Significance
Poxviruses cause human and epizootic diseases and are employed as vaccine vectors. The present investigation provides insights into a key step in poxvirus replication, the assembly of infectious virus particles. Enveloped viruses acquire membranes from cellular organelles; nevertheless, the source of the poxvirus membrane has been an enigma. Poxvirus assembly occurs in cytoplasmic factories, and the first recognizable structures are membrane crescents without discernible connections to cellular membranes. The key to demonstrating connections was isolation of vaccinia virus mutants that are defective in assembly. Electron tomographic analyses of cells infected with the mutants unambiguously demonstrated continuity between viral membranes and the endoplasmic reticulum and suggested that viral proteins induce or stabilize membrane scissions during a normal infection.
Keywords: membrane dynamics, membrane disruption, poxvirus assembly, endoplasmic reticulum dynamics, endoplasmic reticulum breakage
Abstract
The long-standing inability to visualize connections between poxvirus membranes and cellular organelles has led to uncertainty regarding the origin of the viral membrane. Indeed, there has been speculation that viral membranes form de novo in cytoplasmic factories. Another possibility, that the connections are too short-lived to be captured by microscopy during a normal infection, motivated us to identify and characterize virus mutants that are arrested in assembly. Five conserved vaccinia virus proteins, referred to as Viral Membrane Assembly Proteins (VMAPs), that are necessary for formation of immature virions were found. Transmission electron microscopy studies of two VMAP deletion mutants had suggested retention of connections between viral membranes and the endoplasmic reticulum (ER). We now analyzed cells infected with each of the five VMAP deletion mutants by electron tomography, which is necessary to validate membrane continuity, in addition to conventional transmission electron microscopy. In all cases, connections between the ER and viral membranes were demonstrated by 3D reconstructions, supporting a role for the VMAPs in creating and/or stabilizing membrane scissions. Furthermore, coexpression of the viral reticulon-like transmembrane protein A17 and the capsid-like scaffold protein D13 was sufficient to form similar ER-associated viral structures in the absence of other major virion proteins. Determination of the mechanism of ER disruption during a normal VACV infection and the likely participation of both viral and cell proteins in this process may provide important insights into membrane dynamics.
Members of the poxvirus family have large double-stranded DNA genomes and replicate entirely within the cytoplasm of infected cells (1). The infectious virus particles, which have a complex inner core and a lipoprotein envelope, are assembled in viral factories. Five successive stages of assembly have been discerned by transmission electron microscopy: crescents, immature virions (IVs), mature virions (MVs), wrapped virions (WVs), and extracellular virions (EVs). The most intriguing question regarding virion assembly concerns the origin of the crescent membrane, the precursor of the IV. Crescents and IVs were originally thought to arise de novo based on electron micrographs showing single bilayer open-ended membranes without discernible connections to cellular organelles (2), observations that have been confirmed by numerous investigators. Subsequent studies, however, demonstrated cotranslational insertion of the major vaccinia virus (VACV) membrane proteins A17 and A14 into microsomal membranes in vitro and their colocalization with the endoplasmic reticulum (ER) and post-ER membranes as well as crescents, IVs, and MVs in vivo (3–7). In addition, smooth membranes labeled with ER markers and viral proteins were detected in close proximity to crescents (8, 9). Biochemical studies focused attention on the ER as a possible source of the viral membrane by demonstrating that transport beyond the ER was unnecessary for IV formation (10). Nevertheless, direct connections between the ER and crescent membranes during a normal VACV infection have yet to be reported.
Insights into the mode of formation of the viral membrane have come largely from studies of VACV proteins and conditional lethal mutants (11). The A17 and A14 transmembrane proteins (3, 7, 12–14) and the D13 scaffold protein (15, 16) are major components of IVs. When expression of A17 or A14 is repressed, small vesicles and large electron-dense structures containing core proteins accumulate. Recent studies suggest that A17 is a reticulon-like protein that promotes membrane curvature (17). Both A17 and A14 are phosphorylated by the F10 protein kinase (7, 18–20), and evidence for a role of the kinase at a very early stage of membrane formation has been obtained with temperature-sensitive and inducible mutants (20–22). D13 trimers have structural homology with the double-barrel capsid proteins of adenovirus and other lipid-containing icosahedral viruses (23, 24). When expression of D13 is repressed, or the drug rifampicin that targets D13 is added, irregular membranes comprised of single bilayers with free ends form instead of IVs (15, 25–27).
Recently, five additional viral proteins that are present in low amounts or absent from MVs were found to be required for IV formation (Table 1). These sequence-unrelated proteins, which are conserved in all chordopoxviruses and have no homologs elsewhere, were dubbed viral membrane assembly proteins or VMAPs (11). Limited information is available regarding the structure and functions of these proteins. L2 and A30.5 are small proteins that interact with each other, have C-terminal hydrophobic domains, and associate with the ER as well as the edges of crescent membranes (28, 29). The N- and C-termini of L2 face the cytoplasmic and luminal sides of the ER, respectively, whereas both termini of A30.5 are cytoplasmic (30). A11 also has a C-terminal hydrophobic domain and localizes at the edges of crescents but associates with the ER only within virus factories and in the presence of other viral proteins (31–33). In contrast, neither H7 (34–36) nor A6 (37) have hydrophobic domains, and the proteins are distributed throughout the cytoplasm in addition to the viral factory. A6 has a two-domain architecture and weakly associates with A11 (32, 38). The atomic structure of H7 revealed a phosphoinositide binding site, which could enhance membrane association (36).
Table 1.
Virus membrane assembly proteins
| Protein | kDa | Hydrophobic domain | Synthesis | Localization | Association |
| L2 | 10 | C-terminal | E | Crescents, ER | A30.5 |
| A30.5 | 5 | C-terminal | PR | Crescents, ER | L2 |
| A11 | 40 | C-terminal | PR | Crescents, ER* | A6 |
| A6 | 43 | None | PR | Cytoplasmic | A11 |
| H7 | 40 | None† | PR | Cytoplasmic | — |
E, early; PR, postreplicative.
Only associates with ER in virus factory.
Has phosphoinositide-binding site.
Inducible and deletion mutants have been used to study the roles of the VMAPs. When expression of any VMAP is severely reduced or prevented, normal-looking IVs are absent, and large, electron-dense inclusions comprised of core proteins are formed (28, 31, 34, 35, 37, 39, 40). Nevertheless, the phenotypes of the various VMAP mutants differ in other ways. With some mutants, short crescents are juxtaposed to the outer surface of the electron-dense inclusions (29, 34, 37, 39) but not in others (31, 35, 38). The most intriguing finding is the presence of crescent membranes that appear by transmission electron microscopy (TEM) to be connected to the ER membrane and bud into expanded ER lumens where electron lucent circular IV-like (IV-L) structures accumulate (29, 33, 39). An important caveat, however, is that continuity of viral and ER membranes had not been verified by electron tomography (ET). In addition, although these structures were seen in cells infected with the L2 or A30.5 deletion mutant and the A11 inducible mutant, they were not noted in cells infected with A6 or H7 mutants (35, 38). Variations in the described phenotypes of the individual mutants may be due to different roles of the VMAPs or to technical reasons such as incompletely repressed synthesis of an inducible VMAP, use of different cell types, and alternative methods of processing samples for TEM.
Ideally, to compare the roles of the individual VMAPs, the studies should all be done with true null mutants, which recently became possible with construction of H7 (35), A6 (38), and A11 (to be described elsewhere) deletion mutants in addition to the earlier L2 (29) and A30.5 (39) deletion mutants. Furthermore, ET is needed to determine the 3D structures of the IV-L particles and unambiguously demonstrate connections between viral and cellular membranes. In addition, while knockout studies are useful for determining the proteins necessary for viral membrane formation, expression studies are required to determine which are sufficient to modify the ER. Here we show that the membranes of virus-like particles are continuous with the ER in cells infected with each of the VMAP deletion mutants and that similar structures form when the A17 transmembrane protein and the D13 scaffold protein are coexpressed in the absence of other major virion components. Our model for the formation of the crescent membrane involves remodeling of the ER by the A17 protein, binding of D13 to A17 to increase curvature, and participation of the VMAPs, likely in collaboration with cellular proteins, for scission of the ER membrane and stabilization of the free ends.
Results
Analysis of L2 and A30.5 Deletion Mutants.
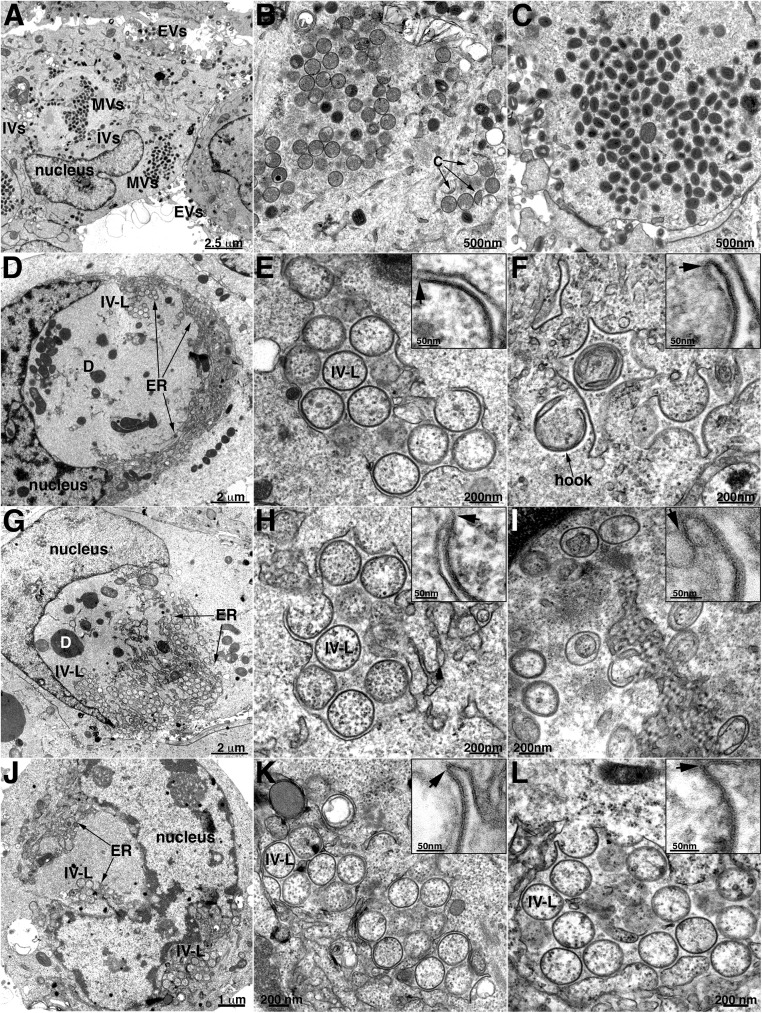
Representative images of normal VACV assembly are shown (Fig. 1 A–C) to appreciate the difference in cells infected with VMAP deletion mutants. At low magnification (Fig. 1A), the cytoplasmic factories were recognized by their fine-granular appearance, location near the nucleus, scarcity of cellular organelles, and presence of circular IVs and denser MVs. Clusters of IVs and MVs and a smaller number of crescents are shown at higher magnification in Fig. 1 B and C, respectively. The outer surface of the IV is covered by a honeycomb lattice comprised of trimers of the D13 scaffold protein, which is removed during the transition to the MV. This scaffold increases the electron density of the IV membrane and may appear as a brush border or as spicules by TEM, depending on the thickness, staining, and magnification of the section. Importantly, neither the crescents nor the IVs are closely associated with cellular membranes.
Fig. 1.
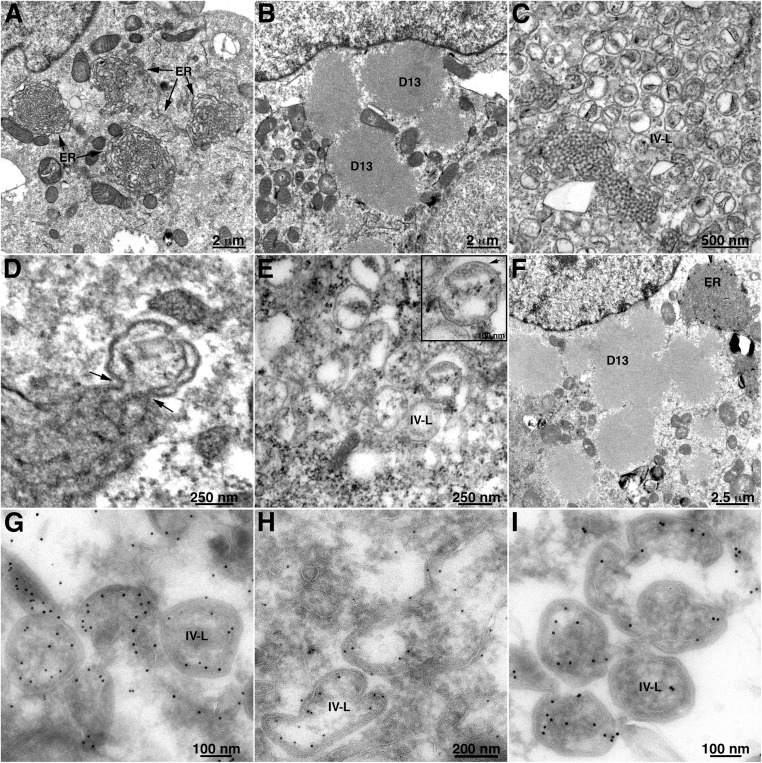
TEM Images of crescent and IV-L structures in cells infected with VACV L2, A30.5, or A6 deletion mutants. RK13 cells were infected with the wild-type VACV (A–C) or VACV A30.5 (D–F), L2 (G–I) or A6 (J–L) deletion mutant and prepared for TEM. (A–C) Low- and high-magnification images of cells infected with wild-type VACV, showing factory regions of cytoplasm containing clusters of circular immature virions or dense brick-shaped mature virions and few individual crescents and extracellular virions. (D–F) Low- and high-magnification images of cells infected with A30.5 deletion mutant, showing dense inclusions and clusters of IV-L structures enclosed within a membrane sac in the factories. Arrows in the Inset point to the junction of smooth and spicule-coated membranes. A structure resembling a hook is labeled. (G–I) Similar to previous panels except cells were infected with the L2 deletion mutant. (J–L) Similar to previous panels except cells were infected with the A6 deletion mutant. Scale bars are shown. Abbreviations: C, crescent; D, dense body consisting of aggregate of viral core proteins; ER, endoplasmic reticulum; EVs, extracellular virions; IV-L, immature virion-like structure; IVs, immature virions; MVs, mature virions.
Although TEM images of cells infected with L2 and A30.5 deletion mutants were previously shown (39), we prepared new ones for direct comparison with the other mutants and to provide a link to the ET analysis. These two proteins interact with each other, and the images of rabbit kidney RK13 cells infected with the A30.5 gene deletion mutant (Fig. 1 D–F) were distinct from the wild-type virus infection but indistinguishable from those infected with the L2 gene deletion mutant (Fig. 1 G–I). At low magnification (Fig. 1 D and G), the fine-granular factory areas were readily recognized, although there was a noticeable absence of MVs and presence of large, amorphous, electron-dense inclusions that frequently form when there is interference with virus assembly and have been shown to contain core proteins (31, 34, 41). Most interesting are the many circular forms termed IV-Like (IV-L) because of their resemblance in size and shape to IVs, within and adjacent to membranes near the boundaries of the factories. At higher magnification (Fig. 1 E and H), clusters of the circular forms appeared to be enclosed within a sac comprised of a smooth membrane. Furthermore, the smooth membrane appeared to be connected to crescent-like incomplete circular structures that appear to be budding into the lumen of the sac (Fig. 1 E and H, Insets). We previously showed by immunogold labeling that the smooth membranes comprising the sac have ER markers, and the circular membranes contain the viral A17 membrane protein and the D13 trimer scaffold (29). In contrast to typical IVs, the circular IV-L structures lacked the electron-dense interior comprised of core proteins, which instead are aggregated in the dense inclusions outside of the ER. Some membranes appeared as half-moon or hook structures; however, one side always exhibited a dense coat, indicating a viral membrane (Fig. 1 F and I).
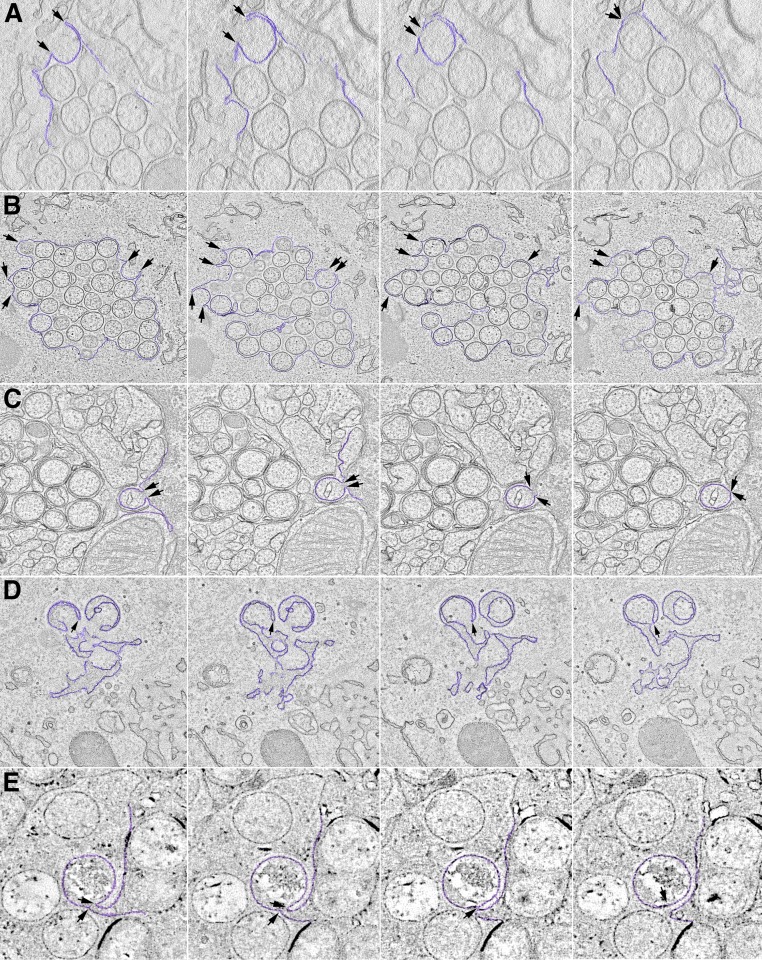
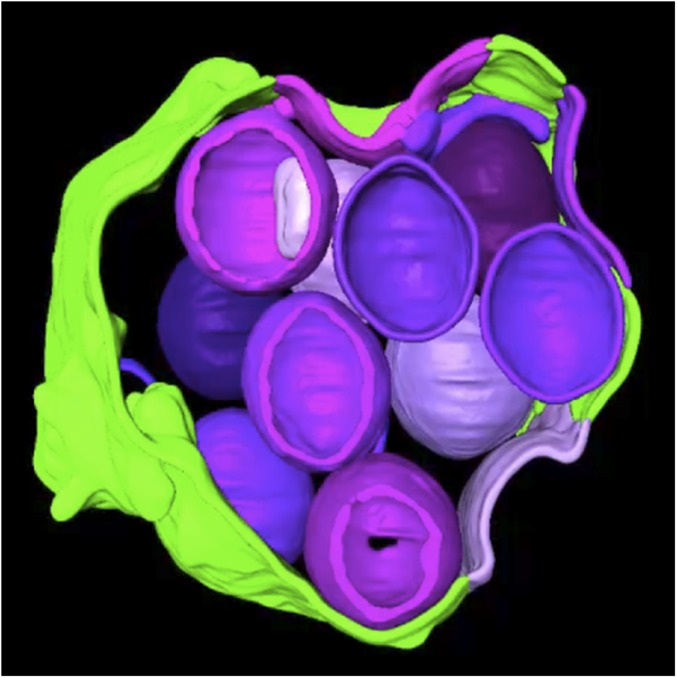
Although the TEM images suggested continuity between the curved viral membranes and the smooth ER membrane, in some cases the junctions appeared out of focus, and in others the membranes might have been merely overlapping. ET was carried out to resolve these issues and further characterize the structures. In this procedure, the electron beam projects through the cell section, and a series of tilted images are acquired. The data are subsequently assembled into 3D constructions. Movie S1 shows a region of the cytoplasm with a cluster of circular structures surrounded by a smooth membrane from a cell infected with the A30.5 deletion mutant. The images and volumetric rendering clearly reveal that the circles are sections through spherical IV-L particles and that the crescents are cupules continuous with the surrounding ER membrane. Several individual frames of an ET analysis similar to that of Movie S1 are shown in Fig. 2A. The arrows in each panel of the tilt series point to the same location and show the continuity between the ER and viral membranes. The 3D rendering shown at the end of Movie S1 is reproduced in Fig. 3. The smooth membranes comprising the sac are colored green, and the curved membranes continuous with the latter and sections through IV-L particles are violet (Fig. 3).
Fig. 2.
Visualization of viral structures by ET. Frames from tomographic datasets are shown. Rows: (A) A30.5 deletion mutant from a movie similar to Movie S1; (B) A6 deletion mutant corresponding to Movie S2; (C) H7 deletion mutant corresponding to Movie S3; (D) A11 deletion mutant corresponding to Movie S4. (E) Coexpression of A17 transmembrane protein and D13 scaffold protein corresponding to Movie S5. Arrows and colored areas reveal connections between viral membranes and smooth ER.
Fig. 3.
Three-dimensional rendering of a cluster of IV-L structures within ER lumen. Curved viral membranes are colored violet, and smooth ER is green. From Movie S1 of cell infected with A30.5 deletion mutant.
Analysis of the A6 Deletion Mutant.
Previous TEM images of African green monkey BS-C-1 cells infected by the A6 deletion mutant revealed dense inclusions with an absence of associated crescents, but ER-associated structures were not noted (38), perhaps because of poor preservation of the membranes due to the formaldehyde/glutaraldehyde fixation treatment. The low magnification of a RK13 cell infected with the A6 deletion mutant in Fig. 1J shows viral factory regions with clusters of IV-L structures on either side of the nucleus. At a higher magnification, crescents connected to smooth membranes and IV-Ls were clearly seen (Fig. 1 K and L). Without prior knowledge, the images of cells infected with the A6 deletion mutant could not be distinguished from the A30.5 and L2 deletion mutants. The virtual identity of the mutant phenotypes extended to ET analysis. Movie S2 shows a cluster of IV-L particles surrounded by a smooth membrane in cells infected with the A6 deletion mutant. There is clear continuity between the smooth membrane and crescent structures, which appear as cupules in three dimensions. The colored 3D rendering highlights the connection between an IV-L particle and the peripheral membranes. Individual frames from Movie S2, shown in Fig. 2B, reveal that some structures appear as an open crescent in one frame and a closed circle in others, depending on the tilt.
Analysis of the H7 Deletion Mutant.
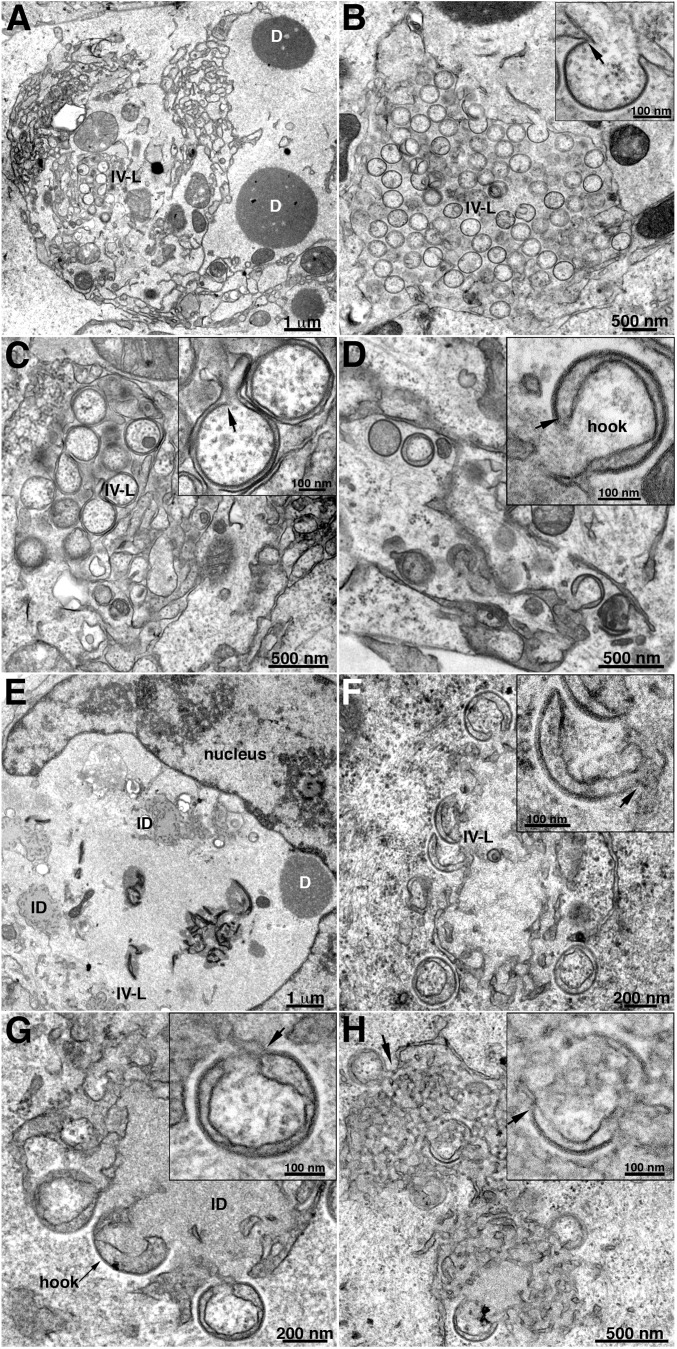
Previous TEM images of BS-C-1 cells infected by an H7-inducible mutant revealed dense inclusions, some of which had associated crescents (34), whereas no crescents were seen in cells infected with an H7 deletion mutant, indicating greater stringency (35). Nevertheless, viral membrane structures associated with the ER were not described in either of those studies. The low-magnification TEM image of RK13 cells infected with the H7 deletion mutant revealed dense inclusions and membranes at the periphery of the factory (Fig. 4A) similar to that found with the other VMAP deletion mutants. At higher magnification, crescents with apparent connection to smooth membranes and clusters of IV-L structures appeared (Fig. 4B, Inset), also like those seen in cells infected with the A30.5, L2, and A6 deletion mutants. However, in many images the surrounding smooth membrane appears to protrude inward to secondarily wrap around individual IV-L particles that are still connected to the ER (Fig. 4C, Inset). There were more examples of this secondary wrapping in the images of the H7 deletion mutant than with the other VMAP deletion mutants as well as hook-like structures (Fig. 4D, Inset). The continuity between smooth membranes and viral particles and the interleaving of membranes between the particles were verified by ET (Movie S3). The colored 3D rendering at the end of Movie S3 depicts the multiple membranes. Individual frames from Movie S3 are shown in Fig. 2C.
Fig. 4.
TEM images of crescent and IV-L structures in cells infected with VACV H7 or A11 deletion mutant. (A–D) RK13 cells were infected with VACV H7 deletion mutant and prepared for TEM. (A) Low-magnification image, showing viral factory regions containing IV-Ls and dense inclusions. (B) Field of IV-Ls resembling those formed in cells infected with the VACV A30.5, L2, and A6 deletion mutants. (C) Higher-magnification image, showing secondary wrapping of IV-Ls by smooth membrane to form a double layer. (D) Higher magnification image, showing hook-like structures. (E–H) RK13 cells infected with VACV A11 deletion mutant. (E) Low-magnification image, showing viral factory region containing dense inclusions, intermediate dense bodies, and IV-Ls near periphery. (F) Higher magnification showing IV-Ls and hook-like structures. (G and H) Arrows point to junctions of coated viral and smooth ER membranes and to a hook structure. Abbreviations: D, dense inclusion; ID, intermediate dense body; IV-L, IV-like particles.
Analysis of the A11 Deletion Mutant.
Low-magnification TEM images of cells infected with the A11 deletion mutant confirmed the absence of normal-looking IVs or more mature forms. Both electron-dense inclusions, comprised of core proteins, and intermediate-density bodies characteristic of the D13 protein intermixed with ribbons of ER (31) were found (Fig. 4E). Similar D13-ER inclusions were also seen with the other VMAP mutants (not shown). IV-L structures were less frequent with the A11 deletion mutant compared with the other mutants, and many had a hook shape or appeared to be double-wrapped with an electron-dense convex membrane (Fig. 4 F–H). Apparent connections to smooth membranes are marked by arrows. IV-L particles similar to those formed by the A30.5, L2, and A6 deletion mutants were found in RK13 cells infected with an inducible A11 mutant in the absence of inducer (33, 42). ET revealed that the hook and double-wrapped IV-L particles seen in thin sections are all different views of crescent and IV-L structures that are connected to the peripheral smooth membranes (Movie S4). The colored 3D rendering at the end of Movie S4 shows the membrane connections. The individual movie frames in Fig. 2D illustrate how the membrane connections appear and disappear, depending on the tilt. Thus, using the same protocol, we found viral membrane structures in continuity with smooth ER for all five deletion mutants.
Formation of IV-L Structures by Coexpression of the A17 Transmembrane Protein and the D13 Scaffold Protein.
The IV-L structures that form in the absence of VMAPs contain both the A17 transmembrane protein and the D13 scaffold protein. We previously showed that intracellular expression of the A17 transmembrane protein in the absence of other viral structural proteins transformed the ER into aggregated 3D tubular networks (17). We now wanted to determine whether coexpression of the D13 scaffold protein with A17 would further transform the ER into structures that resemble the forms made during VACV infection in the absence of the VMAPs. Our strategy was to infect cells with a recombinant VACV that expresses T7 RNA polymerase in the presence of the DNA synthesis inhibitor AraC and transfect plasmids encoding A17 and D13 regulated by the T7 promoter. In this way, transcription would occur in the cytoplasm, and AraC would prevent synthesis of DNA, formation of viral factories, and the intermediate and late structural proteins. Of the five VMAPs, only L2 is expressed early in the presence of AraC. TEM images show the tight ER networks that formed when the A17 plasmid alone was transfected (Fig. 5A), the D13 depots that formed when the D13 plasmid alone was transfected (Fig. 5B), and structures resembling IV-Ls that formed when both were transfected (Fig. 5C). Perhaps because the viral proteins were overexpressed, IV-L structures rather than crescents predominated at the time of analysis. The electron-dense border characteristic of D13 was visible on the convex side of the IV-L membranes with apparent connections to ER (Fig. 5D). The structures were somewhat irregular, with diameters from about 150 to 350 nm, suggesting that they are not uniformly coated with D13 trimers. The diameters are similar to those of the structures formed in vitro when D13 was incorporated into liposomes via a six-histidine tag (23). Immunogold staining with antibodies to A17 (Fig. 5G), D13 (Fig. 5H), and protein disulfide isomerase (Fig. 5I) verified the presence of the viral and ER proteins in association with the membrane structures. The resemblance between the A17/D13 structures and IV-Ls that formed when cells are infected with VMAP deletion mutants was verified by ET, which showed connections with the smooth membranes (Movie S5) that are also seen in the individual frames of Fig. 2E.
Fig. 5.
Modification of ER membranes by expression of VACV A17 and D13 proteins. BS-C-1 cells were infected with a recombinant VACV expressing T7 RNA polymerase (vTF7-3) in the presence of the DNA replication inhibitor AraC and transfected with plasmids with T7 promoters regulating (A) full-length A17 alone, showing tightly curved ER networks; (B) D13 alone, showing protein inclusions; (C and D) D13 and full-length A17, showing IV-L structures with arrows pointing to membrane junctions; (E) D13 and C-terminal truncated A17, showing IV-L structures; (F) D13 and N-terminal truncated A17, showing D13 inclusions and tightly curved ER networks in the same cell. The bottom row contains images of BS-C-1 cells infected with vTF7-3 in the presence of AraC and transfected with plasmids expressing D13 and full-length A17. Cells were cryosectioned and stained with antibody to (G) A17, (H) D13, and (I) protein disulfide isomerase and then protein A conjugated to 10 nm gold.
Previous experiments determined that the N terminus of A17 is necessary for association with D13 (7, 43). Using the protocol above, plasmids containing D13 plus A17 with C-terminal or N-terminal truncations were transfected into cells infected with VACV expressing T7 RNA polymerase in the presence of AraC. Structures indistinguishable from those of full-length A17 plus D13 were seen when the C terminus of A17 was truncated (Fig. 5E), whereas these structures were not seen when the N terminus of A17 was truncated; instead, separate A17-modified ER networks and D13 depots formed in the same cells (Fig. 5F).
Discussion
The inability to discern connections between nascent VACV membranes and cellular organelles during a normal infection may be due to their transient nature or to rupture of the cellular membranes at a very early stage. Our approach to overcome this problem employed mutant viruses that are unable to complete the assembly process. We showed previously by TEM that there is an accumulation of viral membranes that appeared to be connected to ER membranes in cells infected with the VACV L2, A30.5, and A11 mutants. The viral membranes were identified by immunogold labeling with antibody to the A17 transmembrane protein and the D13 scaffold protein, and the ER was identified by labeling with antibody to calnexin. ET was employed in the present study to further characterize the viral structures and establish that the viral and ER membranes are truly continuous rather than overlapping. Another goal was to analyze the viral structures that form when cells were infected with each of the five VMAP deletion mutants, which were not all previously available for a comparative study. This was of special interest because IV-L structures had not previously been noted with mutants of A6 or H7, which lack hydrophobic domains, in contrast to the other VMAPs. In the last part of this study we took an opposite approach by expressing individual VACV proteins to determine the minimal number needed to modify ER membranes.
Using ET, we showed that the crescent structures, which formed when cells were infected with the A30.5 and L2 mutants, were indeed continuous with the ER membrane and appeared as cupules in three dimensions. The circular structures within the expanded ER lumen were sections of IV-L spheres. The finding that virtually identical viral structures formed in the absence of A30.5 or L2 was not surprising because the proteins form a heterodimer (39). However, the viral structures that formed in the absence of A6 were also indistinguishable from those that formed when A30.5 or L2 was not expressed, suggesting functional, if not physical, associations. Although some images of cells infected with the H7 deletion mutant also appeared indistinguishable from the others, there was more extensive ER wrapping and additional particles that looked like hooks and double-wrapped IVs by conventional TEM. Similar structures were also frequently seen in cells infected with the A11 mutant. However, ET revealed that they also represented incomplete spherical or crescent structures with connections to the ER. Thus, there is a commonality in function of all five VMAP proteins: deletion of any VMAP gene resulted in continuity between viral and ER membranes. A physical interaction between L2 and A30.5 was demonstrated by cross-linking (39), and A11 was shown to coprecipitate with A6, suggesting interaction (32). Interestingly, A11 was diminished when L2 or A30.5 was deleted (29, 39), but not when H7 was deleted (35). Whether higher-order complexes containing all five VMAPs exist is not yet known.
Our finding that the crescents and IV-like structures bud into the lumen of the ER rather than into the cytoplasm was unexpected and implies that the N terminus of A17, which is the binding site for D13, faces the luminal side of the ER membrane and that some breaks in the ER allow D13 to enter this space even in the absence of individual VMAPs. However, the core proteins are excluded and aggregate in the cytoplasm to form dense inclusions. If the N- and C-termini of A17 face the luminal side of the ER, then the corresponding segments of the A14 membrane protein may face the cytoplasm, which is opposite to the proposed orientation based on intermolecular disulfide bonds (7). A cytoplasmic orientation, however, would explain why the C terminus is not glycosylated during a normal VACV infection. More definitive studies of the topology of both A17 and A14 membrane proteins are warranted.
Although studies with mutant viruses can identify proteins that are needed for the formation of viral membranes, additional approaches are required to determine which proteins are sufficient. Because the IV-L structures contain both the A17 and D13 proteins, which are known to interact with each other, we wanted to determine whether together they could enable remodeling of the ER membranes. The A17 protein shares topological features with cellular reticulon proteins (17). The latter cell proteins promote membrane curvature and contribute to the tubular structure of the ER. When purified A17 protein was incorporated into liposomes, 25-nm vesicles and tubules formed, depending on the protein concentration (17). Cellular overexpression of A17 in the absence of other viral structural proteins transformed the ER into aggregated 3D tubular networks (17). In other in vitro experiments, six-histidine–tagged D13 trimers were added during the formation of liposomes that had been doped with nickel-chelating lipids to bind the trimers. Closed 150-nm to 350-nm particles formed as well as open-ended curved lattices (23). Thus, both A17 and D13 have membrane-remodeling properties. In the present study, when A17 and D13 were simultaneously overexpressed in cells, irregular IV-L structures formed that were labeled with antibodies to both proteins. Furthermore, electron tomography confirmed that these structures were continuous with smooth membranes. In contrast, when the two proteins were expressed separately, A17 induced the formation of tubular networks, and D13 aggregated in cytoplasmic depots. It was previously shown that D13 interacts with the N terminus of A17 (7, 43). When an A17 C-terminal truncation mutant was overexpressed with D13, the structures appeared identical to that of full-length A17, consistent with the retention of the interaction site. In contrast, when the N-terminal truncation mutant was overexpressed with D13, separate foci of ER tubulation and D13 depots were seen in the same cell. Similar structures formed when the A14 transmembrane protein and the F10 kinase were coexpressed with A17 and D13 (data not shown), suggesting that the VMAPs and likely additional proteins are needed for completion of IV formation.
The ER is a continuous membrane system comprised of the nuclear envelope and a network of peripheral tubules and sheet-like cisternae with a common luminal space that is continuously undergoing dynamic changes, including breakage and fusion (44, 45). The dynamin family GTPase atlastin has a key role in homotypic fusion of ER, including the formation of three-way junctions (46, 47). Less is known about ER breakage. Fusion and fission of mitochondrial membranes are also carried out by dynamin GTPases (48). The VMAPs lack predicted GTPase-active sites that potentially could mediate breakage, suggesting the possibility that other viral proteins are needed or, alternatively, that the VMAPs collaborate with cellular proteins to make nicks. A role for cellular proteins might help explain why the numbers of IV-L structures formed by VMAP mutants are more numerous in RK-13 cells than BS-C-1 cells (39). It is known from in vitro studies that the edges of lipid bilayers are very unstable, and the role of the VMAPs may be to stabilize breaks, which are normally quickly sealed by cellular proteins. Indeed, we previously showed an association of VMAP proteins with the edges of crescents during a normal infection (33, 42). Even so, it is puzzling why five viral proteins are required.
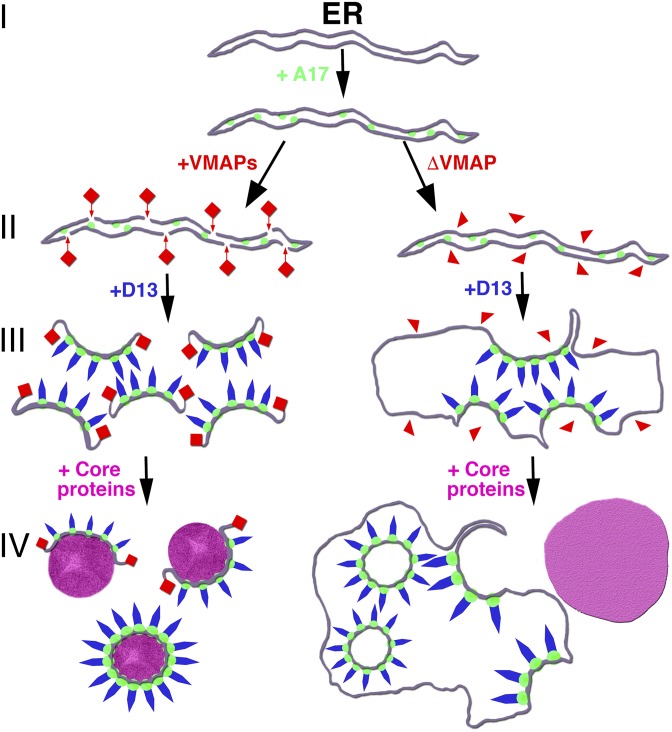
In conclusion, studies with the VMAP deletion mutants and expression of A17 and D13 support a model for the formation of viral crescent membranes during an infection with wild-type virus that is depicted on the left side of Fig. 6: (I) modification of the ER by insertion of the reticulon-like A17 transmembrane protein; (II) breakage of the ER membrane and stabilization of the breaks by VMAPs in collaboration with other cellular or viral proteins; (III) association of the D13 trimers with the N-terminal region of A17 providing additional curvature; and (IV) extension of the viral membrane around core proteins and DNA to form crescents and IVs. We infer from studies with the drug rifampicin and a D13-inducible mutant that step II can precede the association of D13 (9, 15, 25–27), although steps II and III may occur simultaneously during a natural infection. As depicted on the right side of Fig. 6, step II fails to occur, and the crescents remain attached to the ER in the absence of any individual VMAP. This simple model will undoubtedly be further refined in the future.
Fig. 6.
Model of viral membrane formation from ER. The first step (I) consists of modification of the ER by insertion of the A17 transmembrane protein, represented by green spheres. In the presence of all five VMAPs, represented by red rectangles, membrane scission occurs, and the edges of the sheets are stabilized, as shown on the Left (II). D13, represented by blue shapes, associates with the N terminus of A17 to increase curvature-forming crescent structures (III), which extend around core proteins (purple) to form the spherical IVs (IV). VMAPs missing one component (ΔVMAP), represented by red triangles, are unable to induce membrane scissions, the crescents remain attached to the ER, and empty IV-L particles bud into the lumen, as shown on the Right. The core proteins form dense inclusion aggregates (purple) outside of the ER.
Materials and Methods
Viruses and Plasmids.
VACV strain Western Reserve (WR) with deletions of A30.5, L2, A6, or H7 were described previously and propagated in complementing cell lines (29, 35, 38, 39). The A11 deletion mutant was made similarly and will be described elsewhere. MVs were purified by sedimentation through a 36% sucrose cushion and in some cases by an additional 25–40% sucrose gradient as described previously (49). Plasmids expressing D13 and full-length and truncated forms of A17 regulated by the T7 promoter were previously described (43).
Transfection.
BS-C-1 cells were grown on coverslips in 24-well plates. After reaching 80% confluency the cells were infected with 5 PFU per cell of vTF7-3 (50) and after 1 h were transfected with plasmids. Lipofectamine 2000 (Invitrogen Life Sciences) was used for transfection according to instructions of the provider. The cells received fresh medium at 6 h after transfection. At 16 h after infection the cells were fixed and prepared for electron microscopy.
TEM.
RK13 cells were infected with 1–5 PFU of VACV for 20 h. The cells were fixed with 2% glutaraldehyde and embedded in EmBed-812 resin (Electron Microscopy Sciences) as described (28). Cryosectioning and immunogold staining was carried out as described previously (51). Specimens were viewed with a Tecnai Spirit transmission electron microscope (FEI).
ET.
For cells infected with the A30.5 deletion mutant, 200-nm-thick sections were collected on glow-discharged carbon grids, and 10-nm colloidal gold fiducial markers were applied. Using a linear tilt scheme and a Tecnai BioTwin Spirit TEM (FEI) operated at 120 kV, a series of single-axis tilt images were collected. Images captured over a tilt range of ±60° (1° increments) at a 1-μm defocus level were recorded using an UltraScan 1000 Gatan charge-coupled–device (CCD) camera (2,048 by 2,048 pixels) and automated tomography acquisition software (Xplore 3D; FEI). The resulting images had a binning factor of 1 and a pixel size of 0.9075 nm. Tilt series were then initially aligned using IMOD’s Batchruntomo and refined in ETOMO before reconstructing weighted back-projection tomograms (version 4.8.37). Voltex volume renderings were created from Gaussian-filtered tomograms with a 3D kernel value of 1.75 with inverted contrast by thresholding selected areas of interest using the Amira Visualization Package (version 6.3.0; FEI). All manual segmentation was done on unfiltered tomograms with inverted contrast. For cells infected with all other deletion mutant viruses or transfected with plasmids, 150-nm-thick sections were collected on Formvar-coated slot grids, and 10-nm colloidal gold fiducial markers were applied. Dual-axis, serial section tilt-series were acquired using SerialEM (52). Automated electron microscope tomography using robust prediction of specimen movements (52) on a Tecnai BioTwin Spirit TEM (FEI) operated at 120 kV was used. Images were captured over a tilt range of ±60° (1° increments) at a 200-nm defocus on a Gatan US4000 CCD camera (4K × 4K) at 11,000× magnification (0.98-nm pixel). Tomograms were reconstructed using IMOD (53).
Supplementary Material
Acknowledgments
We thank Catherine Cotter for preparation of cells and viruses and Kaytlyn Menk for aiding in segmentation, rendering, and movie generation. The work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), NIH, and Grant AI079217 from NIAID (to Y.X.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716255114/-/DCSupplemental.
References
- 1.Moss B. Poxviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Vol 2. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 2129–2159. [Google Scholar]
- 2.Dales S, Siminovitch L. The development of vaccinia virus in Earle’s L strain cells as examined by electron microscopy. J Biophys Biochem Cytol. 1961;10:475–503. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolffe EJ, Moore DM, Peters PJ, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krijnse-Locker J, et al. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J Biol Chem. 1996;271:14950–14958. doi: 10.1074/jbc.271.25.14950. [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez JR, Risco C, Carrascosa JL, Esteban M, Rodríguez D. Characterization of early stages in vaccinia virus membrane biogenesis: Implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J Virol. 1997;71:1821–1833. doi: 10.1128/jvi.71.3.1821-1833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betakova T, Moss B. Disulfide bonds and membrane topology of the vaccinia virus A17L envelope protein. J Virol. 2000;74:2438–2442. doi: 10.1128/jvi.74.5.2438-2442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unger B, Mercer J, Boyle KA, Traktman P. Biogenesis of the vaccinia virus membrane: Genetic and ultrastructural analysis of the contributions of the A14 and A17 proteins. J Virol. 2013;87:1083–1097. doi: 10.1128/JVI.02529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husain M, Weisberg AS, Moss B. Existence of an operative pathway from the endoplasmic reticulum to the immature poxvirus membrane. Proc Natl Acad Sci USA. 2006;103:19506–19511. doi: 10.1073/pnas.0609406103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chlanda P, Carbajal MA, Cyrklaff M, Griffiths G, Krijnse-Locker J. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host Microbe. 2009;6:81–90. doi: 10.1016/j.chom.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Husain M, Moss B. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J Virol. 2003;77:11754–11766. doi: 10.1128/JVI.77.21.11754-11766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moss B. Poxvirus membrane biogenesis. Virology. 2015;479–480:619–626. doi: 10.1016/j.virol.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez D, Esteban M, Rodríguez JR. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez JR, Risco C, Carrascosa JL, Esteban M, Rodríguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traktman P, et al. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: Construction and characterization of a tetracycline-inducible recombinant. J Virol. 2000;74:3682–3695. doi: 10.1128/jvi.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]
- 16.Szajner P, Weisberg AS, Lebowitz J, Heuser J, Moss B. External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J Cell Biol. 2005;170:971–981. doi: 10.1083/jcb.200504026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erlandson KJ, et al. Poxviruses encode a reticulon-like protein that promotes membrane curvature. Cell Rep. 2016;14:2084–2091. doi: 10.1016/j.celrep.2016.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betakova T, Wolffe EJ, Moss B. Regulation of vaccinia virus morphogenesis: Phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J Virol. 1999;73:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer J, Traktman P. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J Virol. 2003;77:8857–8871. doi: 10.1128/JVI.77.16.8857-8871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szajner P, Weisberg AS, Moss B. Evidence for an essential catalytic role of the F10 protein kinase in vaccinia virus morphogenesis. J Virol. 2004;78:257–265. doi: 10.1128/JVI.78.1.257-265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traktman P, Caligiuri A, Jesty SA, Liu K, Sankar U. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J Virol. 1995;69:6581–6587. doi: 10.1128/jvi.69.10.6581-6587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Punjabi A, Traktman P. Cell biological and functional characterization of the vaccinia virus F10 kinase: Implications for the mechanism of virion morphogenesis. J Virol. 2005;79:2171–2190. doi: 10.1128/JVI.79.4.2171-2190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyun JK, et al. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Pathog. 2011;7:e1002239. doi: 10.1371/journal.ppat.1002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19:1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimley PM, Rosenblum EN, Mims SJ, Moss B. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: Accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moss B, Rosenblum EN, Katz E, Grimley PM. Rifampicin: A specific inhibitor of vaccinia virus assembly. Nature. 1969;224:1280–1284. doi: 10.1038/2241280a0. [DOI] [PubMed] [Google Scholar]
- 27.Nagaya A, Pogo BGT, Dales S. Biogenesis of vaccinia: Separation of early stages from maturation by means of rifampicin. Virology. 1970;40:1039–1051. doi: 10.1016/0042-6822(70)90150-9. [DOI] [PubMed] [Google Scholar]
- 28.Maruri-Avidal L, Domi A, Weisberg AS, Moss B. Participation of vaccinia virus l2 protein in the formation of crescent membranes and immature virions. J Virol. 2011;85:2504–2511. doi: 10.1128/JVI.02505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruri-Avidal L, Weisberg AS, Bisht H, Moss B. Analysis of viral membranes formed in cells infected by a vaccinia virus L2-deletion mutant suggests their origin from the endoplasmic reticulum. J Virol. 2013;87:1861–1871. doi: 10.1128/JVI.02779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyun SI, Maruri-Avidal L, Moss B. Topology of endoplasmic reticulum-associated cellular and viral proteins determined with split-GFP. Traffic. 2015;16:787–795. doi: 10.1111/tra.12281. [DOI] [PubMed] [Google Scholar]
- 31.Resch W, Weisberg AS, Moss B. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J Virol. 2005;79:6598–6609. doi: 10.1128/JVI.79.11.6598-6609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, et al. Vaccinia virus virion membrane biogenesis protein A11 associates with viral membranes in a manner that requires the expression of another membrane biogenesis protein, A6. J Virol. 2012;86:11276–11286. doi: 10.1128/JVI.01502-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maruri-Avidal L, Weisberg AS, Moss B. Association of the vaccinia virus A11 protein with the endoplasmic reticulum and crescent precursors of immature virions. J Virol. 2013;87:10195–10206. doi: 10.1128/JVI.01601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satheshkumar PS, Weisberg A, Moss B. Vaccinia virus H7 protein contributes to the formation of crescent membrane precursors of immature virions. J Virol. 2009;83:8439–8450. doi: 10.1128/JVI.00877-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng X, Wu X, Yan B, Deng J, Xiang Y. Analysis of the role of vaccinia virus H7 in virion membrane biogenesis with an H7-deletion mutant. J Virol. 2013;87:8247–8253. doi: 10.1128/JVI.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolli S, et al. Structure-function analysis of vaccinia virus H7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication. J Virol. 2015;89:2209–2219. doi: 10.1128/JVI.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng X, et al. Vaccinia virus A6 is essential for virion membrane biogenesis and localization of virion membrane proteins to sites of virion assembly. J Virol. 2012;86:5603–5613. doi: 10.1128/JVI.00330-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng X, Rose L, Han Y, Deng J, Xiang Y. Vaccinia virus A6 is a two-domain protein requiring a cognate N-terminal domain for full viral membrane assembly activity. J Virol. 2017;91:e02405-16. doi: 10.1128/JVI.02405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruri-Avidal L, Weisberg AS, Moss B. Direct formation of vaccinia virus membranes from the endoplasmic reticulum in the absence of the newly characterized L2-interacting protein A30.5. J Virol. 2013;87:12313–12326. doi: 10.1128/JVI.02137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suarez C, Hoppe S, Pénard E, Walther P, Krijnse-Locker J. Vaccinia virus A11 is required for membrane rupture and viral membrane assembly. Cell Microbiol. 2017;19:e12756. doi: 10.1111/cmi.12756. [DOI] [PubMed] [Google Scholar]
- 41.Szajner P, Weisberg AS, Wolffe EJ, Moss B. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J Virol. 2001;75:5752–5761. doi: 10.1128/JVI.75.13.5752-5761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maruri-Avidal L, Weisberg AS, Moss B. Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins. J Virol. 2011;85:12431–12441. doi: 10.1128/JVI.05573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bisht H, Weisberg AS, Szajner P, Moss B. Assembly and disassembly of the capsid-like external scaffold of immature virions during vaccinia virus morphogenesis. J Virol. 2009;83:9140–9150. doi: 10.1128/JVI.00875-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pendin D, McNew JA, Daga A. Balancing ER dynamics: Shaping, bending, severing, and mending membranes. Curr Opin Cell Biol. 2011;23:435–442. doi: 10.1016/j.ceb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Novick P, Ferro-Novick S. ER structure and function. Curr Opin Cell Biol. 2013;25:428–433. doi: 10.1016/j.ceb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 48.van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5:a011072. doi: 10.1101/cshperspect.a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Earl PL, Moss B, Wyatt LS, Carroll MW. 2001. Generation of recombinant vaccinia viruses. Curr Protoc Mol Biol Chap 16, Unit 16.17.1–16.17.19.
- 50.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Senkevich TG, Wyatt LS, Weisberg AS, Koonin EV, Moss B. A conserved poxvirus NlpC/P60 superfamily protein contributes to vaccinia virus virulence in mice but not to replication in cell culture. Virology. 2008;374:506–514. doi: 10.1016/j.virol.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.