Significance
Aminoglycosides are well known as antibiotics that target the bacterial ribosome. However, they also impact the eukaryotic translation mechanism to promote read-through of premature termination codons (PTCs) in mRNA. Aminoglycosides are therefore considered as potential therapies for PTC-associated human diseases. Here, we performed a comprehensive study of the mechanism of action of aminoglycosides in eukaryotes by applying a combination of structural and functional approaches. Our findings reveal complex interactions of aminoglycosides with eukaryotic 80S ribosome caused by their multiple binding sites, which lead to inhibition of intersubunit movement within the human ribosome that impact nearly every aspect of protein synthesis.
Keywords: ribosome, aminoglycosides, PTC read-through, translation, protein synthesis
Abstract
Aminoglycosides are chemically diverse, broad-spectrum antibiotics that target functional centers within the bacterial ribosome to impact all four principle stages (initiation, elongation, termination, and recycling) of the translation mechanism. The propensity of aminoglycosides to induce miscoding errors that suppress the termination of protein synthesis supports their potential as therapeutic interventions in human diseases associated with premature termination codons (PTCs). However, the sites of interaction of aminoglycosides with the eukaryotic ribosome and their modes of action in eukaryotic translation remain largely unexplored. Here, we use the combination of X-ray crystallography and single-molecule FRET analysis to reveal the interactions of distinct classes of aminoglycosides with the 80S eukaryotic ribosome. Crystal structures of the 80S ribosome in complex with paromomycin, geneticin (G418), gentamicin, and TC007, solved at 3.3- to 3.7-Å resolution, reveal multiple aminoglycoside-binding sites within the large and small subunits, wherein the 6′-hydroxyl substituent in ring I serves as a key determinant of binding to the canonical eukaryotic ribosomal decoding center. Multivalent binding interactions with the human ribosome are also evidenced through their capacity to affect large-scale conformational dynamics within the pretranslocation complex that contribute to multiple aspects of the translation mechanism. The distinct impacts of the aminoglycosides examined suggest that their chemical composition and distinct modes of interaction with the ribosome influence PTC read-through efficiency. These findings provide structural and functional insights into aminoglycoside-induced impacts on the eukaryotic ribosome and implicate pleiotropic mechanisms of action beyond decoding.
Aminoglycosides are broad-spectrum, bactericidal antibiotics of critical importance to the treatment of life-threatening infections. Despite their proven clinical utility, these therapeutics can lead to potential toxic side effects, including ototoxicity and nephrotoxicity, and an increased prevalence of resistance (1, 2). The most heavily employed and extensively investigated aminoglycosides contain a central 2-deoxystreptamine (2-DOS) ring. This class is comprised of both natural products (such as neomycin and paromomycin) and semisynthetic derivatives (such as dibekacin and amikacin).
The 2-DOS aminoglycosides effectively inhibit protein synthesis in bacteria by targeting the mechanisms of translation elongation, termination, and recycling (3–6). These activities have, in part, been distilled to the capacity of the 2-DOS rings to engage chemical features within the deep and narrow major groove of the 16S rRNA secondary structure (7). Structural insights into the mechanisms of 2-DOS aminoglycoside action were first obtained through chemical footprinting methods (8) and later using RNA fragments of the bacterial ribosome (7, 9) and isolated Thermus thermophilus 30S ribosomal subunits (10–12). These investigations revealed that aminoglycosides interact within the major groove of a conserved, asymmetric internal loop within the helix 44 (h44) decoding center of 16S rRNA within the small ribosomal subunit to affect the decoding mechanism.
Structural investigations using isolated 30S ribosome subunits led to the hypothesis that the universally conserved A1492, A1493, and G530 residues within the h44 decoding center actively “monitor” the interaction between the tRNA anticodon and the mRNA codon (12). To do so, A1492/A1493 must extrude from the helical axis of h44 to “recognize” the codon–anticodon helix through A-minor groove interactions. This local conformational change then couples to global conformational changes in the ribosome (domain closure) that enable tRNA accommodation. It was also suggested that the inability of mismatched near-cognate tRNA to form proper A-minor groove interactions prevents domain closure, thereby favoring tRNA rejection (11). The binding of paromomycin and neomycin to helix 44 in the crystals of isolated 30S subunits also extrudes both decoding nucleotides A1492 and A1493, leading to the hypothesis that stabilization of extrahelical A1492/A1493 positions is directly related to the misincorporation of near- and noncognate tRNAs into the ribosome during translation (10, 12). However, later studies of the fused-ring 2-DOS aminoglycoside apramycin demonstrated that extrahelical A1492/A1493 positions alone are insufficient to induce miscoding (13). The non-DOS aminoglycoside streptomycin, which also promotes translation errors, exerts distinct conformational changes in the decoding site such that residues A1492 and A1493 remain intercalated within the h44 helical axis (14).
Recent structural studies of the functional 70S ribosome in complex with mRNA and tRNAs in the P- and E-sites (peptidyl- and exit-tRNA–binding sites, respectively) show that the A1493 nucleotide adopts an extrahelical positon in the absence of tRNA within the decoding site (15). By contrast, the decoding-specific changes in positions of nucleotides A1492 and G530 and domain closure require the binding of either cognate or misincorporated near-cognate tRNAs (16, 17). In the context of the 70S ribosome, paromomycin binding to the decoding center was also shown to elicit moderate structural rearrangements in the A-site tRNA-binding pocket, which may influence translation accuracy (16, 18, 19).
Binding of 2-DOS aminoglycosides to the ribosome has also been documented within the major groove of Helix 69 (H69) of the large ribosomal subunit, which forms a critical intersubunit bridge (B2a) that directly contacts the h44 decoding site of the small subunit (3). Paromomycin or neomycin binding to H69 alters the conformation of bridge B2 and the process of small subunit rotation with respect to the large subunit that accompanies nearly every aspect of the translation mechanism (4, 20). These impacts also hinge on interactions of the 6′-OH group of h44-bound aminoglycosides with the universally conserved A1913 residue located within the apical tip of the H69 stem loop (21).
The basis of 2-DOS aminoglycoside-class antibiotic selectivity is understood to arise from structural differences in the h44 decoding sites of bacterial and eukaryotic ribosomes. In eukaryotes, the presence of A1408G and G1491A base substitutions (bacterial numeration) within h44 (Fig. 1A) alter key binding interactions mediated by aminoglycoside rings I and II (22, 23). Nonetheless, specific 2-DOS aminoglycosides such as geneticin (G418) retain the capacity to bind eukaryotic ribosomes. G418 belongs to the 4,6-linked aminoglycoside class that contains a ring I 6′-OH group (Fig. S1). Its interactions with the eukaryotic h44 decoding region are accommodated by conformational plasticity within both the target and drug that enables a network of specific, stabilizing interactions (24).
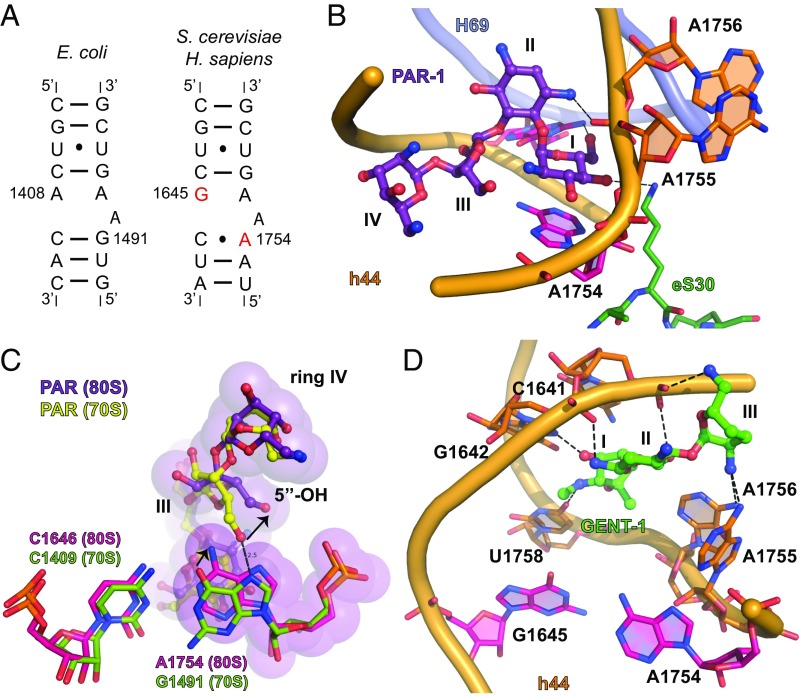
Fig. 1.
Aminoglycosides target the decoding center of the 80S ribosome in a different ways. (A) Secondary structure of h44 of the small ribosomal subunit from bacteria (E. coli) and eukaryotes (identical in S. cerevisiae and Homo sapiens). Substituted nucleotides implicated in the selectivity of aminoglycosides are marked in red. (B) Binding of paromomycin (PAR-1) to h44 in the 80S ribosome from S. cerevisiae. Paromomycin is colored violet, and rings I, II, III, and IV of paromomycin are marked. Ring I is in stacking with A1754. Residues A1754, and G1645 are colored pink. h44 is shown in orange; H69 of the large ribosomal subunit is shown in light blue; and the eukaryote-specific protein eS30 is shown in green. Oxygen atoms are colored red, and nitrogen atoms are colored blue. (C) Comparison of PAR-1 binding to h44 in the 70S ribosome from T. thermophilus (PDB ID code 5EL6) and the 80S ribosome from S. cerevisiae. Paromomycin in complex with 70S is shown in yellow; residues of 16S rRNA of 70S are in green; other color-coding is as in B. The shift in the position of A1754 and the movement of 5′-OH group in ring III of paromomycin are marked with arrows. (D) Binding of gentamicin (GENT-1) to h44 in the 80S ribosome from S. cerevisiae. Gentamicin is shown in green; other color-coding is as in B. Rings I, II, and III of gentamicin are marked, and atoms located at a hydrogen bonding distance are connected by dashed lines.
Investigations of aminoglycoside activity in both human cells and the wheat embryo system revealed that aminoglycosides such as paromomycin and G418, which both possess a 6′-OH group in ring I, are efficient in promoting missense errors during protein synthesis (25, 26). Paromomycin and, with much less efficiency, neomycin were also recognized as being effective suppressors of nonsense mutations (27). Since that time, gentamicin, G418, tobramycin, and amikacin, which possess either a 6′-OH or 6′-NH2 moiety in ring I, have all been shown to induce suppression of premature termination codons (PTCs) (28). Mutations that introduce PTCs are understood to be causative in ∼11% of the >5,000 human genetic diseases identified to date, including sporadic cancers arising from mutations in tumor-suppressor genes such as TP53 (29, 30). Consequently, aminoglycosides are regarded as potential therapies for the treatment of human disease.
The application of aminoglycosides for suppression therapies has been limited in practice by their toxicities and their low efficiencies of stop-codon read-through (31, 32). Despite these shortcomings, aminoglycosides have been enrolled in clinical trials for the treatment of cystic fibrosis (33) and Duchenne muscular dystrophy (34) and have shown therapeutic potential for the treatment of dystrophic epidermolysis bullosa (35) and Werner syndrome (36) as well as specific cancers (37). The neomycin derivative TC007 has also been tested in the context of spinal muscular atrophy (SMA) in both human fibroblasts (32) and mouse models of disease (38).
As the molecular basis of aminoglycoside action against eukaryotic ribosomes is currently lacking, we have examined the interactions of aminoglycosides with 80S eukaryotic ribosomes using X-ray crystallography and single-molecule FRET (smFRET) imaging. X-ray structural analyses of Saccharomyces cerevisiae 80S ribosomes in complex with paromomycin, G418, gentamicin, and TC007 reveal that aminoglycosides interact at multiple sites within both 18S and 28S rRNA. Multivalent aminoglycoside–ribosome interactions were further corroborated by the impact of aminoglycosides on the spontaneous dynamics within the human ribosome and their propensities to promote errors in tRNA selection. These investigations further revealed that TC007 exhibited distinct modes of interaction with and miscoding and structural impacts on the eukaryotic ribosome at ∼20-fold lower concentrations than observed for G418, paromomycin, or gentamicin. These findings are consistent with the efficacy profiles of TC007 in human cells and with this compound’s unique capacity to promote PTC read-through.
Results
Aminoglycoside Binding to the Eukaryotic h44 Decoding Site.
To investigate the binding mode of aminoglycosides with low affinity to the 80S ribosome, we first sought alternative experimental conditions for S. cerevisiae 80S ribosome crystal treatment. Osmium hexamine, which is normally present in this procedure, binds rRNA, including the aminoglycoside-binding site within the h44 decoding region (39). Hence, to examine the binding of aminoglycosides that exhibit low affinities for the 80S ribosome, we removed osmium hexamine and instead treated crystals of the ribosome in the presence of high concentrations (up to 4 mM) of aminoglycosides (Materials and Methods).
Under these conditions, we obtained a structure of the 4,6-linked aminoglycoside G418 bound to the S. cerevisiae 80S ribosome at 3.7-Å resolution (Table S1). A similar structure had been previously solved in the presence of osmium hexamine [Protein Data Bank (PDB) ID code 4U4O] (24), where it was shown that G418 can compete with osmium hexamine for h44 binding. In both structures, G418 was found to exhibit canonical binding to the h44 decoding site with ring I of the antibiotic stacking on nucleotide A1754 (G1491 in bacterial numbering), displacing the conserved nucleotides A1755 (A1492) and A1756 (A1493) away from the helical axis (Fig. S2). There, the 6′-OH group of ring I is positioned within hydrogen bonding distance (2.7 Å) of the N2 atom of G1645 (A1408) (Fig. S2). Replacement of the 6′-OH group in ring I with a hydrogen bond acceptor, such as a 6′-NH2 group, could create repulsion from the N2 atom of G1645 (A1408) precluding aminoglycoside binding. Likewise, the presence of adenosine instead of guanosine at position 1754 (1491) would disrupt Watson–Crick base pairing with C1646 (C1409), a structural component of the bacterial h44 decoding site that is essential to aminoglycoside binding (7, 24, 40).
Using similar osmium hexamine-free conditions, we solved an 80S–paromomycin structure at 3.3-Å resolution (I/σ = 0.96, CC1/2 = 34.9) (Table S1). Like G418, the 4,5-linked aminoglycoside paromomycin contains a 6′-OH group in ring I (Fig. S1). As for G418, at high drug concentration (4 mM for soaking of paromomycin), we observed weak, positive electron density for paromomycin (PAR-1) in the h44 decoding site (Fig. 1B). Within this site, rings I and II of paromomycin adopted a position globally similar to that of G418, wherein ring I of the antibiotic stacked on nucleotide A1754 (G1491 in bacterial numbering), and the contact between the 6′-OH group in ring I and the N2 atom of G1645 (A1408) was maintained (3.3 Å distance) (Fig. S3A). To avoid steric clash with the 6-NH2 group of A1754 (G1491), ring III of paromomycin was found to be rotated compared with the structure of paromomycin bound to the bacterial 70S ribosome (Fig. 1C) (18). This repositioning reoriented ring IV within the major groove, breaking the hydrogen bond between the 5′-OH in ring III and the N7 atom of G1491 present in the 70S–paromomycin complex. Interestingly, the 4′-OH group in ring I of paromomycin is positioned within hydrogen bonding distance (3.5 Å) of the ζ-NH2 group of lysine 3 of the eukaryote-specific protein eS30, which approaches h44 from the minor groove face (Fig. 1B). This interaction, which is absent in both the paromomycin–70S ribosome complex and the 80S–G418 structures (18, 24), may partially compensate for the reductions in binding affinity arising from sequence changes within the h44 decoding site (Fig. 1A) so that low levels of miscoding are maintained (25, 26).
Gentamicin, a 4,6-linked aminoglycoside produced as a mixture of C1, C1a, C2, and C2a isoforms (29) and which contains amine along with a few methyl groups in the 6′ position of ring I (Fig. S1) (41), does not induce errors in translation but competes with paromomycin-induced miscoding (26). These data suggest a common binding site within h44 but a unique mode of interaction with the 80S ribosome. As for paromomycin, the crystal structure of the 80S ribosome in complex with gentamicin, solved at 3.4 Å resolution (I/σ = 0.94, CC1/2 = 33.5) (Table S1), exhibits positive electron density in h44 (GENT-1) at a high drug concentration (4 mM) (Fig. S3B). Under these conditions, gentamicin’s ring I does not stack upon A1754 (G1491), and nucleotides A1755 (A1492) and A1756 (A1493) adopt only semiextruded positions relative to the axis of h44 so that their N2 atoms are within hydrogen bonding distance of the 2′-NH2 group of ring III. Thus, positioned, rings I–III also make sequence-specific contacts with O4 of U1758 (U1495) and N7 of G1642 (G1405) (Fig. 1D and Fig. S3C). This noncanonical pose, which appears to be enforced by repulsion between the N1 and N2 atoms of G1645 (A1408) and the NH2-group in the 6′ position of ring I, rationalizes gentamicin’s capacity to compete with paromomycin binding while being unable to support miscoding.
Interactions of TC007 with the 80S Ribosome and Implications for PTC Read-Through.
SMA, a leading genetic cause of infantile death, is an autosomal recessive disease for which there is currently no cure (42). SMA is associated with the loss of full-length SMN protein (43). Initial screens for small molecule-mediated suppression therapies for potential treatment of SMA led to the discovery of TC007, a three-ring 4,5-linked aminoglycoside bearing a 6′-NH2 group on ring I (Fig. S1) (32). TC007 testing in fibroblasts from SMA patients and in SMA mouse models showed good toxicity profiles and prolonged the lifespan of SMA mice while partially lessening the severity of disease (44).
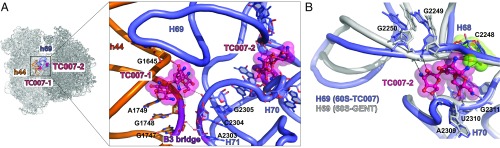
To gain insight into TC007 interactions with the 80S ribosome, we solved the X-ray structure of the 80S–TC007 complex at 3.7-Å resolution (I/σ = 0.98, CC1/2 = 36.1) (Table S1). Consistent with a steric clash between the 6′-NH2 and the G1645 (A1408) residue (24), TC007 was not observed to bind the small subunit decoding center, despite being present at 4 mM concentration. Accordingly, no electron density was evidenced for the disordered nucleobases of residues A1755 (A1492) or A1756 (A1493). However, positive electron density was observed for TC007 (TC007-1) immediately below the canonical decoding site and above intersubunit bridge B3, spanning between h44 of the small subunit and Helix 71 (H71) of the large subunit at the intersubunit space (Fig. 2A and Fig. S4A). A second TC007 molecule (TC007-2) was also evidenced between Helix 68 (H68) and Helix 70 (H70) in the large ribosomal subunit, proximal to the highly conserved H69 element of intersubunit bridge B2. This binding site is close to (within ∼10 Å) but is distinct from the noncanonical aminoglycoside-binding site observed within the H69 major groove in the bacterial ribosome (3, 20, 21) and distorts the H69 conformation (Fig. 2 A and B and Fig. S4B).
Fig. 2.
Interactions of the aminoglycoside derivative TC007 with the 80S ribosome. (A, Left) The view from the A-site of the ribosome is indicated. (Right) Enlarged view showing binding sites of TC007-1 and TC007-2 in close proximity to h44 of the small ribosomal subunit and H69 of the large ribosomal subunit, respectively. The regions of h44 of the small ribosomal subunit and H71 of the large ribosomal subunit, which form intersubunit bridge B3, are colored violet. The 40S subunit is shown in orange. Contacts of TC007 and rRNA residues are depicted by dashed lines. The 60S subunit is shown in blue, and TC007 is in magenta. Oxygen atoms are colored red, and nitrogen atoms are colored blue. (B) Close-up view of the second binding site of TC007-2 located between helices H68, H69, and H70 of the large ribosomal subunit. Local alignment demonstrates that TC007-2 would clash with the phosphate group of C2248 if H69 adopted the conformation characterized by the different rotational state of the ribosomal subunits as observed in the 80S–gentamicin structure. The 60S subunit from the 80S–gentamicin structure is colored gray; residue C2248 is colored lime and is marked with spheres; other color-coding is as in A.
Interactions of TC007 with Bacterial 70S Ribosome.
To shed light on the selectivity of TC007, we solved the structure of the bacterial 70S ribosome from T. thermophilus cocrystallized in complex with three tRNAs, mRNA, and TC007 at 2.95 Å (I/σI = 1.38, CC1/2 = 47.1) (Table S1), where the drug was at 50-fold excess over the ribosome (Materials and Methods). As expected, we observed strong positive electron density in the h44 decoding site, where rings I and II were observed to overlap with those of paromomycin (Fig. S4C). In contrast to ring III of paromomycin, which interacts with the Hoogsteen face of G1491 (Escherichia coli numbering), the O7 atom of ring III of TC007 is positioned within hydrogen bonding distance of N4 of C1407. When TC007 was soaked into preformed crystals of the same 70S–tRNA–mRNA complex (500 µM for 24 h), we were able to obtain a structure at 3.05 Å resolution (I/σI = 1.25, CC1/2 = 39.9) (Table S1) in which TC007 was bound in an identical position within h44 (TC007–h44) (Fig. S4D). As for neomycin, gentamicin, and paromomycin binding to H69 within the Escherichia coli 70S ribosome (3, 20, 21), TC007 was observed to make an array of hydrogen bonding contacts with bases lining the H69 major groove, including sequence-specific contacts with residues G1921 and G1922 (Fig. S4 B and E). Remarkably, the orientation of TC007 is different from that in a previously reported 70S–neomycin structure (21): The position of ring II is similar in the two antibiotics, but the positions of rings I and III are swapped. As for gentamicin, intersubunit bridging contacts present in both paromomycin and neomycin structures were not observed due to the absence of ring IV. As H69 is sterically accessible within the eukaryotic 80S ribosome, we attribute the observed differences in TC007 binding to Ψ2264 (G1921) and C2265 (G1922) substitutions present in the 80S ribosome (Fig. S4 B and F), which likely disrupt the drug’s capacity to pack tightly against the floor of the major groove. These findings confirm that the TC007-binding sites in the T. thermophilus 70S ribosome are distinct from those found in the 80S ribosome of S. cerevisiae (Fig. S4B).
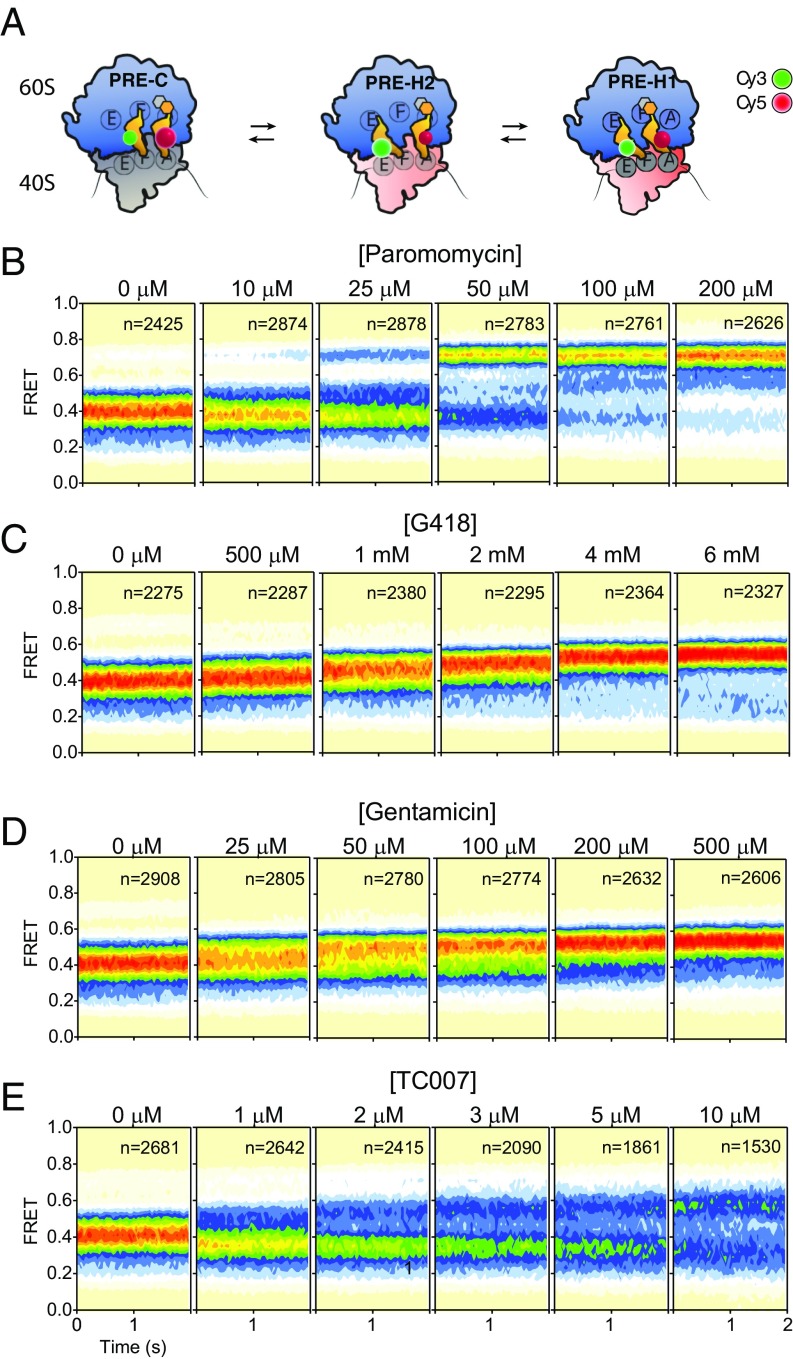
Aminoglycosides Inhibit Intersubunit Rotation of the Eukaryotic Ribosome.
To assess the impact of aminoglycosides on the dynamics of subunit rotation within functional 80S ribosome complexes, we performed smFRET imaging on surface-immobilized human 80S pretranslocation (PRE) complexes in the absence and presence of paromomycin, G418, gentamicin, and TC007 (45). As previously reported (45), in the absence of drug, human PRE complexes predominantly exhibited a mixture of lower-FRET (H2: ∼0.2 and H1: ∼0.4) hybrid state configurations in which the 3′-CCA end of deacylated tRNA occupies the large subunit E-site and the 3′-CCA end of peptidyl-tRNA occupies either the aminoacyl (A)- or P-site, respectively (A/A and P/E; A/P and P/E) (Fig. 3 A and B, Left). The addition of paromomycin to the 80S PRE complex first lowered the average FRET value of the hybrid-state tRNA configurations, suggesting a relative stabilization of the H2 hybrid state in which peptidyl-tRNA returns to its classic (A/A) position while deacylated tRNA remains in its hybrid (P/E) state (46). This impact was maximized at a drug concentration of ∼10 µM. Above this concentration, we observed the appearance of an intermediate-FRET (∼0.55) state, followed by stabilization of a high-FRET, classic (C) PRE complex conformation (Fig. 3B). The estimated EC50 of this effect was ∼35 µM. These data are consistent with a multivalent impact of paromomycin binding to the 80S PRE complex, which first stabilizes peptidyl-tRNA in its classic position within the A-site, followed by a shift of deacylated P-site tRNA from its hybrid to its classic position. By contrast, G418, which also bears a ring I 6′-OH substituent, only stabilized at an intermediate-FRET state (∼0.55) (Fig. 3C). Notably, consistent with the concentration range used to inhibit mammalian cell culture growth (47), the EC50 of this impact was ∼2 mM. By analogy to bacterial systems, the intermediate-FRET state may reflect a reversal of subunit rotation from the P/E hybrid state, which promotes a chimeric, intersubunit hybrid configuration of deacylated P-site tRNA associated with the global inhibition of translation factor binding (20, 21).
Fig. 3.
Aminoglycoside-induced changes in human 80S PRE complex conformation. (A) Schematic showing classic (C) and hybrid (H2 and H1) states of the human PRE ribosome complex. Large (60S) and small (40S) subunits (unrotated, gray; rotated, pink), tRNAs (orange), and sites of donor (Cy3, green) and acceptor (Cy5, red) fluorophore labeling on tRNA in the P- and A-sites, respectively, are indicated. (B–E) Population FRET histograms showing the impact of paromomycin (B), G418 (C), gentamicin (D), and TC007 (E) on the equilibrium distribution of FRET states exhibited by the 80S PRE complex. The concentration of antibiotic is indicated; n, number of single-molecule observations made in each experiment.
Analogous investigations of gentamicin, which contains a 6′-NH2 group on ring I, also led to intermediate-FRET (∼0.55) state stabilization, but the EC50 of its impact was approximately an order of magnitude lower (∼100 µM) than that of G418 (Fig. 3D). TC007, which also has a 6′-NH2 group on ring I, exhibited impacts on the 80S PRE complex that shared characteristics of G418 and gentamicin as well as paromomycin, in which drug binding was bimodal in nature. At concentrations below 2 µM, TC007 predominantly promoted a lower-FRET PRE complex conformation, in line with H2 hybrid state stabilization (A/A; P/E). Higher TC007 concentrations increasingly stabilized an intermediate-FRET (∼0.55) state, similar to that evidenced in the presence of subsaturating paromomycin and saturating G418 and gentamicin concentrations (Fig. 3E). At 10 µM, both H2 and intermediate-FRET PRE complex conformations persisted. Above this concentration the FRET distribution broadened substantially, consistent with an increase in dynamic PRE complex behaviors.
Aminoglycoside-Induced Errors in tRNA Selection on the Eukaryotic Ribosome.
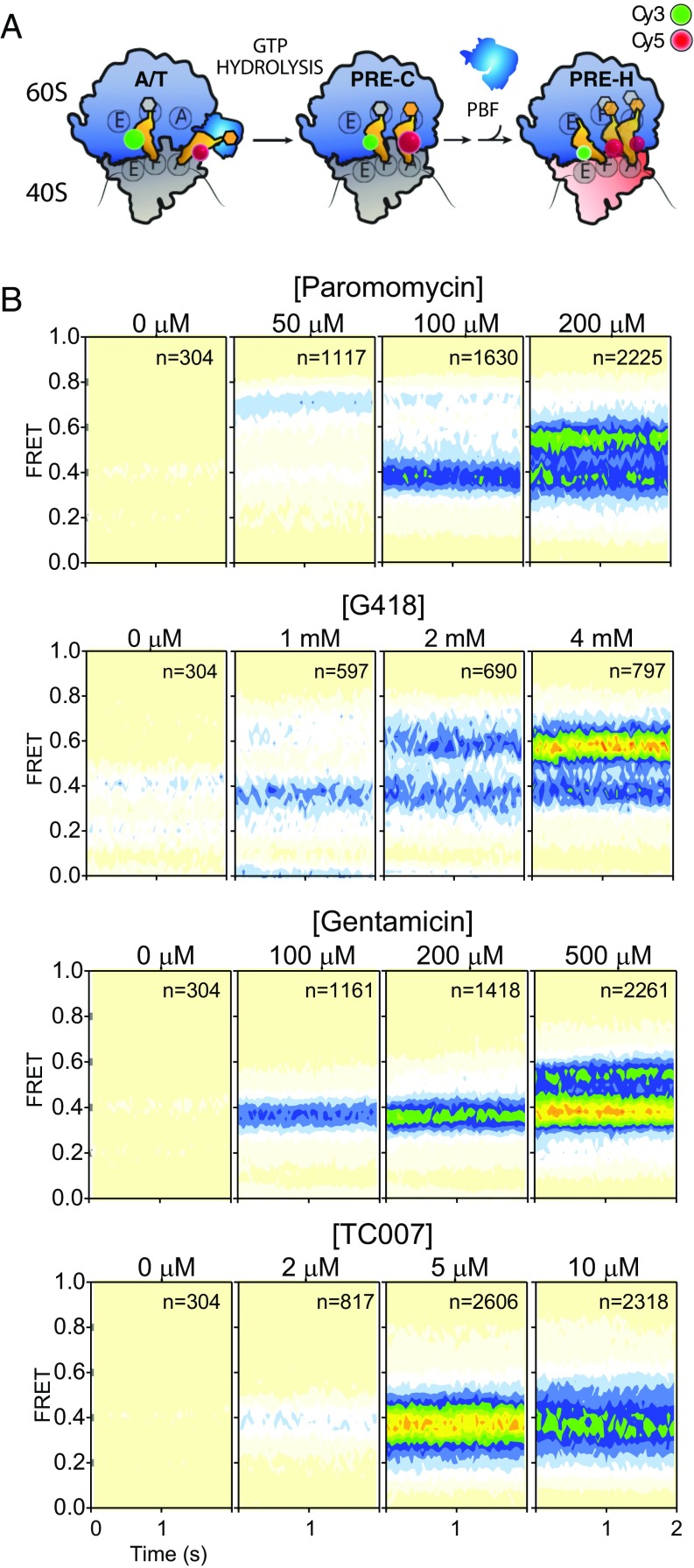
To evaluate the impact of aminoglycosides on aminoacylated tRNA (aa-tRNA) miscoding at the A-site, we examined the extent of aa-tRNA misincorporation of Phe-tRNAPhe into surface-immobilized 80S initiation complexes (ICs) containing (Cy3)Met-tRNAiMet in the P-site, in which the A-site codon was changed from the cognate UUU codon to the near-cognate UCU codon. To provide adequate time for miscoding, the eEF1A(GTP)-(Cy5)Phe-tRNAPhe ternary complex (20 nM) was incubated with surface-immobilized ribosome complexes for 2 min (Materials and Methods) followed by buffer exchange to remove unbound ternary complexes. Using this approach, we could specifically examine the extent of PRE complex formation in the absence of convoluting signals arising from transient ternary complex binding events. Consistent with a high-fidelity decoding mechanism, near-cognate ternary complexes were efficiently rejected from the A-site in the absence of drug so that little or no PRE complex formed (Fig. 4 A and B, Left). By contrast, increasing levels of A-site miscoding were observed as a function of paromomycin, G418, gentamicin, and TC007 concentration. In each case, the extent of miscoding correlated with the concentrations of drug, where the EC50 of miscoding closely mirrored that observed for drug binding to the cognate PRE complex (Figs. 3 and 4). Strikingly, the distribution of FRET values exhibited by precomplexes bearing near-cognate tRNA at the A-site were distinct from those bearing cognate tRNAs; the near-cognate complexes were found to exhibit multiple distinct FRET states (compare far right panels in Figs. 3 and 4). Inspection of individual FRET trajectories of PRE complexes bearing near-cognate tRNA in the A-site showed that the FRET states observed were in dynamic exchange. These findings suggest that the nature of the codon–anticodon pair in the A-site influences the ribosome’s interaction with antibiotics and tRNA in a manner that affects both the conformation and dynamics of the PRE complex.
Fig. 4.
Aminoglycoside-induced miscoding during tRNA selection on the human ribosome. (A) Schematic showing the process of tRNA selection, in which the ternary complex of eEF1A (blue), GTP, and aa-tRNA (orange) enters the A-site of the 80S ribosome. The process of tRNA selection proceeds through the A/T state in which codon–anticodon pairing on the small subunit occurs while the 3′-aminoacylated CCA-end of tRNA remains bound to eEF1A. GTP hydrolysis by eEF1A facilitates the aa-tRNA release from eEF1A and the accommodation of its ′CCA end into the large subunit A-site on the classically (C) configured (unrotated) PRE complex. Peptidyl- and aminoacyl-tRNAs in the P- and A-sites, respectively, then undergo peptide bond formation (PBF), enabling deacylated P-site tRNA and peptidyl-tRNA to achieve hybrid (H2 and H1) states. Large (60S) and small (40S) subunits (unrotated, gray; rotated, pink), tRNAs (orange), and sites of donor (Cy3, green) and acceptor (Cy5, red) fluorophore labeling on tRNA in the P- and A-sites, respectively, are indicated. (B) Population FRET histograms showing that aminoglycoside-induced errors in tRNA selection lead to the accumulation of PRE ribosome complexes bearing near-cognate tRNA in the A-site. The concentration of antibiotic is indicated; n, number of single-molecule observations made in each experiment.
Secondary Binding Sites: Hot Spots Targeted in the Eukaryotic 80S Ribosome by Aminoglycosides.
Aminoglycosides are positively charged and are well known to bind a diverse range of RNA molecules, including catalytic RNAs such as ribonuclease P, self-splicing introns, and ribozymes (48–50). Crystal structures of the isolated small ribosomal subunit as well as intact 70S ribosomes from bacteria have all shown evidence of multiple aminoglycoside-binding sites (3, 13, 14, 20, 21). The positively charged anticancer drug cisplatin also has several binding sites in the 70S ribosome (51).
As high aminoglycoside concentrations were used in each of our crystallographic investigations under conditions of relatively low ionic strength (∼130 mM salt concentration) and neutral pH (7.0–7.5), secondary binding sites were observed for each of the aminoglycosides examined. While it is difficult to link the binding sites observed with the impairment of specific ribosome functions due to the presence of numerous copies of rRNA genes in the genome, we note that several of the secondary aminoglycoside-binding sites map to key functional centers of the ribosome (Fig. 5A). These findings are consistent with the known propensities of aminoglycosides to show diverse impacts on the bacterial translation mechanism (20, 52).
Fig. 5.
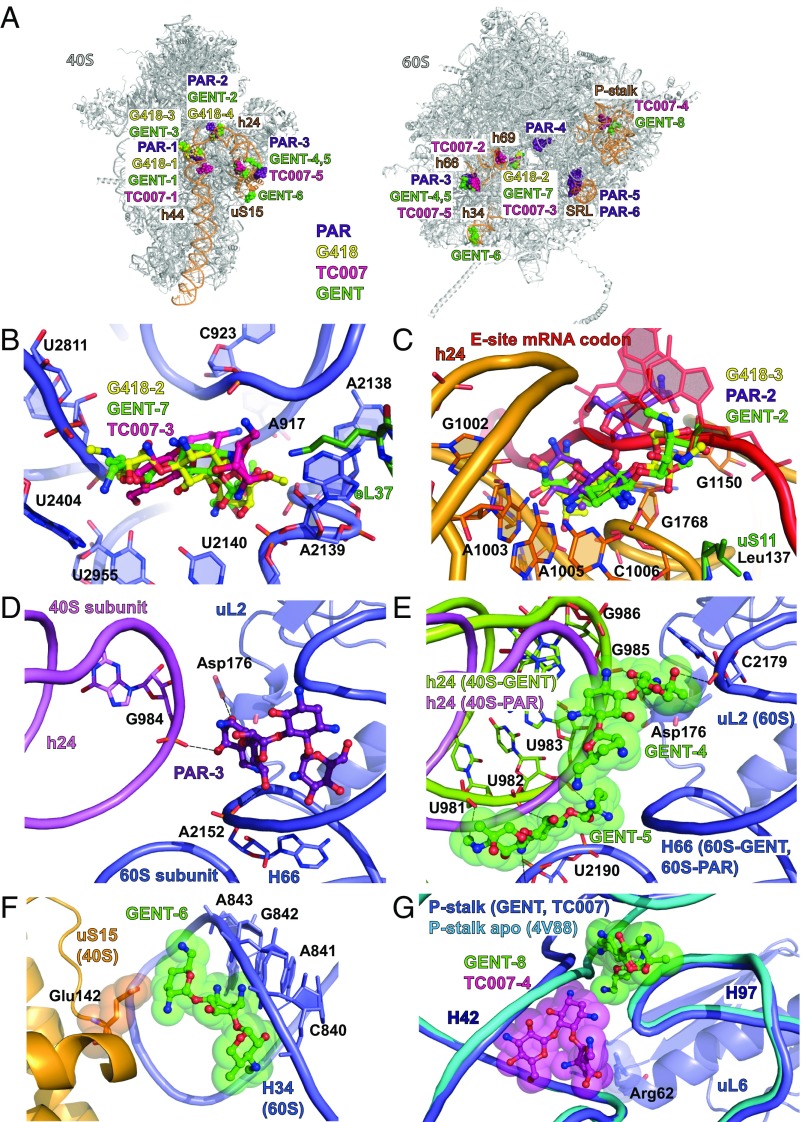
Overview of the secondary binding sites of aminoglycosides in 80S ribosome. (A) Binding sites of gentamicin (GENT), G418, TC007, and paromomycin (PAR) in the 80S ribosome from S. cerevisiae. All structures were aligned either on 18S rRNA or on 28S rRNA, for small and large subunits, respectively, in the 80S–gentamicin structure. The ribosome is colored light gray; elements of the binding pockets of aminoglycosides are colored light orange; gentamicin is colored green; G418 is colored yellow; TC007 is colored magenta; and paromomycin is colored violet. (B) Binding of GENT-7, G418-2, and TC007-3 to the peptide exit tunnel of the 80S ribosome. G418 is colored yellow; gentamicin is colored light green; TC007 is colored magenta; the large ribosomal subunit is colored blue; and the eukaryote-specific protein eL37 is colored green. Similar poses are observed for G418-2 and GENT-7, but the orientation of TC007-3 is different. Oxygen atoms are colored red, and nitrogen atoms are colored blue. (C) Binding sites of PAR-2, GENT-2, and G418-3 in the E-site of the small ribosomal subunit overlapping the position of the mRNA. Structures of the 80S ribosome in complex with paromomycin, gentamicin, and G418 were locally aligned on the structure of the 70S ribosome from T. thermophilus in complex with tRNAs and mRNA (PDB ID code 5EL6). mRNA is colored red; elements of the 70S ribosome are omitted for clarity. The 40S subunit is colored orange; paromomycin is colored violet; the universal protein uS11 is colored green; and other color-coding is as in A. (D) Interactions of PAR-3 with the elements of the intersubunit bridge B2c. Contacts made between paromomycin and G984 of the 40S subunit, A2152 of the 60S subunit, and Asp176 of uL2 are marked with dashed lines. The 40S subunit is colored light pink; paromomycin is colored violet; the 60S subunit is colored blue; and the universal protein L2 (uL2) is colored blue. Oxygen atoms are colored red, and nitrogen atoms are colored blue. (E) GENT-4 and GENT-5 stabilize particular conformations of bridge B2c. The 80S–paromomycin structure was aligned on the 80S–gentamicin structure based on the 28S rRNA. The alignment demonstrates that rearrangement of the bridge B2c would be blocked by gentamicin due to a clash with h24 of the 40S subunit in the 80S–paromomycin structure (colored in pink). 80S–gentamicin contacts are marked with dashed lines. The 40S subunit from 80S–gentamicin structure is colored lime; other color-coding is as in A–C. (F) GENT-6 targets bridge B4 formed by the universal protein uS15 protein and H34 of the large subunit. uS15 is shown in yellow; other color-coding is as in D. Glutamine 142, which interacts with GENT-6, is depicted as spheres. (G) Interactions of GENT-8 and TC007-4 with the elements of the ribosomal P-stalk. Different conformation of the helices 42 and 97 of the P-stalk are shown in blue for 80S-GENT and 80S-TC007 structures and in cyan for the 80S-apo structure (PDB ID code 4V88). H42 in the apo conformation would clash with GENT-8. Arg62 in the uL6 protein approaching TC007 is shown as spheres. Color-coding is as in A–E.
The peptide exit tunnel, which spans the peptidyl-transferase center to the solvent side of the ribosome, is subjected to translation regulation by small molecules and peptides that induce translational stalling (53). We find that gentamicin, G418, and TC007 each bind to the peptide exit tunnel (Fig. 5B). Paromomycin was not found in the exit tunnel, suggesting that only aminoglycosides with a maximum of three rings can be accommodated within this pocket. Superposition with the structure of the 50S ribosome subunit from Haloarcula marismortui complexed with erythromycin shows that aminoglycosides and erythromycin bind opposite sides of the exit tunnel wall (Fig. S5A) (54). Interestingly, although each drug interacts with the same 25S rRNA residues, the orientations of G418 and gentamicin within the exit tunnel, which are very similar, are distinct from TC007 (Fig. 5B). These distinctions likely reflect the unique ring topologies of the 4,6-linked (G418 and gentamicin) and 4,5-linked (TC007) aminoglycosides. Alignment of each of these structures with that of the Escherichia coli ribosome containing the SecM stalling peptide (55) suggests that aminoglycoside occupancy within the exit tunnel wall is unlikely to be sufficient to block the path of nascent peptides lacking bulky amino acid side chains (Fig. S5B) (56, 57).
The E-site of the small ribosomal subunit is a potential point of regulation of eukaryotic protein synthesis (45, 58, 59). We observe that paromomycin (PAR-3), geneticin (G418-3), and gentamicin (GENT-2) bind in the space normally occupied by the E-site mRNA codon and exhibit similar ring I and II positions (Fig. 5C). Aminoglycoside binding in this region may have important impacts on tRNA occupation within the translating ribosome (45) and/or alter the mRNA-binding and -scanning mechanism required for translation initiation (60). Similar to the binding mode of streptomycin to the bacterial 30S subunit (14), gentamicin (GENT-3) was also found within <4 Å of the mRNA backbone wedged between helices 1, 44, 18, and 27 within the A-site, where it interacts with the phosphate groups of residues U9 and A11 (Fig. S6 A–C). In contrast to streptomycin, however, gentamicin does not appear to induce substantial conformational changes in h44 and h45. This same pocket was also occupied by geneticin (G418-4) (Fig. S6C). In addition to potential impacts on the initiation mechanism, binding in such proximity to the decoding region may influence tRNA selection and translocation of the mRNA–tRNA module during the elongation phase of protein synthesis.
Aminoglycosides interact extensively with the intersubunit region of the ribosome. Paromomycin (PAR-3) provides additional contacts between the subunits in the vicinity of bridge B2c formed by h24 of the small subunit and H66 of the large subunit (Fig. 5D). Gentamicin (GENT-4, 5) is also observed to bind bridge B2c (Fig. 5E), and GENT-6 binds bridge B4, a protein–RNA bridge comprised of H34 of the large subunit and protein uS15 of the small subunit (Fig. 5F). The interactions of the GENT-5 molecule with bridge B2c are expected to preclude bridge B2c rearrangements relative to h24 in the small subunit (Fig. 5E). These findings, together with the observation that TC007 binds close to bridge B2c (Fig. S6D) and B3 (Fig. 2A), are consistent with aminoglycosides interfering with intersubunit rotation in distinct ways to affect the translation mechanism. Such distinctions may impact the mechanism of translocation in particular, which requires dramatic remodeling events within the central bridge B2 domain (20, 21), as well as rearrangements in B4 (61, 62).
Additional impacts on the elongation mechanism may also arise from aminoglycoside binding in proximity to the peptidyl-transferase center. Paromomycin (PAR-4) is located just 3 Å away from the phosphate groups of the catalytic residues A2820 (A2450) and C2821 (C2451) and 3.4 Å away from C75 of the A-site tRNA (Fig. S7A). Aminoglycosides are also observed to bind the base of the P-stalk and sarcin–ricin loop (H95), elements of the large ribosomal subunit that interact with translation elongation factors (Fig. 5G and Fig. S7B).
Discussion
The structural and functional insights into aminoglycoside interactions with the 80S eukaryotic ribosome obtained through the present investigations serve as a foundation for exploring the molecular mechanisms of aminoglycoside action in eukaryotes. Although the impairment of mitochondrial translation is considered one of the main causes of side effects produced by aminoglycosides in eukaryotic cells (63, 64), emerging evidence suggests that aminoglycosides also exert effects on cytosolic ribosomes to alter translation elongation and termination in a manner that induces read-through of PTCs (27, 65). These findings have led to their consideration as potent drugs to treat human diseases caused by PTCs (28). Aminoglycosides also operate against eukaryotic human pathogens, including Leishmania and Trypanosoma families (66, 67), due to sequence variations in their canonical h44 decoding sites (Fig. S8).
The concentrations of aminoglycosides required for eukaryotic cell growth inhibition and the EC50 values measured for the inhibition of eukaryotic translation by distinct aminoglycosides in vitro generally correlate with their affinities for the canonical h44 decoding region within the small subunit A-site (23, 68). As illustrated by crystal structures of the 80S ribosome with gentamicin and TC007, which adopt noncanonical poses in the vicinity of the h44 decoding site, the eukaryotic-specific G1645 residue (equivalent to A1408 in bacteria) within h44 tends to preclude the binding of aminoglycosides containing a 6′-NH2 substituent in the ring I (Figs. 1D and 2A). Paromomycin and G418, which contain a 6′-OH substituent, can achieve canonical interactions with the h44 decoding site, but the absence of Watson–Crick base pairing between C1646 and A1754 impedes drug-binding interactions that hinge on ring I interactions with the floor of the aminoglycoside-binding site (Fig. 1B and Fig. S1). Human mitochondrial ribosomes contain an adenosine at the 1408 position (bacterial numbering), making it a good target for aminoglycosides with both 6′-OH and 6′-NH2 substituents in ring I (63). However, aminoglycoside affinity to mitochondrial ribosomes is likely to be attenuated by two consecutive noncanonical base pairs [C1494–A1555 (A1410–U1490) and C1493–C1556 (C1409–G1491)] that are likely to strongly disrupt ring I interactions (Fig. S8). Consistent with this notion, the reestablishment of Watson–Crick interactions in the floor of the aminoglycoside-binding site by the naturally occurring mutations A1555G or C1494T leads to aminoglycoside hypersusceptibility in humans (69, 70).
Our smFRET experiments indicate that each of the aminoglycosides tested increases the error rate of A-site decoding (Fig. 4). However, the behaviors of the miscoded 80S precomplexes bearing near-cognate tRNA in the A-site are unique for each drug. These findings suggest that aminoglycosides may promote PTC read-through by distinct mechanisms. Aminoglycosides containing 6′-OH substituent in ring I likely induce miscoding and PTC read-through due to residual binding to the canonical binding site in h44 of the eukaryotic ribosome. In this case, near-cognate or noncognate tRNA may efficiently accommodate at the stop codon-programmed A-site to compete with the termination factors. Aminoglycosides containing a 6′-NH2 constituent in ring I, including gentamicin and TC007, do not bind h44 in a canonical fashion. Their impacts on PTC read-through may entail alternative mechanisms, including intersubunit rotation effects that are anticipated to hamper RF1 interactions with the classically configured ribosome (Fig. S9) (71).
The propensity of eukaryotic ribosomes to adopt rotated states and the impact of aminoglycosides in enforcing closer interactions (higher FRET) between deacylated and peptidyl-tRNA within the 80S human PRE complex suggest that aminoglycoside binding to intersubunit regions of the ribosome (bridges B2c and B4 for gentamicin and bridge B3 for TC007) facilitate conformational changes in the PRE 80S–ribosome complexes that shift peptidyl tRNA toward the P-site. Such impacts may relate to the stabilization of partially rotated ribosome configurations that move deacylated tRNA toward the A-site (20, 21).
The nature of the observed aminoglycoside interactions with the eukaryotic ribosome hint at potentially multiple modes of action on the translation mechanism. These insights also provide an important framework for understanding the diversity of aminoglycoside interaction sites and drug-binding modes with the 80S ribosome. The combined perspectives afforded by X-ray crystallography and direct imaging of aminoglycoside impacts on functional ribosome complexes using smFRET has the potential to facilitate the design of new antibiotic derivatives and may be particularly suited for the identification of compounds capable of mediating efficient PTC read-through. Such efforts will be greatly aided by in-depth functional investigations of a diversity of functional ribosome complexes relevant to termination. In this regard, the present findings suggest that nonspecific impacts on decoding may be reduced by avoiding aminoglycoside scaffolds bearing a ring I 6′-OH moiety, which exhibit generally higher affinity for the h44 decoding site, and instead focusing on aminoglycosides, other compounds, or mixtures of compounds that give rise to stop codon-specific miscoding in the absence of dominant-negative downstream impacts.
Materials and Methods
Yeast and Bacterial Ribosome Purification, Crystallization, and Crystal Treatment.
Ribosomes from S. cerevisiae were purified and crystallized as described (39). The crystal treatment procedure was modified based on the procedure described previously (39). Briefly, crystals were transferred to the solution containing 80 mM Tris⋅acetate (pH 7.0), 70 mM KSCN, 10 mM Mg(OAc)2, 20% (vol/vol) glycerol, 5% (wt/vol) PEG 20,000, 6.5 mM spermidine, 7.5 mM NH4OAc, 1.4 mM N,N'-bis-(3-D-gluconamidopropyl)deoxycholamide (Deoxy Big Chap), 2 mM DTT, and stepwise increasing concentrations of PEG 4000, PEG 3350, or PEG 2000 MME up to 20%. The crystals were incubated for 1 h or 4 h and were flash-frozen in a stream of liquid nitrogen. All manipulations were performed at 4 °C. Aminoglycosides G418, paromomycin, and gentamicin were ordered from Sigma. TC007 was obtained as described in ref. 32. High-concentration stocks of aminoglycosides were prepared and introduced during the last steps of treatment. We observed that soaking in high concentrations of paromomycin or gentamicin improves the diffraction of the crystal. For example, crystals prepared in the same conditions and soaked in 2 mM of paromomycin diffracted up to 3.7-Å resolution. Ribosomes from T. thermophilus were purified and crystallized as described in ref. 72. TC007 was added for cocrystallization in 50-fold excess over the ribosome concentration (70S = 1.25 µM; TC007 = 62.5 µM). The crystal treatment was performed as described. If necessary, TC007 was added for soaking during all steps of the crystal treatment procedure at a concentration of 500 µM.
Purification of 40S and 60S Ribosomal Subunits from Human Cells.
Preparation of small (40S) and large (60S) human ribosomal subunits was adapted from refs. 45 and 73. Specific deviations implemented for the purification of polysome fractions from human tissue culture are described here. Cell pellets were resuspended in lysis buffer [20 mM Tris HCl (pH 7.5), 2.5 mM MgCl2, 10 mM KCl, and 1 mM freshly prepared DTT] with the RNase inhibitor RNase Out (Invitrogen), EDTA-free Halt Protease Inhibitor (Thermo Scientific), and cycloheximide (Sigma) at 100 μg/mL (∼350 µM). The solution was incubated on ice for 10 min before centrifugation in a Microfuge 22R Refrigerated Centrifuge (Beckman Coulter) at 14,000 rpm for 10 min at 4 °C to pellet cell debris. The supernatant was loaded onto precooled 10–50% sucrose density gradients and spun at 35,000 rpm for 3 h at 4 °C in an Optima l-100 XP ultracentrifuge (Beckman Coulter). The gradients were then fractionated using a BR-186-1 Fractionator and a UA-6 UV/Vis detector (Teledyne Isco). Fractions corresponding to polysomes were collected and subsequently pelleted and dissociated into subunits according to ref. 73. Pelleted subunits were resuspended with storage buffer [30 mM Hepes (pH 7.5), 15 mM MgCl2, 50 mM NH4Cl, 2 mM spermidine, 5 mM putrescine, 1 mM DTT, and 6% sucrose] for stable, long-term storage in liquid nitrogen.
Data Collection, Crystal Structure Determination, and Analysis.
Diffraction data were collected at 90 K using 0.05° oscillation on beamline PROXIMA I at the Soleil synchrotron (Saint-Aubin, France) equipped with a Pilatus 6M detector (Dectris) or on the PXI beamline at the Swiss Lightsource synchrotron (Villigren, Switzerland) equipped with an Eiger 16M detector (Dectris). Two to eight crystals were used for each dataset. Data were reduced and scaled using the XDS suite (74). Coordinates of vacant 80S ribosome from S. cerevisiae from PDB 4V88 were used to determine structures of 80S–paromomycin, 80S–gentamicin, and 80S–TC007. Phenix software was used for structure refinement, starting with several rounds of a rigid body refinement, and validation (75, 76). P-stalk elements were disordered and were removed from the structures except for P-stalk rRNA in the 80S–paromomycin structure and P-stalk rRNA and protein L12 in the 80S–gentamicin structure. Due to weak electron density, protein S31 was removed from the 80S–paromomycin structure, and a few structural elements were remodeled, in particular, amino acids 103–113 of protein uL16 and residues 80–87 of 5.8S rRNA. An unbiased difference electron density map (Fobs − Fcalc) was used to locate the binding sites of aminoglycosides. Ligand fitting was performed in Coot (77). Geometry restraints for antibiotics were generated with the help of Grade web server (Global Phasing, grade.globalphasing.org/cgi-bin/grade/server.cgi). Peaks of positive electron density maps were inspected manually to add magnesium ions, with coordinated water molecules often replacing osmium hexamine molecules. Manual corrections were followed by several iterations of reciprocal space refinement of atomic coordinates, B-factors (one isotropic B-factor per residue), and occupancies (one occupancy value per ligand and individual occupancies for magnesium ions). Real-space refinement in Phenix was applied to fit rotamer outliers. Finally, translation–libration–screw-rotation (TLS) refinement was performed with two TLS groups. For structure determination of 70S–tRNA–mRNA–TC007 complexes, coordinates of the 70S ribosome from PDB 4WSM and tRNAs and mRNAs coordinates from PDB 4V6F were used for two rounds of rigid-body refinements. The electron density maps were inspected manually, and the molecules of TC007 were localized in the peaks of positive electron density. Secondary binding sites of TC007 (four in total) were located on the periphery of the ribosome far from the functional centers. Additionally, one binding site of TC007 to the intersubunit region was detected. Here TC007 interacts with the low part of h44 of the 30S subunit and the junction of helices 62 and 64 in the 25S rRNA of the 50S subunit between intersubunit bridges B5 and B6. Geometry restraints for TC007 were generated with the help of Grade web server (Global Phasing, grade.globalphasing.org). Manual modeling was done in Coot and was followed by several rounds of reciprocal space refinement of atomic coordinates and B-factors. Crystallographic statistics are reported in Table S1. All figures were generated using PyMOL 1.5 ( https://pymol.org/2/) (Schrödinger). Local structure alignments were performed in Coot (77). Ribosomal proteins are named throughout the paper according to the newly established nomenclature (78). Atomic coordinated and structure factors for structures of 80S–paromomycin, 80S–gentamicin, 80S–TC007, 80S–geneticin (G418), 70S-tRNAs-mRNA–TC007 (cocrystallization), and 70S–tRNAs–mRNA–TC007 (soaking) have been deposited in the Protein Data Bank (https://www.rcsb.org/pdb/home/home.do) under ID codes 5NDV (80S–paromomycin), 5OBM (80S–gentamicin), 5NDW (80S–TC007), 5NDG [80S–geneticin (G418)], 5NDK (70S–tRNA–mRNA–TC007 cocrystallized), and 5NDJ (70–tRNA–mRNA–TC007 soaked).
Preparation of Native and Fluorescently Labeled tRNAs.
E. coli tRNAfMet and tRNAPhe were purified as previously described (79, 80). Aminoacylation and fluorescent labeling of tRNAs (tRNAiMet at 4sU8 and tRNAPhe at acp3 U47 positions) were performed following established protocols.
In Vitro Reconstitution of 80S ICs.
As previously described (45), 80S ICs were assembled following a procedure that bypasses the need for exogenous initiation factors (81). Purified 40S subunits were mixed with an equal volume of 80S association buffer [30 mM Hepes (pH 7.5), 5 mM MgCl2, 50 mM NH4Cl, 2 mM spermidine, 5 mM putrescine, 1 mM DTT] and then were heat activated at 42 °C for 5 min. Fourfold excess of mRNA with the sequence 5′-CAA CCU AAA ACU UAC ACA CCC UUA GAG GGA CAA UCG AUG UUU UUU UUU UUU UUU UUU UUU-3′ (Dharmacon) (henceforth referred to as “MFF mRNA”) or 5′-CAA CCU AAA ACU UAC ACA CCC UUA GAG GGA CAA UCG AUG UCU UUC UUC UUC UUC UUC UUC-3′ (henceforth referred to as “MFF near-cognate mRNA”) was added, heated to 37 °C for 10 min and subsequently cooled on ice. To this mixture, a twofold excess of fluorescently labeled Met-tRNAiMet (prepared as described in ref. 82) was added, and the reaction was heated and cooled as above. At this time, equimolar amounts of 60S subunits were heat activated at 42 °C for 5 min. The 60S subunits were then added to the mixture of 40S/tRNA/mRNA. After an additional heating and cooling cycle, the MgCl2 concentration of the reaction was raised to 15 mM, and the mixture remained on ice for 5 min. It was then loaded on a 10–30% sucrose gradient in 80S association buffer and was ultracentrifuged in a Beckman SW41 rotor at 35,000 rpm for 1.5 h at 4 °C before fractionation. The peak corresponding to 80S complexes was collected and aliquoted before storage in liquid nitrogen. To achieve surface immobilization, the mRNA was hybridized to a double-stranded, biotinylated DNA oligonucleotide (sequence 1: 5′-GTA AGT TTT AGG TTG CCC CCC TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT-3′; sequence 2: 5′-AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA-3′) before its mixture with the 40S subunit.
Formation of the eEF1A(GTP)–aa-tRNA Ternary Complex.
aa-tRNAs (tRNAPhe, tRNAMet) were first generated as previously described (82) and were mixed with 1 mM GTP, 6 mM phosphoenolpyruvate, 12 units/mL pyruvate kinase, and 12 units/mL myokinase. A twofold excess of eEF1A isolated from rabbit reticulocyte lysate (45), which bears 100% sequence identity with human eEF1A, was then added, and the mixture was incubated at 37 °C for 5 min to form the ternary complex.
Single-Molecule Fluorescence Imaging.
Complexes were surface-immobilized via the biotin–streptavidin interaction in PEG-passivated quartz chambers. All imaging experiments were performed in Hepes (KOH)-Polymix buffer (pH 7.5) containing 5 mM MgCl2, 50 mM NH4Cl, 2 mM spermidine, and 5 mM putrescine, as well as an oxygen scavenging system (2 mM protocatechuic acid, 50 nM protocatechuate 3,4-dioxygenase) together with a mixture of solution additives (1 mM Trolox, 1 mM cyclooctatetraene, 1 mM nitrobenzyl-alcohol) (83) to reduce photobleaching. As previously described (82), single-molecule fluorescence imaging was performed using a custom prism-based total internal reflection fluorescence microscope. Cy3 fluorophores were illuminated with an Opus 532-nm solid-state laser (Laser Quantum), and fluorescence emissions from Cy3 and Cy5 fluorophores were collected using a 60×, 1.27 NA Plan-Apo water immersion objective (Nikon) and were spectrally separated using a MultiCam-LS device (Cairn) equipped with a T635lpxr-UF2 dichroic mirror (Chroma) and imaged onto ORCA-Flash 4.0 v2 sCMOS cameras (Hamamatsu). Data were acquired at 40-ms time resolution using custom software implemented in LabVIEW (National Instruments).
Single-Molecule tRNA Selection Assay.
As previously described (45), the process of tRNA selection on the ribosome was performed by stopped-flow injection of a 20-nM solution of ternary complex [eEF1A(GTP)–aa-tRNA] containing (Cy5)Phe-tRNAPhe into surface-immobilized ribosome complexes containing (Cy3)tRNAiMet in the P-site. Here, 80S ICs were formed with the near-cognate UCU mRNA codon in the A-site, and the period of incubation with the ternary complex was extended from 30 s to 2 min. To prevent spurious, nonenzymatic binding of deacylated tRNAPhe to the E-site, tRNA selection experiments were performed in the presence of 500 µM cyclohexamide. Subsequent steady-state imaging of PRE complexes was performed following buffer exchange into a solution lacking the ternary complex.
Analysis of smFRET Data.
Analysis of single-molecule fluorescence data was performed using the SPARTAN analysis software package MATLAB (84). Single-molecule fluorescence traces were extracted from wide-field movies and were corrected for background, spectral crosstalk, and unequal apparent brightness (85). FRET trajectories were calculated as EFRET = IA/(IA + ID), where IA and ID are the acceptor and donor fluorescence intensities at each frame, respectively. Traces were selected for further analysis according to the following criteria: (i) single-step photobleaching; (ii) signal-to-background noise ratio >8; (iii) fewer than four donor blinking events; and (iv) >0.12 FRET efficiency for at least 50 frames (2 s). FRET histograms were calculated from the first 50 frames of all individual molecules passing the aforementioned criteria from each dataset with bin sizes of 0.03.
Supplementary Material
Acknowledgments
We thank Daniel S. Terry for helpful discussions and support with smFRET data analysis; the French Infrastructure for Integrated Structural Biology ANR-10-INSB-05-01 and Instruct, which is part of the European Strategy Forum on Research Infrastructures; all staff members of SOLEIL synchrotron PROXIMA-1 beamline; and all staff members of Swiss Lightsource synchrotron PX-1 beamline. This work was supported by French National Research Agency (ANR) Grant ANR-15-CE11-0021-01 (to G.Y.); Fondation ARC pour la Recherché sur le Cancer (G.Y.); La Fondation pour la Recherche Médicale Grant DBF20160635745, France (to G.Y.); European Research Council Advanced Grant 294312 (to M.Y.); the Russian Government Program of Competitive Growth of Kazan Federal University (M.Y.); an Association Française Contre les Myopathies Telethon Postdoctoral Fellowship (I.P.); and Grant ANR-10-LABX-0030-INRT from a French State Fund managed by the ANR under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02 (to M.D.). M.D. is an International PhD Program fellow of the Institute of Genetics and Molecular and Cellular Biology supported by LabEx Integrative Biology: Nuclear Dynamics, Regenerative and Translational Medicine funds. Funding support for S.C.B., R.B.A., and A.F. was provided by NIH Grant GM079238 and the Tri-Institutional Stem Cell Initiative supported by the Starr Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinated and structure factors for crystal structures have been deposited in the Protein Data Bank, https://www.rcsb.org/pdb/home/home.do {PDB ID codes 5NDV (80S–paromomycin), 5OBM (80S–gentamicin), 5NDW (80S–TC007), 5NDG [80S–geneticin (G418)], 5NDK (70S–tRNA–mRNA–TC007 cocrystallized), and 5NDJ (70–tRNA–mRNA–TC007 soaked)}.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715501114/-/DCSupplemental.
References
- 1.Fosso MY, Li Y, Garneau-Tsodikova S. New trends in aminoglycosides use. MedChemComm. 2014;5:1075–1091. doi: 10.1039/C4MD00163J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 3.Borovinskaya MA, et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 4.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2010;6:244. doi: 10.1038/nchembio0310-244c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Davies J, Gorini L, Davis BD. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol Pharmacol. 1965;1:93–106. [PubMed] [Google Scholar]
- 7.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 8.Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 9.Recht MI, Fourmy D, Blanchard SC, Dahlquist KD, Puglisi JD. RNA sequence determinants for aminoglycoside binding to an A-site rRNA model oligonucleotide. J Mol Biol. 1996;262:421–436. doi: 10.1006/jmbi.1996.0526. [DOI] [PubMed] [Google Scholar]
- 10.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 11.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 12.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 13.Matt T, et al. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc Natl Acad Sci USA. 2012;109:10984–10989. doi: 10.1073/pnas.1204073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demirci H, et al. A structural basis for streptomycin-induced misreading of the genetic code. Nat Commun. 2013;4:1355. doi: 10.1038/ncomms2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 16.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 17.Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. New structural insights into translational miscoding. Trends Biochem Sci. 2016;41:798–814. doi: 10.1016/j.tibs.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Rozov A, et al. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun. 2016;7:10457. doi: 10.1038/ncomms10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozov A, Westhof E, Yusupov M, Yusupova G. The ribosome prohibits the G•U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res. 2016;44:6434–6441. doi: 10.1093/nar/gkw431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, et al. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasserman MR, et al. Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions. Nat Commun. 2015;6:7896. doi: 10.1038/ncomms8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Recht MI, Douthwaite S, Puglisi JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan-Minogue H, Bedwell DM. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA. 2008;14:148–157. doi: 10.1261/rna.805208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garreau de Loubresse N, et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 25.Wilhelm JM, Jessop JJ, Pettitt SE. Aminoglycoside antibiotics and eukaryotic protein synthesis: Stimulation of errors in the translation of natural messengers in extracts of cultured human cells. Biochemistry. 1978;17:1149–1153. doi: 10.1021/bi00600a002. [DOI] [PubMed] [Google Scholar]
- 26.Wilhelm JM, Pettitt SE, Jessop JJ. Aminoglycoside antibiotics and eukaryotic protein synthesis: Structure—function relationships in the stimulation of misreading with a wheat embryo system. Biochemistry. 1978;17:1143–1149. doi: 10.1021/bi00600a001. [DOI] [PubMed] [Google Scholar]
- 27.Palmer E, Wilhelm JM, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee HL, Dougherty JP. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol Ther. 2012;136:227–266. doi: 10.1016/j.pharmthera.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Baradaran-Heravi A, et al. Gentamicin B1 is a minor gentamicin component with major nonsense mutation suppression activity. Proc Natl Acad Sci USA. 2017;114:3479–3484. doi: 10.1073/pnas.1620982114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 31.Xue X, et al. Synthetic aminoglycosides efficiently suppress cystic fibrosis transmembrane conductance regulator nonsense mutations and are enhanced by ivacaftor. Am J Respir Cell Mol Biol. 2014;50:805–816. doi: 10.1165/rcmb.2013-0282OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattis VB, et al. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum Genet. 2006;120:589–601. doi: 10.1007/s00439-006-0245-7. [DOI] [PubMed] [Google Scholar]
- 33.Keeling KM, Wang D, Conard SE, Bedwell DM. Suppression of premature termination codons as a therapeutic approach. Crit Rev Biochem Mol Biol. 2012;47:444–463. doi: 10.3109/10409238.2012.694846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner KR, et al. Gentamicin treatment of Duchenne and Becker muscular dystrophy due to nonsense mutations. Ann Neurol. 2001;49:706–711. [PubMed] [Google Scholar]
- 35.Cogan J, et al. Aminoglycosides restore full-length type VII collagen by overcoming premature termination codons: Therapeutic implications for dystrophic epidermolysis bullosa. Mol Ther. 2014;22:1741–1752. doi: 10.1038/mt.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agrelo R, et al. A novel Werner syndrome mutation: Pharmacological treatment by read-through of nonsense mutations and epigenetic therapies. Epigenetics. 2015;10:329–341. doi: 10.1080/15592294.2015.1027853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bordeira-Carriço R, Pêgo AP, Santos M, Oliveira C. Cancer syndromes and therapy by stop-codon readthrough. Trends Mol Med. 2012;18:667–678. doi: 10.1016/j.molmed.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Mattis VB, Tom Chang CW, Lorson CL. Analysis of a read-through promoting compound in a severe mouse model of spinal muscular atrophy. Neurosci Lett. 2012;525:72–75. doi: 10.1016/j.neulet.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 40.Pfister P, et al. Mutagenesis of 16S rRNA C1409-G1491 base-pair differentiates between 6‘OH and 6’NH3+ aminoglycosides. J Mol Biol. 2005;346:467–475. doi: 10.1016/j.jmb.2004.11.073. [DOI] [PubMed] [Google Scholar]
- 41.Byrne KM, Kershner AS, Maehr H, Marquez JA, Schaffner CP. Separation of gentamicin C-complex into five components by Craig distribution. J Chromatogr A. 1977;131:191–203. doi: 10.1016/s0021-9673(00)80933-4. [DOI] [PubMed] [Google Scholar]
- 42.Iannaccone ST, Smith SA, Simard LR. Spinal muscular atrophy. Curr Neurol Neurosci Rep. 2004;4:74–80. doi: 10.1007/s11910-004-0016-6. [DOI] [PubMed] [Google Scholar]
- 43.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 44.Mattis VB, Ebert AD, Fosso MY, Chang CW, Lorson CL. Delivery of a read-through inducing compound, TC007, lessens the severity of a spinal muscular atrophy animal model. Hum Mol Genet. 2009;18:3906–3913. doi: 10.1093/hmg/ddp333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferguson A, et al. Functional dynamics within the human ribosome regulate the rate of active protein synthesis. Mol Cell. 2015;60:475–486. doi: 10.1016/j.molcel.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bar-Nun S, Shneyour Y, Beckmann JS. G-418, an elongation inhibitor of 80 S ribosomes. Biochim Biophys Acta. 1983;741:123–127. doi: 10.1016/0167-4781(83)90018-0. [DOI] [PubMed] [Google Scholar]
- 48.Stage TK, Hertel KJ, Uhlenbeck OC. Inhibition of the hammerhead ribozyme by neomycin. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- 49.von Ahsen U, Davies J, Schroeder R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 50.Mikkelsen NE, Brännvall M, Virtanen A, Kirsebom LA. Inhibition of RNase P RNA cleavage by aminoglycosides. Proc Natl Acad Sci USA. 1999;96:6155–6160. doi: 10.1073/pnas.96.11.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melnikov SV, Söll D, Steitz TA, Polikanov YS. Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome. Nucleic Acids Res. 2016;44:4978–4987. doi: 10.1093/nar/gkw246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weisblum B, Davies J. Antibiotic inhibitors of the bacterial ribosome. Bacteriol Rev. 1968;32:493–528. [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta P, et al. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat Chem Biol. 2016;12:153–158. doi: 10.1038/nchembio.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, et al. Mechanisms of ribosome stalling by SecM at multiple elongation steps. Elife. 2015;4:09684. doi: 10.7554/eLife.09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arenz S, et al. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat Commun. 2016;7:12026. doi: 10.1038/ncomms12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsui WH, et al. Dual effects of MLS antibiotics: Transcriptional modulation and interactions on the ribosome. Chem Biol. 2004;11:1307–1316. doi: 10.1016/j.chembiol.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Budkevich T, et al. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson DN, Nierhaus KH. The E-site story: The importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell Mol Life Sci. 2006;63:2725–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 61.Bock LV, Blau C, Vaiana AC, Grubmüller H. Dynamic contact network between ribosomal subunits enables rapid large-scale rotation during spontaneous translocation. Nucleic Acids Res. 2015;43:6747–6760. doi: 10.1093/nar/gkv649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Q, Fredrick K. Contribution of intersubunit bridges to the energy barrier of ribosomal translocation. Nucleic Acids Res. 2013;41:565–574. doi: 10.1093/nar/gks1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hobbie SN, et al. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc Natl Acad Sci USA. 2008;105:20888–20893. doi: 10.1073/pnas.0811258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.El-On J, Bazarsky E, Sneir R. Leishmania major: In vitro and in vivo anti-leishmanial activity of paromomycin ointment (Leshcutan) combined with the immunomodulator Imiquimod. Exp Parasitol. 2007;116:156–162. doi: 10.1016/j.exppara.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 67.Hobbie SN, et al. Genetic reconstruction of protozoan rRNA decoding sites provides a rationale for paromomycin activity against Leishmania and Trypanosoma. PLoS Negl Trop Dis. 2011;5:e1161. doi: 10.1371/journal.pntd.0001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hobbie SN, et al. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 2007;35:6086–6093. doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao H, et al. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prezant TR, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 71.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. Structural insights into the translational infidelity mechanism. Nat Commun. 2015;6:7251. doi: 10.1038/ncomms8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bommer UA, et al. In: Ribosomes and Polysomes. Graham JM, Rickwood D, editors. IRL at Oxford Univ Press; Oxford: 1996. pp. 271–300. [Google Scholar]
- 74.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ban N, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang L, Altman RB, Blanchard SC. Insights into the molecular determinants of EF-G catalyzed translocation. RNA. 2011;17:2189–2200. doi: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burgess AB, Mach B. Formation of an initiation complex with purified mammalian ribosomal subunits. Nature. 1971;233:209–210. [PubMed] [Google Scholar]
- 82.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Juette MF, et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods. 2016;13:341–344. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.