Significance
Bacterial flagella are nanomachines rotated by powerful motors and drive motility that is often associated with virulence. Regulators that inhibit motor function are potentially reversible and arrest flagellar rotation either as a brake, to increase resistance, or as a clutch, to cut power to the machine, analogous to their counterparts in a car. Here we show that MotI of Bacillus subtilis acts like a molecular clutch by interacting with the power-transducing protein MotA and separating it from the drivetrain protein FliG. We further demonstrate that partial motility inhibition by clutch proteins decreases swimming speed.
Keywords: flagella, motility, MotA, c-di-GMP, YcgR
Abstract
Stator elements consisting of MotA4MotB2 complexes are anchored to the cell wall, extend through the cell membrane, and interact with FliG in the cytoplasmic C ring rotor of the flagellum. The cytoplasmic loop of MotA undergoes proton-driven conformational changes that drive flagellar rotation. Functional regulators inhibit motility by either disengaging or jamming the stator–rotor interaction. Here we show that the YcgR homolog MotI (formerly DgrA) of Bacillus subtilis inhibits motility like a molecular clutch that disengages MotA. MotI-inhibited flagella rotated freely by Brownian motion, and suppressor mutations in MotA that were immune to MotI inhibition were located two residues downstream of the critical force generation site. The 3D structure of MotI bound to c-di-GMP was solved, and MotI-fluorescent fusions localized as transient MotA-dependent puncta at the membrane when induced at subinhibitory levels. Finally, subinhibitory levels of MotI expression resulted in incomplete inhibition and proportional decreases in swimming speed. We propose a model in which flagellar stators are disengaged and sequestered from the flagellar rotor when bound by MotI.
Bacteria swim in liquid and swarm over solid surfaces by assembling and rotating helical flagella. The bacterial flagellum is structurally complex, requires tens of thousands of subunits from over 30 different proteins, and is temporally regulated to ensure that each machine is assembled in the correct order (1, 2). Furthermore, flagella are integrated in the cell envelope and are seldom degraded, perhaps because flagellar synthesis is costly in the consumption of metabolic building blocks and in the multigenerational time needed for their assembly (3–5). Nonetheless, flagellar motility is conditionally advantageous, and reversible regulators of motor function inhibit motility while preserving the investment in the machine (6–9).
Flagellar rotation occurs by the interaction between flagellar stator complexes and the rotor (10). Stator complexes are composed of two copies of MotB and four copies of MotA, and as many as 11 complexes surround the Escherichia coli flagellum (11, 12). The rotor is a gear-like disk of the protein FliG, and below it the C-ring complex proteins FliM and FliN are connected to the cytoplasmic face of the flagellar basal body (13–16). When protons flow through the stator complex, MotA changes conformation, electrostatically interacts with FliG, and turns the rotor to generate torque (17, 18). The interaction between MotA and FliG can be regulated in at least three ways to control rotation. First, stator subunits interact dynamically with the rotor, and flagellar power increases with the number of stator units bound (19, 20). Second, molecular clutch proteins, such as the Bacillus subtilis functional regulatory protein EpsE, bind and disengage the rotor from the stators to cut power to the flagellum (6). Third, molecular brake proteins, such as the Rhodobacter sphaeroides chemotaxis protein CheY and the E. coli functional regulatory protein YcgR, can lock rotation by increasing resistance between the rotor and the stators (7–9, 21). Functional regulation of the flagellum appears to be controlled in response to flagellar load, cellular interaction with surfaces, and the formation of biofilms (5).
B. subtilis also encodes a homolog of YcgR, called “DgrA” (formerly YpfA). DgrA inhibits motility and abolishes a flagellar-mediated form of surface migration known as “swarming motility” when it is overexpressed and bound to c-di-GMP. Metabolism of c-di-GMP in B. subtilis is relatively simple, as three diguanylate cyclases (DgcK, DgcP, and DgcW) synthesize detectable levels of c-di-GMP when the phosphodiesterase (PdeH) is inactivated (22, 23). The mechanism by which DgrA inhibits motility is thought to be related to the flagellar stators, as DgrA and MotA interacted in a bacterial two-hybrid assay (22). Whether DgrA inhibits motility at the level of flagellar rotation or at some other step is unknown. Furthermore, the environmental conditions that cause c-di-GMP to naturally accumulate and produce DgrA are unknown, and thus the biological function of DgrA is unclear (22, 23).
DgrA was originally named “diguanylate receptor” because it binds c-di-GMP, and here we rename it “motility inhibitor” (MotI) based on its ability to inhibit flagellar rotation at the mechanistic level of the MotA stator protein. Using suppressor analysis, we found MotI-resistant mutants altered at a single residue of MotA located two residues downstream of a critical force-generating site (24). Tethering studies indicated that MotI-inhibited flagella rotated freely by Brownian motion and resembled flagella inhibited by the clutch protein EpsE or a strain in which the MotAB stator complex had been deleted. Furthermore, we resolved the 3D structure of MotI bound to c-di-GMP, to generate functional fluorescent fusions for cell biological studies. MotI localized as transient, MotA-dependent puncta but only when induced at subinhibitory levels, and partial inhibition by MotI reduced cell swimming speeds. We conclude that MotI acts like a molecular clutch on the flagellar motor by interacting with MotA and preventing contact with FliG.
Results
MotI Inhibits Flagellar Rotation.
Overexpression of MotI (DgrA) in the presence of high levels of c-di-GMP due to the absence of the phosphodiesterase PdeH (i.e., in a MotI-inhibitory background) resulted in complete inhibition of swarming motility atop soft agar surfaces (Fig. 1A) (22, 23). Swarming motility is flagellar-dependent but has additional requirements beyond the swimming motility that occurs in 3D liquid environments (25). To determine whether MotI function also inhibited swimming motility, video microscopy was performed on wild-type and MotI-inhibitory cells. Whereas wild-type cells swam in a powered manner, MotI-inhibitory cells appeared to move only by Brownian motion (Fig. 1 B and C). We conclude that, when artificially expressed in the presence of high levels of c-di-GMP, MotI inhibits both swimming and swarming motility.
Fig. 1.
MotI inhibits swimming and swarming postflagellar assembly. (A) Quantitative swarm expansion assay for wild-type strain 3610 (open circles) and a MotI-inhibitory strain NPS235 (MotIInh) (closed circles) in which the c-di-GMP–degrading phosphodiesterase PdeH is deleted and MotI is overexpressed from a constitutive promoter. (B and C) Ten frames of time-lapse images taken over 2 s of swimming motility in wet-mount tunnel slides of individual wild-type strain 3610 (B) and MotIInh strain NPS235 (C) cells were combined into single averaged panels. (Magnification: 400×.) Motile cells appear as dashed lines in B and are not found in C. (D and E) Fluorescence microscopy images of wild-type strain DS9294 (D) and MotIInh strain NPS394 (E) expressing the Phag-GFP reporter (false-colored green) that indicates expression of the hag gene encoding flagellin. (F and G) Fluorescence microscopy images of wild-type strain DS8062 (F) and MotIInh strain NPS309 (G) expressing the HagT209C allele and stained with a fluorescent maleimide dye (false-colored green) that indicates flagellar filament assembly. In D–G the membrane was stained with FM4-64 (false-colored red). (Scale bar: 4 µm.)
To determine whether MotI inhibited motility at the level of flagellar expression or synthesis, a reporter was generated to the most downstream flagellar gene hag, in which the promoter (Phag) was transcriptionally fused to the gfp gene (26). The wild type and a MotI-inhibitory strain were similarly fluorescent, suggesting that MotI did not impair flagellar gene expression (Fig. 1 D and E) (6). Furthermore, both the wild type and a MotI-inhibitory strain containing an allele of the Hag flagellar filament (HagT209C), which when assembled can be stained with a cysteine-reactive fluorescent dye, synthesized flagellar filaments normally (Fig. 1 F and G). We conclude that MotI does not inhibit motility at the level of flagellar gene expression or flagellar assembly.
To determine the mechanism of motility inhibition, spontaneous suppressor mutations were isolated that restored motility to a MotI-inhibitory strain that was sensitized genetically in the following ways. To overcome mutations that directly disrupted motI, two redundant copies of motI were introduced in the chromosome and expressed from an isopropyl β-d-thiogalactopyranoside (IPTG)-inducible and a constitutive promoter, respectively. To overcome mutations that disrupt c-di-GMP synthesis, a transgenic diguanylate cyclase, DgcA, from Clostridium difficile was cloned downstream of a constitutive promoter and integrated in the chromosome. Finally, the biofilm activator protein RemA was mutated to decrease the contribution of a passive form of surface migration called “sliding” and increase selective pressure for swarming (27).
MotI inhibited swarming motility in the sensitized strain such that the cells remained in the center of the swarm agar plate. Upon prolonged incubation, however, spontaneous suppressor mutations arose that abolished MotI inhibition and self-segregated from the rest of the colony as a motile flare. Only five spontaneous soi (suppressor of MotIinh) suppressor mutation flares emerged after 48 h of induction on 50 parallel and independent swarming motility assay plates; the long duration and low frequency suggested that the soi mutations were rare. Each soi mutant was found to contain the same missense mutation, a glutamate-to-lysine substitution at position 92 (E92K) in the stator protein MotA, located two residues downstream of the critical force-generating residue R90 (24, 28). The MotAE92K allele was reconstructed and was fully motile in both the wild-type and MotI-inhibitory backgrounds (Fig. S1). We conclude that MotI inhibits motility through MotA.
MotI Acts as a Molecular Clutch on the Flagellar Stator to Inhibit Motility.
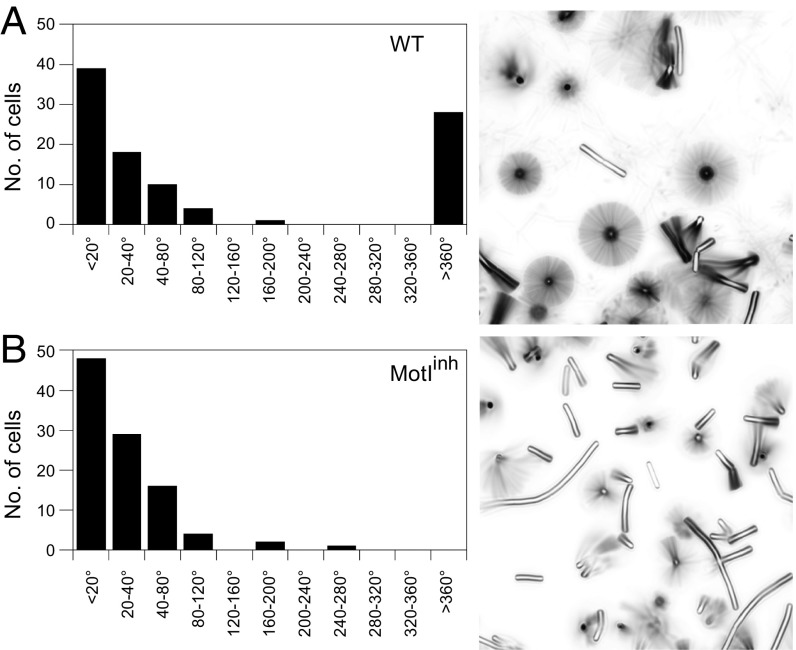
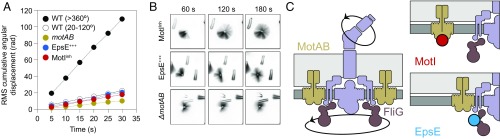
The mechanism by which MotI inhibits motility through MotA could work in one of two different ways. In one mechanism, MotI could function as a molecular brake by adding resistance between the stator and the rotor until flagellar rotation becomes locked (7–9, 16). Mechanically locked flagellar rotation is seen in E. coli as a 3° angular deviation about a tethered flagellum (6, 29, 30). Alternatively, MotI could function as a molecular clutch by separating stators from the rotor, causing flagella to be depowered but free to rotate by Brownian motion (6). To determine how MotI impairs MotA function, flagella-tethering studies were undertaken. Briefly, flagella were mechanically sheared to generate flagellar stubs, the stubs were attached to glass slides, and cells were monitored microscopically for 60 s. While many cells did not rotate, likely due to being tethered by multiple stubs, a subpopulation of wild-type cells rotated more than 360° over the course of 60 s (Fig. 2A). No cells expressing MotI rotated 360° over 60 s, but these cells were not restricted to a 3° angle of rotation. Instead, cells rotated between 20° and 120°, similar to a second rotating subpopulation of wild-type cells, cells mutated for MotAB stator proteins, and cells expressing the flagellar clutch protein EpsE, consistent with the rmsd predicted for angular displacement by Brownian motion (Fig. 2B and Fig. S2) (6). The angular displacement of all cell types that rotated more than 20° but less than 360°, including MotI-inhibited cells, was constant in rate, suggesting that no rotational resistance was present (Fig. 3A) (31). Finally, threefold-longer observation showed examples of MotI-inhibited, EpsE-inhibited, and MotAB-deleted cells rotating 360°; thus the flagella were unlikely to be locked and rotating simply by torsional compliance of the flagellum (Fig. 3B). We conclude that flagellar motors were not restrained by MotI expression and that MotI functions mechanistically as a molecular clutch to disable motility (Fig. 3C).
Fig. 2.
MotI abolishes powered flagellar rotation. Wild-type strain DS8022 cells (A) and MotIInh strain DK1995 cells (B) were tethered by flagellar stubs and monitored for 60 s in time-lapse microscopy. (Left) The angles of rotation of 100 cells were binned and expressed as a frequency histogram. (Right) Time-lapse composite imaged of sample fields. Sample movies used to generate these data are included as Movie S1 (wild-type) and Movie S2 (MotIInh) in Supporting Information.
Fig. 3.
MotI-inhibited flagella freely rotate by Brownian motion. (A) Cumulative angular rmsd displacement of strain DS8022 wild-type cells rotating >360° (black circles), strain DS8022 wild-type cells rotating 20°–360° (open circles), strain DK1995 MotIInh cells rotating 20°–360° (red circles), strain DS3317 EpsE-induced cells rotating 20°–360° (blue circles), and strain DS3318 motAB mutant cells rotating 20°–360° (brown circles) as a function of time measured every 5 s. An arbitrary value of 2 rad was added to each data point of WT (20°–360°, white circles), so that the line would be visible. (B) Time-lapse composite images taken at the end of each minute for cells expressing the flagellar clutch protein EpsE, cells expressing MotI (MotII), and cells mutated for MotAB stator proteins. Approximately 3–5% of each cell type rotated a full 360°. (C) Cartoon of flagellar rotation and models for clutch inhibition. (Left) Flagella rotate when proton motive force, gated by MotB, causes a conformational change in MotA (stator, light brown), which in turn clutches with FliG (rotor, dark brown) to create torque on the flagellum (purple). Clutch proteins separate MotA from FliG, thereby separating the power source from the drivetrain. (Upper Right) MotI binds to MotA to separate flagellar stators from contact with the rotor. (Lower Right) EpsE binds to FliG to separate the flagellar rotor from contact with the stators.
MotI Localizes as Transient MotA-Dependent Puncta.
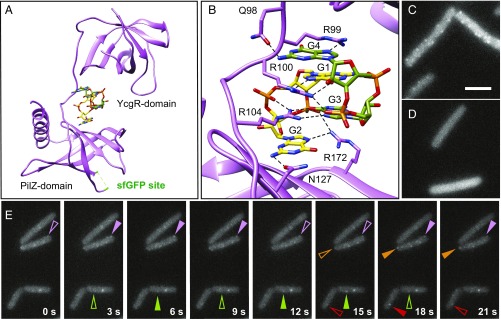
One way in which cytoplasmic MotI might inhibit MotA is by binding MotA at the membrane. To determine the location of MotI in living cells, a series of constructs was generated in which fluorescent proteins were fused to either the N terminus or C terminus with and without flexible domain-breaking linkers. In no case were the fluorescent fusions interpretable, as they were nonfunctional for motility inhibition, nonfluorescent, or exhibited cleavage of the fluorescent protein from MotI in vivo. To aid in the generation of a functional fluorescent fusion to MotI, the 3D structure of MotI was determined. When MotI was expressed and purified from E. coli, it resolved as two bands on a SDS/PAGE gel. Mass spectrometry analysis indicated that the smaller band represented a proteolyzed product of MotI in which the last four amino acids had been cleaved (MotIΔ4). A mixed population of full-length and truncated forms of MotI would complicate crystallization, and thus MotIΔ4, which was found to be functional in B. subtilis, was cloned and expressed for purification instead (Fig. S1B). Crystals of purified MotIΔ4 and MotIΔ4•SeMet were grown in the presence of c-di-GMP. Initial electron density maps were obtained with the MotIΔ4•SeMet crystal datasets using single-wavelength anomalous diffraction (SAD) phasing. Initial phases were combined with native amplitudes from native MotIΔ4 data to solve the structure to 3.2 Å with Rwork and Rfree of 23.6% and 27.7%, respectively (Fig. 4A and Table S4).
Fig. 4.
MotI-structure–enabled GFP fusion indicates that MotI localizes as transient MotA-dependent puncta. (A) 3D structure of MotI∆4 bound to a dimer of c-di-GMP (c-di-GMP monomer carbons are colored green and yellow, respectively). The locations of the N-terminal YcgR domain, the C-terminal PilZ domain, and the sfGFP insertion site (highlighted green in an unstructured loop in the crystal) are indicated. (B) Enhanced view of MotI residues in contact with the c-di-GMP dimer. Hydrogen bonds are found between E98 and the Watson–Crick edge of guanine base G4, between R99 and the Hoogsteen edge of guanine base G4, between R100 and the Hoogsteen edge of guanine base G1, between the phosphate that connects guanine bases G4 and G3, between R104 and the Hoogsteen edge of guanine base G3, and between N127 and the Watson–Crick edge of guanine base G2. Additionally R99 stacks with guanine base G1, R100 stacks with guanine base G4, and R104 stacks guanine base G2. (C and D) Fluorescence micrograph images taken from a time-lapse series of MotIInh-sfGFP induced with 4 µM IPTG in otherwise wild-type (strain NPS913) (C) and motAE92K (strain NPS915) (D) backgrounds. (Scale bar: 8 μm.) (E) Time-lapse series of fluorescence micrograph images of MotIInh-mNeonGreen induced with 2 µM IPTG (DK5142); images were taken every 3 s. Like-colored carets indicate the same puncta; and transience is indicated by open (waning puncta) and filled (waxing puncta) carets.
The MotI structure has an YcgR-like N-terminal domain and a C-terminal PilZ domain, despite the low sequence similarity between MotI and the YcgR protein of E. coli (Fig. S3) (32). Two molecules of c-di-GMP were found interdigitated by base stacking between the N- and C-terminal domains of MotI, similar to that observed in the structures of the homologs PP4397 [from Pseudomonas putida (33)] and Alg44 [from Pseudomonas aeruginosa (34)] (Fig. 4A). MotI interacted with the dimeric c-di-GMP via two binding motifs, QRRQYVR and NISAG (Fig. 4B) (32). To explore the validity of the structure, two adjacent arginine residues in the former motif predicted to interact directly with c-di-GMP, R99 and R100 were mutated to alanine. The MotIR99A,R100A mutant lost the ability to inhibit motility but remained stable in the cytoplasm, suggesting it was made but nonfunctional (Figs. S1C and S4). We conclude that the full-length structure of B. subtilis MotI is similar to other YcgR-like protein structures and that MotI requires bound c-di-GMP to inhibit motility (33). Furthermore, comparison of the two MotI protomers in the asymmetric unit show multiple conformations of the loop connecting the YcgR-like and PilZ domains while maintaining the orientation of these domains relative to one another. Thus, binding of c-di-GMP likely dictates a defined orientation of these domains for proper inhibition.
Using the 3D structure as a guide, we created an internal superfolder GFP (sfGFP) fusion in an exposed loop region of MotI between amino acids 116 and 117, and the resulting fusion construct was functional for motility inhibition (Fig. S1D). When full motility inhibition was induced with 1 mM IPTG, the cytoplasm appeared uniformly green, perhaps due to overexpression and saturation of MotI-sfGFP–binding sites. To minimize the expression of MotI-sfGFP, swarming motility assays were titrated to find the lowest concentration of IPTG necessary for full inhibition (16 µM) (Fig. S1D). When cells were grown in the presence of the fully inhibitory IPTG concentration, 16 µM, fluorescence was still cytoplasmic. When MotI-sfGFP was induced with IPTG concentrations that were subinhibitory (4 µM), however, cell membrane-associated puncta stood out over the cytoplasmic haze (Fig. 4C). These MotI-sfGFP puncta were abolished when expressed in the MotAE92K MotI-resistant mutant background (Fig. 4D). We conclude that MotI forms puncta when it is expressed at a low level and interacts with MotA-containing stator complexes. Finally, use of a photostable MotI-mNeonGreen fusion indicated that the puncta were stationary but transient, so that puncta appeared and disappeared over time (Fig. 4E). We infer that MotI docks at stators associated with basal bodies to form puncta, and puncta dissolution occurs when stators dissociate. We further infer that full inhibition sequesters stators in the membrane and prevents association with basal bodies.
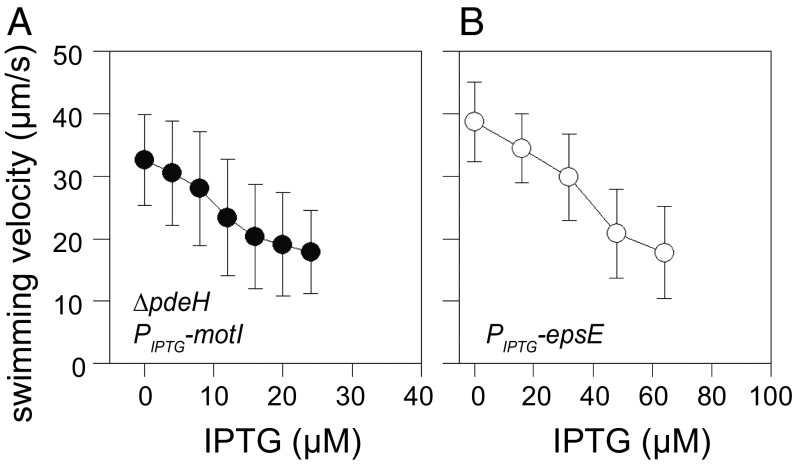
If MotI functions by sequestering individual stator units, then subinhibitory concentrations of MotI expression would reduce swimming speed proportionally. To measure swimming speed, cells containing the IPTG-inducible MotI-inhibitory construct were induced with a range of IPTG concentrations, monitored by phase-contrast microscopy, and tracked in silico using the program MicrobeJ (35). Increasing the level of MotI induction decreased cell swimming speed proportionally (Fig. 5A). Furthermore, the reduction of cell swimming speed by MotI could be counteracted by increasing IPTG induction of MotA and MotB (Fig. S5). Finally, cells containing an IPTG-inducible EpsE construct also showed a correlation between IPTG induction and decreased swimming speed (Fig. 5B). We conclude that clutch proteins fully inhibit flagella at maximal induction but generate intermediate speeds at subinhibitory concentrations.
Fig. 5.
Partial clutch inhibition of the flagellar rotor reduces swimming speed. Wet-mount tunnel microscopy measurement of cell swimming speed in a strain containing either an IPTG-inducible MotI strain (NPS350) (A) or IPTG-inducible EpsE strain (DS1855) (B) shown as a function of IPTG concentration. Speeds of 100 motile cells were measured using MicrobeJ.
Discussion
Flagella are tightly regulated at the level of gene transcription and translation, perhaps due to a high energetic cost of biosynthesis (4). Furthermore, assembly of a flagellum requires more than one generation at high growth rate, which, combined with the resource investment, may explain the need for reversible regulation of motility at the level of flagellar rotation (3, 36). Functional regulators of flagellar rotation are thought to act as either a molecular clutch or a molecular brake. Proteins that act like clutches reduce the interaction between the stators and the rotor, thereby decreasing power (6). Proteins that act like brakes enhance the interaction between the stators and the rotor (21), either by jamming the rotor and stators together or by increasing the resistance between the two. Here we explore motility inhibition by the B. subtilis YcgR-homolog MotI and conclude that MotI inhibits motility by acting as a clutch and not as a brake.
MotI inhibits motility by interacting with MotA. Previous work showed that MotI interacted with MotA in a bacterial two-hybrid assay, and we support this conclusion by titration, suppressor analysis, and localization dependency (22). The motility of cells partially inhibited by MotI was improved by MotA overexpression, indicating that MotA could titrate MotI. Suppressor analysis revealed that substitution of a single residue of MotA with lysine (E92K) restored motility to cells inhibited my MotI. The MotAE92K allele was fully functional in otherwise wild-type cells and abolished MotI subcellular localization, indicating that MotI requires Glu92 for interaction. In the absence of any other candidates, and with the caveat that residues simultaneously essential for both MotI binding and motility, if any, were excluded from the gain-of-motility suppressor selection, we conclude that MotI directly interacts with MotA and very likely does so near Glu92.
MotA Glu92 is in a cytoplasmic loop of MotA that directly interacts with the rotor protein FliG (28). Indeed, Glu92 is only two residues away from MotA residue R90, which is essential for motility in B. subtilis and is homologous to residue R90 of E. coli shown to interact directly with FliG and generate torque (17, 18, 24, 28). If MotI binds to MotA at residue Glu92, as cell biological localization-dependence suggests, MotI would bind between MotA and FliG. Thus, a brake mechanism is unlikely, as it is unclear how MotI interaction adjacent to power-generating residues would increase resistance without also separating MotA and FliG like a clutch. Thus, we conclude that MotI binding to MotA would likely compete with and therefore exclude FliG, as predicted for a clutch mode of action, rather than cause MotA and FliG to interact more tightly, as predicted for a brake.
How does MotI access MotA residue Glu92 when MotA interacts with FliG? Each stator complex contains four subunits of MotA and two subunits of MotB, but only one MotA subunit is considered to be in contact with the rotor instantaneously (37). We infer that the stationary MotI puncta represent the location of flagellar basal bodies, and MotI could form puncta by binding to the three unengaged MotA subunits of a flagellar-associated stator. Furthermore, in E. coli there is a dynamic pool of stators in the membrane that associate with and dissociate from basal bodies, and the equilibrium between the two states changes depending on the rotational load (19, 20, 38). Consistent with dynamism, the MotI puncta were transient, and we interpret puncta dissolution as indicative of stator dissociation. Furthermore, MotI would have unrestricted access to MotA in stators free in the membrane and would prevent subsequent reassociation with basal bodies. We propose a model in which MotI binds to unengaged MotA subunits either at basal bodies or free in the membrane, saturates the stators, and sequesters them from interaction with rotors. MotI could also induce stator dissociation, but thus far we have been unable to observe stator dynamics directly in B. subtilis, as all N-terminal, C-terminal, and internal fluorescent protein fusions to MotA and MotB have either been nonfunctional or nonfluorescent. Whatever the mechanism, we note that puncta transience and the absence of puncta at inhibitory concentrations of MotI are inconsistent with a brake model that would require stator–rotor immobilization.
MotI-inhibited flagella behaved as though they were unpowered. To measure motor output, cells were tethered to a slide by a sheared flagellum, and thus motor behavior was indicated by rotation of the cell body. Flagella that have been tethered and locked for rotation in E. coli by the addition of the cross-linking agent glutaraldehyde rotate only 3° due to torsion on the flagellum (29, 30, 39). When MotI-inhibited cells were tethered by a single flagellum, however, the cell body rotated through angles greater than 3° and resembled cells expressing the flagellar clutch protein EpsE or motAB-mutant cells lacking the stator proteins altogether. Glutaraldehyde cross-linking does not lock rotation in B. subtilis, for unknown reasons (6), and thus the torsional compliance of the B. subtilis flagellum may be greater than that of E. coli. Prolonged (3 min) observation, however, indicated that individual MotI-inhibited cells rotated 360°, an angle that likely excludes a locked motor with high torsional compliance (Fig. 3 A and B). Moreover, the angular displacement of rotation over time was constant, thus providing no evidence of rotational resistance that substantially decreased the rotation speed below that of the stator mutant. Instead, the biophysical analysis indicated that the MotI-inhibited cells had unpowered flagella and cells rotated, with an rmsd consistent with free rotation driven by Brownian motion.
In summary, we propose a model in which MotI inhibits motility like a clutch by binding MotA and antagonizing stator association with the rotor. Titration of MotI resulted in intermediate swimming speeds, likely by the partial sequestration of rotors from stators with an incremental reduction in power (40). We note that a reduction in swimming speed was also observed in YcgR-inhibited E. coli (7). Furthermore, spontaneous suppressors resistant to YcgR in E. coli were also reported in the MotA power-generating loop, and MotA overexpression was shown to titrate YcgR inhibition (9, 41). MotI and YcgR may function differently, however, as YcgR-inhibited flagella still rotated under power but had a rotation bias opposite of that of wild-type flagella (8, 9). Instead, MotI may be more similar to the YcgR homolog of P. aeruginosa, FlgZ, which is thought to sequester one kind of stator complex in favor of another (42). We conclude that molecular clutch proteins are a robust mechanism to arrest motility that can either separate the rotor from stators, in the case of EpsE, or separate stators from the rotor, in the case of MotI (Fig. 3C).
Experimental Procedures
Tethering Studies.
For preparing slides for tethering studies, glass coverslips were treated with 3-aminopropyltriethoxysilane (Thermo Scientific catalog no. 80370) and then were treated with Sulfo-SMCC (Thermo Scientific catalog no. 22322). The coverslips were then placed with the treated side facing down on top of two coverslips (with an ∼1- to 2-cm gap between the coverslips). Cells were grown to OD600 ∼0.6–1.0 and were resuspended in 1 mL of chemotaxis buffer. Flagella were sheared by passing the resuspended cells ∼100 times back and forth between two syringes connected via two 23-gauge needles (BD catalog no. 305145) and tubing (BD Intramedic catalog no. 427410). The cells were then introduced into the tunnel slides and allowed to settle on the treated side by gravity for 1 h. The tunnel slide was washed with chemotaxis buffer to remove any non–cross-linked cells and then was imaged using a Nikon 80i microscope. Movies were recorded for 60 s at 10 frames/s. The movies were then analyzed by MicrobeJ (35) tracking software.
Swimming Velocity Analysis.
Cells were grown to an OD600 of ∼0.6 and then were resuspended to an OD600 of 0.2 in LB medium. Tunnel slides were prepared as before, and cells were introduced into the tunnel slides and imaged using a Nikon 80i microscope. Movies were recorded for 30 s at 5 frames/s and then were analyzed by MicrobeJ (35).
Supporting Information includes SI Extended Experimental Procedures, Figs. S1–S5, Tables S1–S4 listing the strains, primers, and plasmids used in this study and X-ray diffraction data and refinement statistics, respectively, and Movies S1–S5.
Supplementary Material
Acknowledgments
We thank Adrien Ducret for image analysis support; Ethan Garner for the mNeonGreen protein; Guangming Chen, Alisa Klepach, and Paige Matthews for technical support; and Howard Berg and the anonymous reviewers for helpful suggestions. All crystallization experiments were conducted in the Indiana University METACyt Crystallization Automation Facility. Crystallographic data were collected with remote assistance provided by Dr. Jay Nix on beamline 4.2.2 at the Advanced Light Source at the Lawrence Berkeley National Laboratory, part of the Molecular Biology Consortium supported by the US Department of Energy and Indiana University. This work was supported by NIH Grant GM093030 (to D.B.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5VX6).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716231114/-/DCSupplemental.
References
- 1.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukherjee S, Kearns DB. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet. 2014;48:319–340. doi: 10.1146/annurev-genet-120213-092406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlinsey JE, et al. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- 4.Smith DR, Chapman MR. Economical evolution: Microbes reduce the synthetic cost of extracellular proteins. MBio. 2010;1:e00131-10. doi: 10.1128/mBio.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013;37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 7.Boehm A, et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Fang X, Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol. 2010;76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 9.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 11.Braun TF, Al-Mawsawi LQ, Kojima S, Blair DF. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry. 2004;43:35–45. doi: 10.1021/bi035406d. [DOI] [PubMed] [Google Scholar]
- 12.Reid SW, et al. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas DR, Morgan DG, DeRosier DJ. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao R, Amsler CD, Matsumura P, Khan S. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J Bacteriol. 1996;178:258–265. doi: 10.1128/jb.178.1.258-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 16.Hosu BG, Nathan VS, Berg HC. Internal and external components of the bacterial flagellar motor rotate as a unit. Proc Natl Acad Sci USA. 2016;113:4783–4787. doi: 10.1073/pnas.1511691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 19.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. Load-dependent assembly of the bacterial flagellar motor. MBio. 2013;4:e00551-13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci USA. 2013;110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilizota T, et al. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. Proc Natl Acad Sci USA. 2009;106:11582–11587. doi: 10.1073/pnas.0813164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Chai Y, Guo JH, Losick R. Evidence for cyclic Di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol. 2012;194:5080–5090. doi: 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao X, et al. Functional characterization of core components of the Bacillus subtilis cyclic-di-GMP signaling pathway. J Bacteriol. 2013;195:4782–4792. doi: 10.1128/JB.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JM, Guttenplan SB, Kearns DB. Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis. J Bacteriol. 2014;196:740–753. doi: 10.1128/JB.01217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkelman JT, et al. RemA is a DNA-binding protein that activates biofilm matrix gene expression in Bacillus subtilis. Mol Microbiol. 2013;88:984–997. doi: 10.1111/mmi.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Blair DF. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 29.Block SM, Blair DF, Berg HC. Compliance of bacterial polyhooks measured with optical tweezers. Cytometry. 1991;12:492–496. doi: 10.1002/cyto.990120605. [DOI] [PubMed] [Google Scholar]
- 30.Block SM, Blair DF, Berg HC. Compliance of bacterial flagella measured with optical tweezers. Nature. 1989;338:514–518. doi: 10.1038/338514a0. [DOI] [PubMed] [Google Scholar]
- 31.Tavaddod S, Charsooghi MA, Abdi F, Khalesifard HR, Golestanian R. Probing passive diffusion of flagellated and deflagellated Escherichia coli. Eur Phys J E Soft Matter. 2011;34:16. doi: 10.1140/epje/i2011-11016-9. [DOI] [PubMed] [Google Scholar]
- 32.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 33.Ko J, et al. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J Mol Biol. 2010;398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Whitney JC, et al. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem. 2015;290:12451–12462. doi: 10.1074/jbc.M115.645051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guttenplan SB, Shaw S, Kearns DB. The cell biology of peritrichous flagella in Bacillus subtilis. Mol Microbiol. 2013;87:211–229. doi: 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 38.Baker AE, O’Toole GA. Bacteria, rev your engines: Stator dynamics regulate flagellar motility. J Bacteriol. 2017;199:e00088-17. doi: 10.1128/JB.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berg HC. Does the Flagellar Rotary Motor Step? Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1976. [Google Scholar]
- 40.Block SM, Berg HC. Successive incorporation of force-generating units in the bacterial rotary motor. Nature. 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 41.Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 42.Baker AE, et al. PilZ domain protein FlgZ mediates cyclic di-GMP-dependent swarming motility control in Pseudomonas aeruginosa. J Bacteriol. 2016;198:1837–1846. doi: 10.1128/JB.00196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konkol MA, Blair KM, Kearns DB. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J Bacteriol. 2013;195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 46.Guérout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 47.Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- 48.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 49.Bergfors TM. 2009. Protein Crystallization (International University Line, La Jolla, CA), 474 pp.
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 52.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Ordal GW, Goldman DJ. Chemotaxis away from uncouplers of oxidative phosphorylation in Bacillus subtilis. Science. 1975;189:802–805. doi: 10.1126/science.808854. [DOI] [PubMed] [Google Scholar]
- 56.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.