however, responses to several weak DDI compounds were stronger and more robustly measured by nCounter. For example, both busulfan and hydroquinone were predicted as non-DDI by PA, while 2DC and PCA indicated that these are DDI agents in the microarray analysis. The analysis of the nCounter data for these two agents showed consistency across the three classification methods, suggesting that the nCounter system is more sensitive for detecting weak responses to DDI agents. Moreover, all class 4 agents were classified by the nCounter system as non-DDI, which is 100% consistent with CD assay results. This is in contrast to microarray results, in which only 9 out of 10 agents were classified as non-DDI (Table 2). The results of 2DC and PCA analyses for the TGx-DDI nCounter assay are shown in Fig. S4 for each class. The classification of agents in the presence of S9 metabolic activation was also consistent with expectations, demonstrating that the method can be used accurately with S9 (Fig. S6).
Fig. 2.
Performance of TGx-DDI with the nCounter analysis system. (A) Heatmap of NanoString expression analysis using previously tested chemicals. All chemicals were classified as DDI or non-DDI using the same approach used in the DNA microarray analysis. (B) Thirty-eight chemicals were grouped based on mechanistic properties (Table 1). Four chemicals that require metabolic activation were evaluated at different concentrations. Heatmaps are shown for nCounter results for each class and prediction results are displayed above. Three methods were used to predict DDI positivity (yellow), and the overall prediction (Bottom) is based on positive results with any of these three methods. (Top and Middle) Published results from the CD and Ames assays. Yellow and blue indicate positive and negative findings, respectively.
Table 2.
Consistency of TGx-DDI prediction (by microarray and nCounter methods) with CD assay results for selected test classes
| Technology | Class 1 | Class 2 | Class 4 | Class 5 | |
| Topo inhibitor | Antimetabolites | ||||
| Microarray | 100% (8/8) | 80% (4/5) | 40% (2/5) | 90% (9/10) | 9% (1/11) |
| nCounter | 100% (8/8) | 80% (4/5) | 60% (3/5) | 100% (10/10) | 9% (1/11) |
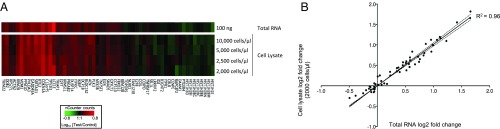
To develop an HTS TGx-DDI nCounter assay, we tested crude cell lysates in addition to isolated RNA with this technology, using solvent- or bleomycin-treated TK6 cells as samples. This method omits RNA extraction steps, allowing it to be coupled to nCounter measurement for a highly automated HTS system. As shown in Fig. 3, nCounter results of cell lysates at various cell concentrations showed comparable results to those for purified RNA from the original bleomycin and solvent control experiments, and yielded correlation coefficients of 0.90–0.96 in fold changes for the TGx-DDI biomarker genes from pure RNA extracts vs. cell lysates.
Fig. 3.
Elimination of the RNA preparation step. (A) Comparison of nCounter results using cell lysate and total RNA methods from cells treated with bleomycin. The number of cells directly analyzed is shown at the right for each row; results with 100 ng of purified RNA are shown above. (B) Representative log2 fold change correlation of genes in TGx-DDI in total RNA and cell lysates. The correlation between results using total RNA and cell lysates was analyzed; the R2 value calculated based on linear regression ranged from 0.90 to 0.96 for each cell concentration. Shown is the comparison of total RNA and cell lysate with a concentration of 2,000 cells/µL.
Discussion
The first objective of the present study was to provide validation data to support the capability of the TGx-DDI biomarker to assess genotoxic hazard and derisking compounds with irrelevant in vitro positive chromosome damage findings. We first showed that the TGx-DDI biomarker performs robustly in predicting DDI and non-DDI agents in our laboratory, and that accurate calls have been made using different cell culture models and platforms in other laboratories (10, 11). We then demonstrated that our integrated TGx-DDI bioinformatic approach has high accuracy for classification of DDI agents and non-DDI agents, and is highly effective in differentiating relevant from irrelevant chromosome damage assay findings.
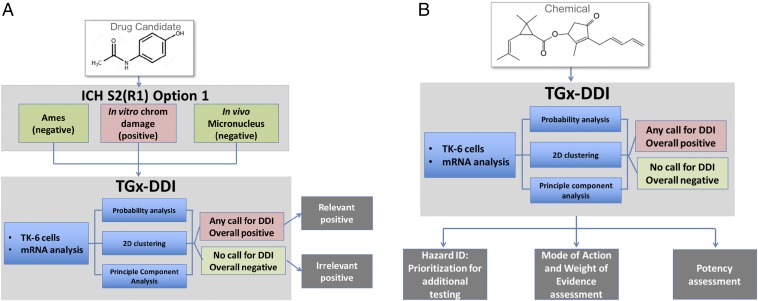
Based on the results of our studies, we developed workflows for application of the TGx-DDI biomarker in genetic toxicology risk assessment (Fig. 4). We propose that the TGx-DDI transcriptomic biomarker can be applied during assessment of pharmaceuticals (Fig. 4A) and environmental/industrial chemicals (Fig. 4B). In pharmaceutical assessments with positive results from in vitro mammalian cell chromosome damage assays, the biomarker provides insight into the relevance of these positive findings for agents that are otherwise negative in Ames and in vivo tests (Fig. 4A). This is important, because the human relevance of a positive in vitro CD finding still necessitates multiple in vivo follow-up studies despite a negative in vivo genotoxicity test (15). Thus, the risk assessment of these positive in vitro findings poses a challenge to industry and regulatory agencies. Our study demonstrates that application of the TGx-DDI transcriptomic biomarker will add significant value to the current genotoxicity testing battery for pharmaceuticals by reducing the need for complicated follow-up in vitro and in vivo tests and streamlining the animal tests that are required.
Fig. 4.
Proposed workflow for applying the TGx-DDI biomarker for genotoxicity assessment of candidates for pharmaceutical drug development (A) or industrial and environmental chemicals (B).
For industrial and environmental chemicals, the TGx-DDI provides a feasible high-throughput approach for detecting and characterizing genotoxicity hazard (Fig. 4B). Specifically, the biomarker could be used in HTS for identifying and prioritizing the agents that may cause DNA damage when large chemical sets require assessment. In addition, as in the pharmaceutical application, the biomarker can be used in parallel with conventional in vitro genotoxicity tests to provide weight of evidence in genotoxicity hazard assessment (16), to aid the differentiation of DDI from non-DDI (i.e., aneugenicity) modes of genotoxic action, and to provide insight into potentially irrelevant positives. Finally, as shown in our published case on use of the TGx-DDI biomarker in chemical assessment, the response of the biomarker genes is useful for determining a chemical’s genotoxic potency when run in parallel with prototype agents (16).
Overall, this transcriptomic biomarker approach has the potential to complement and/or eventually replace standard genotoxicity assays by providing information about biological responses to genotoxic stress that cannot be obtained using current methods. While the standard current in vitro genotoxicity assays, particularly CD and the mouse lymphoma assay (MLA), give phenotypic readouts, the TGx-DDI provides insight into molecular responses by a toxicant. Specifically, a positive response using the TGx-DDI biomarker indicates that sufficient DNA damage was incurred and recognized by the cell to initiate a transcriptional DNA damage response driven by DNA damage response signaling, including p53. Moreover, as described by Clewell and Andersen (18), the pattern of transcription induction by p53 differs among genotoxic agents, and these profiles may be useful in classifying mechanisms of action.
This validation study comprised an assessment of 45 test chemicals across five recommended mechanistic classes using a transcriptomics profiling approach. Fig. 1C summarizes the results of the TGx-DDI toxicogenomic assay and the data from standard genotoxicity testing assays for these 45 test chemicals. The TGx-DDI biomarker data were interpreted using three statistical approaches: 2DC, PCA, and PA. The individual results of each statistical method, as well as the overall call given (positive in any method is a positive overall call) were obtained. The three statistical methods were used for TGx-DDI interpretation to ensure robust data analysis that limited false negatives. The results of the 2DC, PCA, and PA analyses were generally consistent: in 90% of the test cases, the results agreed with one other, with only 5 of the 45 agents showing differing results. Three agents—busulfan, hydroquinone, and diethylstilbestrol—were identified as non-DDI by PA but as DDI by 2DC and/or PCA. These three agents are exceptions, because they induce weaker gene expression responses overall compared with the other positive agents based on visual inspection of the heatmap (Fig. 1C), and are positioned very close to the cutoff line in the PCA plot (Fig. S4 A and C). Two agents, cirprofloxacin and methyl carbamate, were categorized as indeterminate (Fig. 1C). Together with the negative PA result, these data suggest that these agents cause relatively weak genotoxic effects under our test conditions. Thus, to ensure as few false-negative findings as possible in compound screening/assessment and thereby maintain high sensitivity, agents that induced weak TGx-DDI responses were also reported as DDI if at least one analysis was positive. Nevertheless, our approach reduced the irrelevant positives by 90% without increasing false negatives.
As shown in Fig. 1C, the TGx-DDI biomarker classifies all agents in class 1 as DDI, consistent with results for these compounds using in vitro CD and Ames assays. In addition, all of the non-DDI agents in class 4 except methyl carbamate are classified as non-DDI when applying the TGx-DDI transcriptomic biomarker analytical approach, again consistent with the findings of in vitro CD and Ames assays. Norfloxacin, an antibiotic with topoisomerase inhibitory activity, was predicted to be non-DDI by the TGx-DDI biomarker, while the in vitro CD and Ames assay results were positive and negative, respectively. It is known that the fluoroquinolone antimicrobials target bacterial DNA gyrase and topoisomerase IV, and that the effect on eukaryotic topoisomerase is weak and the relevance of genotoxicity depends on the difference in affinity between the bacterial gyrase and mammalian topoisomerase (19, 20). Overall, the mammalian topoisomerase inhibitors were identified by the biomarker.
Three out of five antimetabolites—6-mercaptopurine (6-MP), azidothymidine (AZT), and 5-azacytidine (5AzaC)—were classified as non-DDI using the microarray method, while the other two anti-metabolites, 5-FU and 6-TG, are predicted to be DDI by TGx-DDI. This difference may reflect the different mechanisms of action. Unlike 5-FU and 6-TG, both of which can incorporate into DNA (21) and block DNA synthesis (i.e., a signal adequately detected by TGx-DDI), AZT and 5AzaC interfere with reverse transcriptase and DNA methylation, respectively. 6-MP affects purine nucleotide synthesis by inhibiting phosphoribosyl pyrophosphate amidotransferase, a rate-limiting enzyme for purine synthesis, which leads to genotoxicity, but the effects may not be evident until later time points. Thus, the biomarker may have some limitations in the assessment of antimetabolites. However, in most cases the antimetabolite properties of compounds can be easily predicted based on chemical structure. In the case of two non-DDI kinase inhibitors, imatinib and sorafenib, the biomarker could be triggered by alteration of signaling pathways, which is irrelevant to genotoxic risk. Importantly, as described above, these agents cause genotoxicity only at concentrations that are not physiologically relevant. Finally, the assessment of class 5 responses clearly demonstrates that the classifier is effective in differentiating relevant and irrelevant findings. Indeed, >90% (10 of 11) of the irrelevant CD responses were identified as such in this analysis. Overall, our results indicate that application of the biomarker in genotoxicity testing could significantly increase the efficiency of derisking irrelevant positives in chromosome damage assays.
In the present study, concentration selection was one of the key processes required to ensure robust TGx-DDI assay results, because only one concentration was tested per agent. Indeed, the xenobiotic-induced transcriptional responses can be blunted at very high concentrations, at which transcriptional machinery or cell integrity is compromised (e.g., busulfan in the present study). Moreover, marginal responses at low concentrations can compromise the prediction of toxicity. As shown in Fig. S2, the cytotoxicity varied substantially at the concentrations we selected for these 45 chemicals even within the same class. Thus, we developed a concentration optimization procedure to provide a standardized condition for test agent concentration selection and to decrease the likelihood of false negatives. Our qRT-PCR concentration optimization approach monitors a panel of three well-characterized stress genes—ATF3, CDKN1A, and GADD45A—which serve as indicators for effective transcriptional response to the treatments. While TGx-DDI is a robust biomarker, and accurate genotoxicity prediction can be achieved at a range of different concentrations based on a concentration–response study using TGx-DDI (11), we recommend concentration optimization by qRT-PCR as a standard procedure for this toxicogenomic application to ensure robust response at the test concentrations selected.
Since the TGx-DDI biomarker comprises only 64 genes, it is feasible to use this biomarker in an HTS application, which would make it amenable to routine concentration–response experiments. Table S4 compares systems capable of measuring multiple gene expression. The advantage of global profiling (i.e., microarray or RNAseq) is that it can assess thousands of genes and provide valuable insight into pathways and networks that are activated by specific modes of action. Downsides of this approach are its high cost and low throughput, which limit its use in screening applications. nCounter is a multiplex technology developed to accurately and simultaneously quantify the abundance of up to 800 transcripts. Unlike other multiplex gene expression measurement methods, such as qRT-PCR, nCounter is based on direct multiplexed measurement of gene expression that does not involve reverse transcription or other enzymes or require amplification. Therefore, nCounter achieves high levels of precision, linearity, reproducibility, and sensitivity (<1 copy per cell). These unique characteristics of nCounter make it a well-suited technology platform for developing transcriptomic biomarker-based toxicity screening assay.
The results of the TGx-DDI nCounter assay show that this is an excellent platform for TGx-DDI. First, nCounter is equivalent to microarrays in terms of derisking the agents with irrelevant positive CD results (Table 2). Second, the output of nCounter is more sensitive to small expression changes than that of DNA microarrays. The responses of several weak DDI compounds were greater when measured by nCounter, without compromising specificity. Third, the high-throughput capability of the nCounter system allows the development of a highly automated workflow requiring minimal hands-on time for large-scale multicondition screening. Overall, in contrast to microarray approaches, an HTS approach with the direct use of cell lysates also allows for cost-efficient analyses at multiple doses and conditions.
While important in some cases, extensive standard cross-laboratory validation of specific detection platforms is not necessarily the most appropriate approach for this application, given the many different methods of measuring gene expression changes. Performance evaluation should instead focus on responses measured in the gene set when adopting a new technology. Overall, technologies in this area are constantly evolving, and for new applications it will be critical to confirm that the signature works regardless of the platform. Indeed, we note that this assay is not limited to the specific array platform or technology used in our study, as data collected using other array platforms (22) can also be analyzed using the TGx-DDI biomarker (8, 11). Our consortium has also demonstrated the ability of the TGx-DDI classifier to predict DNA damage in the presence of rat liver S9 in human TK6 cells (10, 11). Interestingly, the TGx-DDI biomarker was able to predict DDI agents (11) using published Affymetrix array data (22) in HepaRG cells, a metabolically competent human liver hepatocyte cell line. Thus, along with confirming the utility of the TGx-DDI biomarker in the presence of S9 and in a different cell line, our findings also provide further validation of the TGx-DDI classifier overall by demonstrating its efficacy in an independent dataset produced in two separate laboratories using different technologies. As the biomarker is enriched in p53-responsive genes (8), the use of p53-competent cells for this assay is mandatory.
TGx-DDI is the first genotoxicity biomarker shown to perform robustly and consistently on different assay platforms. The goal is to continue to develop and use this biomarker in a simple, inexpensive, and rapid method that can be easily integrated into the safety evaluation of compounds and chemical series to identify genotoxic effects in vitro that are relevant to in vivo genotoxicity. Cancer can arise through various modes of action. Genotoxicity is one major risk factor; however, we emphasize that this biomarker cannot be used to argue against the relevance of nongenotoxic modes of action in carcinogenesis. The incorporation of the genomic biomarker in genotoxicity risk assessment would reduce animal testing. Considering that many chemical agents cannot be assessed by animal testing due to either cost or recent legislation (6, 7), the TGx-DDI approach addresses an important need. Furthermore, the strategies and protocols that were used in TGx-DDI identification and its validation can serve as a prototype for the development of genomic biomarkers for other toxicities.
Materials and Methods
Detailed descriptions of the materials and methods used in this study are provided in SI Materials and Methods.
Cell Culture and Treatment.
TK6 cells, a spontaneously transformed human lymphoblastoid cell line, were grown and treated with chemical agents as described previously (8). In brief, exponentially growing cells were treated with the indicated chemical agent for 4 h over a broad dosage range, cells were harvested, and total RNA was isolated. qRT-PCR was carried out with representative indicator genes known to be induced by a broad range of stress agents. For agents requiring metabolic activation, treatment of TK6 cells included S9 rat liver extract as described previously (11). For the cell viability assay, after 4 h of treatment, medium was removed from cells, and cells were washed and recovered in fresh medium for 20 h. Cell viability was measured at the end of recovery period using an MTT Assay Kit (Cayman Chemical).
Microarray Procedures.
RNA samples from the concentration setting experiments of each compound at their selected concentrations were pooled together and analyzed using human whole genome expression long-nucleotide probe microarrays (60 nt long; Agilent Technologies) (8). To ensure consistency with previous results (8), two-color microarrays were used, but comparable results have been obtained with single-color microarrays. Each experiment was run on two arrays, and on each array both treated and reference (vehicle control) samples were hybridized in a dye-swap design. Specifically, the reference and treatment samples were labeled with two different fluorescence dyes, Cy3 and Cy5, and then both samples were hybridized onto one array. To reduce the effects associated with different labeling efficiencies, we used a two-color dye-swapping configuration (23). The results from these two arrays were combined for statistical analysis.
Bioinformatics Analyses.
Gene expression data were exported from GeneSpring based on Entrez Gene identifiers. Posterior PA for test samples was performed given the classifier as described by Tibshirani et al. (17) and implemented in the pamr package for R. 2DC was performed using Euclidean distances with average linkage by Genesis (Genesis@genome.tugraz.at). The DDI and non-DDI agents from the original training set were separated in two main clusters. A chemical clustering with the DDI branch was called DDI, and vice versa for non-DDI agents. PCA was performed using the prcomp function (24) in R Bioconductor. We note that a new tool has been recently introduced that has a user-friendly interface allowing the analysis of new test agents using our two-pronged approach (25).
TGx-DDI nCounter Assay.
The nCounter assay was performed with 100 ng of RNA that had previously been pooled and used in the microarray analysis. Methodological details of the nCounter experiments have been published previously (26). In brief, optimized sequences for genes in the TGx-DDI panel were custom-designed and manufactured by NanoString. The CodeSet included the TGx-DDI gene set and eight housekeeping genes—G6PD, GUSB, HPRT1, LDHA, NONO, PGK1, PPIH, and TFRC—selected based on stability and detectable expression levels. The protocol followed standard nCounter instructions (26). Barcodes were counted for each target, and the data were exported. The counts of each target were analyzed using nSolver Analysis version 3.0 for quality control and normalization. Normalized data were subjected to further analysis.
To develop the HTS assay, 5 × 104 cells per well were seeded in a 96-well plate on the day before the treatment. Cells were treated with bleomycin and its corresponding vehicle control (H2O) for 4 h, rinsed to remove the drug, and then either lysed in RNA lysis buffer (products from Qiagen, Ambion, and Promega were tested and performed comparably) at different concentrations or pelleted for RNA isolation. This treatment was performed in triplicate, after which bioinformatics analyses were conducted as described above.
Supplementary Material
Acknowledgments
We thank Dr. Jay George (Trevigen) for support and encouragement and the following individuals for valuable feedback and advice during this project: Drs. Eric Boitier (Sanofi), Alison Harrill (University of Arkansas for Medical Sciences), Hilla Kedar (Teva Pharmaceutical Industries), Warren KU (Boehringer-Ingelheim), Sandra Truex (Pfizer), Van Vleet (Abbvie), and Jing Yuan (Boehringer-Ingelheim). Portions of these results formed the basis for a provisional patent (application no. 62/465,591). This study was partially supported by the Genomics Consortium of the International Life Sciences Institute’s Health and Environmental Sciences Institute (HESI; hesiglobal.org/who-we-are/), for biomarker validation by microarray, and the National Institute of Environmental Health Sciences (NIEHS; R43-ES026473 01), for the nCounter HTS assay development. Portions of this study were supported by NIEHS Grants R43-ES026473 and 1R01-ES020750. The HESI scientific initiative is supported by in-kind contributions (from public and private sector participants) of time, expertise, and experimental effort. These contributions are supplemented by direct funding from HESI’s corporate sponsors, which largely supports program infrastructure and management. A list of supporting organizations (public and private) is available at hesiglobal.org/application-of-genomics-to-mechanism-based-risk-assessment-technical-committee/.
Footnotes
Conflict of interest statement: Georgetown University has filed a provisional patent application for the technology described in this paper, on which A.J.F. and H.-H.L. are inventors.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE107162).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714109114/-/DCSupplemental.
References
- 1.Ku WW, Aubrecht J, Mauthe RJ, Schiestl RH, Fornace AJ., Jr Genetic toxicity assessment: Employing the best science for human safety evaluation Part VII: Why not start with a single test: A transformational alternative to genotoxicity hazard and risk assessment. Toxicol Sci. 2007;99:20–25. doi: 10.1093/toxsci/kfm147. [DOI] [PubMed] [Google Scholar]
- 2.Li HH, Aubrecht J, Fornace AJ., Jr Toxicogenomics: Overview and potential applications for the study of non-covalent DNA interacting chemicals. Mutat Res. 2007;623:98–108. doi: 10.1016/j.mrfmmm.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Snyder RD, Green JW. A review of the genotoxicity of marketed pharmaceuticals. Mutat Res. 2001;488:151–169. doi: 10.1016/s1383-5742(01)00055-2. [DOI] [PubMed] [Google Scholar]
- 4.Goodsaid FM, et al. Voluntary exploratory data submissions to the US FDA and the EMA: Experience and impact. Nat Rev Drug Discov. 2010;9:435–445. doi: 10.1038/nrd3116. [DOI] [PubMed] [Google Scholar]
- 5.Kirkland D, Aardema M, Henderson L, Müller L. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens I. Sensitivity, specificity and relative predictivity. Mutat Res. 2005;584:1–256. doi: 10.1016/j.mrgentox.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Pauwels M, Rogiers V. Human health safety evaluation of cosmetics in the EU: A legally imposed challenge to science. Toxicol Appl Pharmacol. 2010;243:260–274. doi: 10.1016/j.taap.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 7.EU: Final ban on animal experiments for cosmetic ingredients implemented. ALTEX. 2013;30:268–269. [PubMed] [Google Scholar]
- 8.Li HH, et al. Development of a toxicogenomics signature for genotoxicity using a dose-optimization and informatics strategy in human cells. Environ Mol Mutagen. 2015;56:505–519. doi: 10.1002/em.21941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas RS, et al. Incorporating new technologies into toxicity testing and risk assessment: Moving from 21st century vision to a data-driven framework. Toxicol Sci. 2013;136:4–18. doi: 10.1093/toxsci/kft178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yauk CL, et al. Application of the TGx-28.65 transcriptomic biomarker to classify genotoxic and non-genotoxic chemicals in human TK6 cells in the presence of rat liver S9. Environ Mol Mutagen. 2016;57:243–260. doi: 10.1002/em.22004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buick JK, et al. Integration of metabolic activation with a predictive toxicogenomics signature to classify genotoxic versus nongenotoxic chemicals in human TK6 cells. Environ Mol Mutagen. 2015;56:520–534. doi: 10.1002/em.21940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amundson SA, et al. Stress-specific signatures: Expression profiling of p53 wild-type and -null human cells. Oncogene. 2005;24:4572–4579. doi: 10.1038/sj.onc.1208653. [DOI] [PubMed] [Google Scholar]
- 13.Williams A, et al. A predictive toxicogenomics signature to classify genotoxic versus non-genotoxic chemicals in human TK6 cells. Data Brief. 2015;5:77–83. doi: 10.1016/j.dib.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffat I, et al. Comparison of toxicogenomics and traditional approaches to inform mode of action and points of departure in human health risk assessment of benzo[a]pyrene in drinking water. Crit Rev Toxicol. 2015;45:1–43. doi: 10.3109/10408444.2014.973934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Conference on Harmonisation (ICH) of Technical Requirements for Registration of Pharmaceuticals for Human Use 2012 Guidance on genotoxicity testing and data interpretation for pharmaceuticals intended for human use: S2(R1). Available at www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Safety/S2_R1/Step4/S2R1_Step4.pdf. Accessed November 7, 2017.
- 16.Buick JK, et al. Integration of the TGx-28.65 genomic biomarker with the flow cytometry micronucleus test to assess the genotoxicity of disperse orange and 1,2,4-benzenetriol in human TK6 cells. Mutat Res. 2017;806:51–62. doi: 10.1016/j.mrfmmm.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clewell RA, Andersen ME. Approaches for characterizing threshold dose-response relationships for DNA-damage pathways involved in carcinogenicity in vivo and micronuclei formation in vitro. Mutagenesis. 2016;31:333–340. doi: 10.1093/mutage/gev078. [DOI] [PubMed] [Google Scholar]
- 19.Suto MJ, Domagala JM, Roland GE, Mailloux GB, Cohen MA. Fluoroquinolones: Relationships between structural variations, mammalian cell cytotoxicity, and antimicrobial activity. J Med Chem. 1992;35:4745–4750. doi: 10.1021/jm00103a013. [DOI] [PubMed] [Google Scholar]
- 20.Shimada H, Itoh S. Effects of new quinolone antibacterial agents on mammalian chromosomes. J Toxicol Environ Health. 1996;47:115–123. doi: 10.1080/009841096161825. [DOI] [PubMed] [Google Scholar]
- 21.Noordhuis P, et al. 5-Fluorouracil incorporation into RNA and DNA in relation to thymidylate synthase inhibition of human colorectal cancers. Ann Oncol. 2004;15:1025–1032. doi: 10.1093/annonc/mdh264. [DOI] [PubMed] [Google Scholar]
- 22.Doktorova TY, et al. Transcriptomic responses generated by hepatocarcinogens in a battery of liver-based in vitro models. Carcinogenesis. 2013;34:1393–1402. doi: 10.1093/carcin/bgt054. [DOI] [PubMed] [Google Scholar]
- 23.Patterson TA, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 24.Venables WN, Ripley BD. Modern Applied Statistics with S. Springer; New York: 2002. [Google Scholar]
- 25.Jackson MA, et al. The TGx-28.65 biomarker online application for analysis of transcriptomics data to identify DNA damage-inducing chemicals in human cell cultures. Environ Mol Mutagen. 2017;58:529–535. doi: 10.1002/em.22114. [DOI] [PubMed] [Google Scholar]
- 26.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 27.Kirkland D, Aardema M, Müller L, Makoto H. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens, II: Further analysis of mammalian cell results, relative predictivity and tumour profiles. Mutat Res. 2006;608:29–42. doi: 10.1016/j.mrgentox.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Kirkland D, et al. How to reduce false positive results when undertaking in vitro genotoxicity testing and thus avoid unnecessary follow-up animal tests: Report of an ECVAM workshop. Mutat Res. 2007;628:31–55. doi: 10.1016/j.mrgentox.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Fellows MD, O’Donovan MR, Lorge E, Kirkland D. Comparison of different methods for an accurate assessment of cytotoxicity in the in vitro micronucleus test, II: Practical aspects with toxic agents. Mutat Res. 2008;655:4–21. doi: 10.1016/j.mrgentox.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Dearfield KL, et al. Follow-up actions from positive results of in vitro genetic toxicity testing. Environ Mol Mutagen. 2011;52:177–204. doi: 10.1002/em.20617. [DOI] [PubMed] [Google Scholar]
- 31.Kirkland D, Kasper P, Müller L, Corvi R, Speit G. Recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests: A follow-up to an ECVAM workshop. Mutat Res. 2008;653:99–108. doi: 10.1016/j.mrgentox.2008.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.