Significance
MOR1, the main target of endogenous opioid peptides as well as morphine and other opiate derivatives, is a principal regulator of analgesia. TRPV1, the receptor for capsaicin, is increasingly recognized as a major mediator of mechanical, chemical, and thermal pain sensation. Here we demonstrate that these two pain systems, heretofore thought to operate independently, interact physically and physiologically. TRPV1 activation modulates MOR1 agonist-induced receptor phosphorylation via GRK5 and prevents MOR1 internalization from the cell membrane. TRPV1-mediated regulation of MOR1 thus affords a promising avenue to improve the therapeutic profile of opioid medications.
Keywords: μ-opioid receptor, transient receptor potential vanilloid 1, G protein-coupled receptor kinase 5, opiates, G protein-coupled receptors
Abstract
Opioids are powerful analgesics, but also carry significant side effects and abuse potential. Here we describe a modulator of the μ-opioid receptor (MOR1), the transient receptor potential channel subfamily vanilloid member 1 (TRPV1). We show that TRPV1 binds MOR1 and blocks opioid-dependent phosphorylation of MOR1 while leaving G protein signaling intact. Phosphorylation of MOR1 initiates recruitment and activation of the β-arrestin pathway, which is responsible for numerous opioid-induced adverse effects, including the development of tolerance and respiratory depression. Phosphorylation stands in contrast to G protein signaling, which is responsible for the analgesic effect of opioids. Calcium influx through TRPV1 causes a calcium/calmodulin-dependent translocation of G protein-coupled receptor kinase 5 (GRK5) away from the plasma membrane, thereby blocking its ability to phosphorylate MOR1. Using TRPV1 to block phosphorylation of MOR1 without affecting G protein signaling is a potential strategy to improve the therapeutic profile of opioids.
Opiates are both highly effective analgesics and a significant source of public health concern due to their rewarding and addictive properties coupled with severe, potentially lethal side effects. Opiates signal via MOR1, a member of the G protein-coupled receptor superfamily (GPCRs) (1, 2). GPCRs are a large group of proteins that mediate diverse physiological and pharmacologic events (3). Like other GPCRs, MOR1 can transduce two parallel and dissociable signaling cascades, namely G protein signaling and β-arrestin–mediated signaling (4). MOR1 G protein signaling is responsible for analgesia via MOR1 coupling to G αi subunits, which suppresses cyclic adenosine monophosphate (cAMP) production (2, 5). When metabolism or opiate antagonists reverse sustained G αi signaling, cAMP levels increase above baseline, precipitating opiate withdrawal (6).
Studies in β-arrestin-2 knockout mice have demonstrated that β-arrestin signaling causes several major adverse effects of opiates, notably respiratory depression, constipation, and tolerance (7, 8), and also inhibits G protein signaling-mediated analgesia (5). The genesis of β-arrestin–mediated signaling involves phosphorylation of MOR1 by G protein-coupled receptor kinases (GRKs) at C-terminal amino acid residues. Phosphorylation recruits β-arrestin-2 to the membrane and induces MOR1 internalization and desensitization (9). MOR1 is phosphorylated at human serine 377/mouse serine 375 (S377/S375) after opioid treatment by GRK5, a calcium/calmodulin (Ca2+/CaM)-binding GRK family member (10–15). Mice with a phospho-null mutation of serine to alanine at S375 (S375A) exhibit enhanced opiate analgesia and decreased tolerance with chronic administration of etonitazene, a high-affinity MOR1 agonist, and [d-Ala, N-MePhe, Gly-ol]-enkephalin (DAMGO), an enkephalin analog (15). Taken together, these findings imply that blocking MOR1 phosphorylation could improve the therapeutic profile of opiates.
The discovery of transient receptor potential (TRP) channels as mediators of pain signaling has been an important advancement in the field of pain biology. TRPV1 is a nociceptor activated by noxious heat, protons, and capsaicin, the pungent component of chili peppers. TRPV1 is a nonselective cation channel, but predominantly fluxes calcium ions and is widely expressed in small-diameter C-fiber neurons (16–18). TRPV1 is often coexpressed with MOR1 in peripheral nervous tissues, including dorsal root ganglion cells (DRGs), and is expressed at low levels throughout the brain (2, 19–21).
Despite the well-established roles of both TRPV1 and MOR1 in pain perception and regulation, there has been limited investigation into functional cross-talk between these two signaling systems. In the present study, we investigated whether TRPV1 is a physiological regulator of MOR1. We show that activation of TRPV1 regulates MOR1 function via GRK5. Activating TRPV1 triggers translocation of GRK5 to the nucleus, thereby blocking MOR1 phosphorylation, receptor internalization, and β-arrestin signaling without affecting G protein activation by MOR1. Thus, TRPV1 activation can bias MOR1 signaling, inhibiting β-arrestin signaling while preserving G protein signaling, suggesting a novel therapeutic approach to improving opiate pharmacology.
Results
TRPV1 Binds MOR1 and Regulates Receptor Phosphorylation, but Not G Protein Signaling.
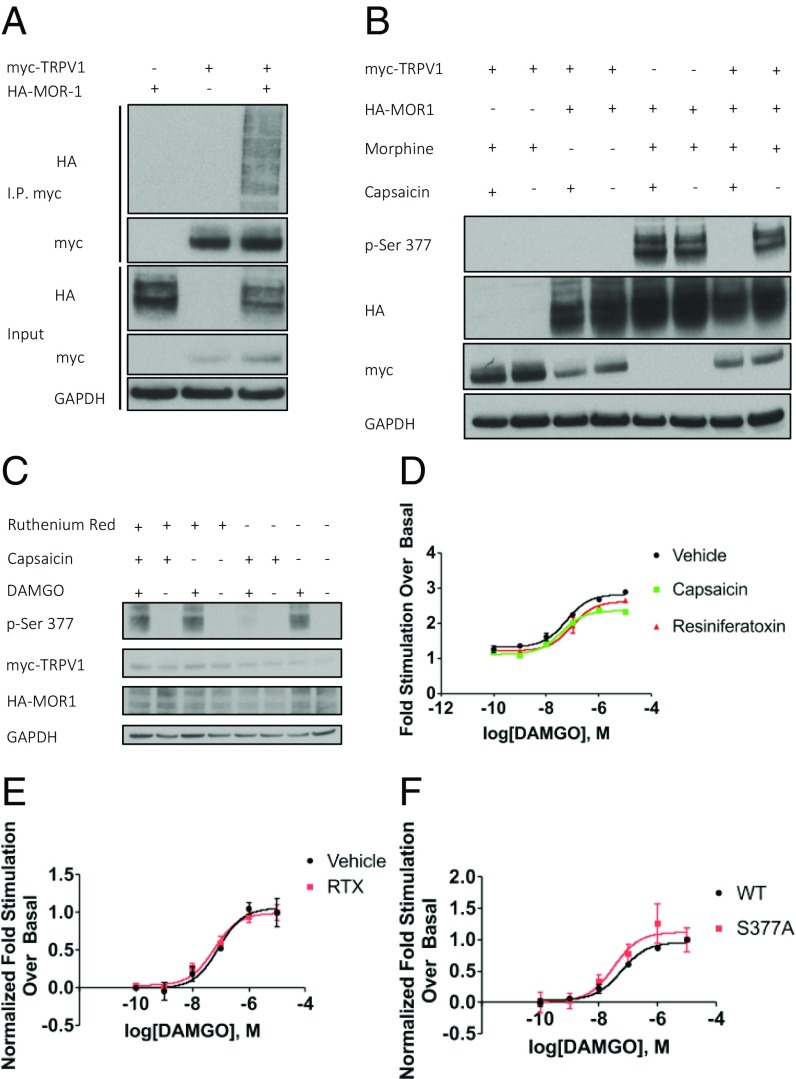
Given the importance of TRPV1 and MOR1 as pain regulators and their coexpression in several regions of the nervous system, we examined whether these two proteins might physically interact. We transiently overexpressed human myc-TRPV1 and HA-MOR1 in HEK293 cells and performed coimmunoprecipitation assays. We observed a robust interaction between TRPV1 and MOR1 (Fig. 1A). MOR1 is well known to be phosphorylated after agonist treatment (15). To determine the functional consequence of the MOR1–TRPV1 interaction, we treated transfected HEK293 cells with capsaicin (1 μM) for 5 min, followed by morphine (10 μM) for 15 min, and subsequently monitored MOR1 phosphorylation with an antibody selective for MOR1, phosphoserine-377 (p-Ser-377) (Fig. 1B). Phosphorylation of MOR1 is abolished by pretreatment with capsaicin (1 μM), but only in cells coexpressing TRPV1. Alternative agonists of MOR1 and TRPV1, DAMGO and resiniferatoxin (RTX), respectively, have the same effect on MOR1 phosphorylation as morphine and capsaicin (Fig. S1A). Furthermore, capsaicin blocks MOR1 phosphorylation induced by morphine or DAMGO equally effectively (Fig. S1B). MOR1 phosphorylation is rescued in the presence of capsaicin by pretreatment with ruthenium red (10 μM), a TRPV1 pore-blocker (16) (Fig. 1C). We performed a time-course analysis to determine whether TRPV1 simply delayed MOR1 phosphorylation, but found instead that capsaicin blocks phosphorylation for up to 60 min of DAMGO exposure (Fig. S1C).
Fig. 1.
TRPV1 binds MOR1 and inhibits MOR1 phosphorylation, but not G protein activation. (A) TRPV1 and MOR1 coimmunoprecipitate in transiently transfected HEK293 cells. (B) Capsaicin (1 µM)-mediated activation of TRPV1 blocks morphine (10 µM)-induced phosphorylation of MOR1 at S377. (C) Ruthenium red (10 µM) pretreatment blocks the effect of capsaicin (1 µM), rescuing MOR1 phosphorylation. (D and E) [35S] GTPγS binding assay of membranes from HEK293 cells expressing MOR1 and TRPV1. (D) Effect of RTX (1 µM) or capsaicin (1 µM) on DAMGO-induced [35S] GTPγS binding to treatment naive crude membrane fraction. (E) Effect of RTX (1 µM) treatment on living cells before membrane fraction isolation on DAMGO-induced [35S] GTPγS incorporation. (F) DAMGO-induced [35S] GTPγS incorporation into membrane preparations containing WT MOR1 and MOR1-S377A. Data for all GTPγS assays are mean ± SEM (n = 3–4).
We wondered whether TRPV1 influences opioid-induced G protein signaling as well as MOR1 phosphorylation. We performed [35S] GTPγS-binding assays to monitor the exchange of GDP for GTP, the first step in the G protein signaling pathway. Crude membrane preparations containing both MOR1 and TRPV1, isolated from HEK293 cells, were incubated with capsaicin, RTX, or vehicle before treatment with a range of DAMGO concentrations. We found that maximal DAMGO stimulation enhances GTP binding by twofold to threefold, and that neither capsaicin nor RTX significantly alters this response (Fig. 1D). Similar results are obtained when RTX is applied to living cells before lysis and membrane isolation (Fig. 1E). Finally, mutation of MOR1 serine 377 to alanine (S377A) does not alter DAMGO-stimulated GTP binding (Fig. 1F). This confirms that phosphorylation at S377 is not required for G protein signaling. These findings demonstrate that TRPV1 binds MOR1 and regulates agonist-dependent MOR1 phosphorylation, but not G protein signaling.
Calcium Influx Through TRPV1 Channels Inhibits MOR1 Phosphorylation and Internalization.
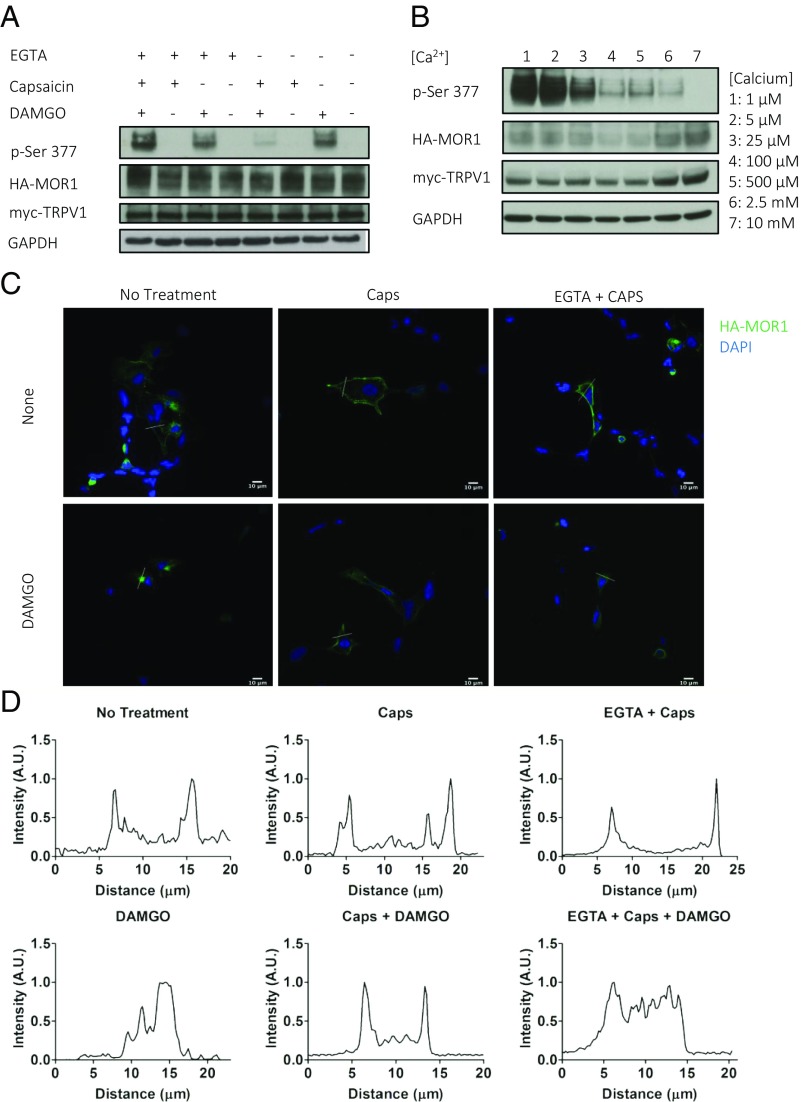
We wanted to characterize the mechanism by which TRPV1 inhibits MOR1 phosphorylation and explore its downstream effects. TRPV1 predominantly conducts calcium (16), thus both depolarizing the cell and increasing intracellular calcium concentrations. Applying ethylene glycol-bis (β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) (5 mM) to cell culture media for 5 min before addition of capsaicin allowed us to remove effectively all extracellular calcium from the media. EGTA completely rescues MOR1 phosphorylation (Fig. 2A). We next monitored MOR1 phosphorylation following DAMGO (1 μM) and capsaicin (1 μM) treatment with increasing concentrations of extracellular calcium, and found that MOR1 phosphorylation decreases in a concentration-dependent manner with increasing calcium concentration (Fig. 2B). Using potassium chloride to depolarize cells (22), we found that depolarization, in either the presence or absence of TRPV1, has no significant effect on MOR1 phosphorylation (Fig. S2A). Taken together, these results suggest that TRPV1 exerts a long-lasting, calcium-dependent inhibition of agonist-induced MOR1 phosphorylation.
Fig. 2.
Calcium influx through TRPV1 blocks MOR1 phosphorylation and internalization. (A and B) Western blots of HEK293 cells coexpressing TRPV1 and MOR1. (A) EGTA (5 mM) chelation of extracellular calcium blocks capsaicin (1 µM) inhibition of MOR1 phosphorylation. (B) Decreasing extracellular calcium from 10 mM to 1 µM rescues MOR1 phosphorylation even with 5 min of TRPV1 activation by capsaicin (1 µM). (C and D) Immunofluorescence of transiently transfected HEK293 cells. (C) Staining of HA-MOR1 in cells coexpressing HA-MOR1-WT and myc-TRPV1 treated with combinations of EGTA (5 mM) for 5 min, capsaicin (1 µM) for 5 min, and DAMGO (1 µM) for 15 min, as indicated. (D) Line plot profile of green intensity (MOR1) along a white line drawn to transect both membranes of cell from associated treatment groups in C. Green, HA-MOR1; blue, DAPI. (Scale bars: 10 µm.)
MOR1 phosphorylation, particularly at S377, is required for receptor internalization (9). Confocal immunofluorescence (IF) microscopy of HEK293 cells transfected with HA-MOR1-WT reveals internalization of MOR1 in response to DAMGO treatment, but not in cells transfected with phosphorylation site mutant HA-MOR1-S377A (Fig. S2B). We performed IF microscopy on cells transiently overexpressing TRPV1 and MOR1 to determine whether TRPV1 activation influences MOR1 localization in addition to phosphorylation. We found that 5 min of capsaicin (1 µM) pretreatment blocks MOR1 internalization elicited by 15 min of DAMGO (1 µM), and that EGTA treatment rescues MOR1 phosphorylation and permits MOR1 receptor internalization (Fig. 2C). We generated line plot profiles of MOR1 staining intensity of cells to more quantitatively analyze MOR1 localization. MOR1 localization at the plasma membrane appears as two distinct peaks, while internalized MOR1 appears as broad plateau or a broad series of peaks (Fig. 2D). Taken together, these data demonstrate that TRPV1 regulates not just MOR1 phosphorylation, but also MOR1 internalization, in a calcium-dependent manner via S377.
TRPV1 Activation Inhibits GRK5 and Prevents MOR1 Phosphorylation by Removing GRK5 from the Cell Membrane.
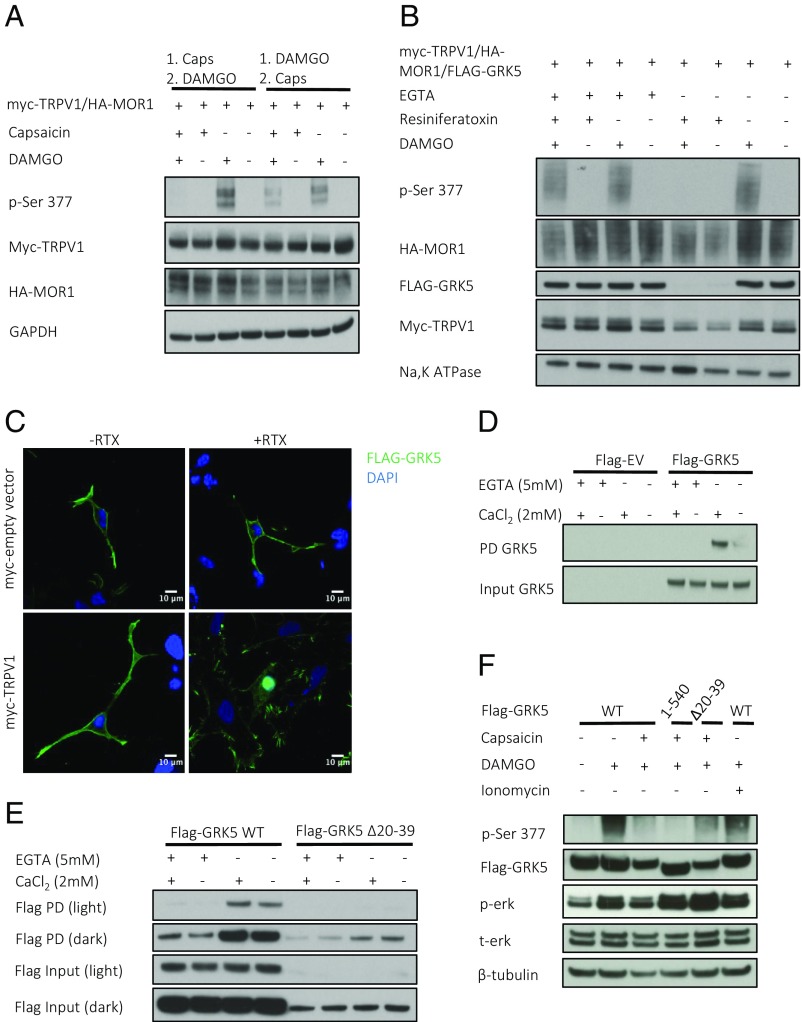
Given that TRPV1 activation inhibits phosphorylation of S377, TRPV1 likely inhibits a particular kinase or activates a phosphatase. If capsaicin treatment activates a phosphatase, then presumably the order of treatment with capsaicin and DAMGO should not affect the final phosphorylation state of MOR1. Accordingly, we assessed whether phosphorylation of S377 was affected by capsaicin (1 µM) treatment before or after a 15-min exposure to DAMGO (1 µM) (Fig. 3A). As in previous experiments, when capsaicin is applied before DAMGO, MOR1 phosphorylation is abolished. Application of DAMGO before capsaicin results in the normal phosphorylation of S377. These findings are consistent with capsaicin inhibiting a kinase responsible for MOR1 phosphorylation. The membrane-associated kinase GRK5 is known to phosphorylate opioid receptors and is the primary kinase for S377 phosphorylation (10, 11, 15). We transiently overexpressed myc-TRPV1, HA-MOR1, and FLAG-GRK5 in HEK293 cells, and combinatorially treated cells with RTX (1 µM), DAMGO (1 µM), and EGTA (5 mM). We then fractionated cells and performed Western blot analysis on the isolated membranes (Fig. 3B). Under basal conditions, GRK5 is abundant in the membrane fraction, while TRPV1 activation by RTX removes it from the membrane. RTX has no effect when extracellular calcium is removed by EGTA (5 mM). We examined the membrane localization of other extraretinal GRKs and found that GRK2, GRK3, and GRK6 are not affected by TRPV1-mediated calcium influx, while GRK4 is partially removed from the cell membrane following TRPV1 activation (Fig. S3A). Confocal IF microscopy of cells coexpressing FLAG-GRK5 and myc-TRPV1 reveal GRK5 translocation from the membrane to the nucleus following RTX treatment (Fig. 3C). These data demonstrate that TRPV1 inhibits MOR1 phosphorylation by removing GRK5 from the membrane in a calcium-dependent manner.
Fig. 3.
TRPV1 activation induces nuclear translocation of GRK5, the Ca2+/CaM-binding kinase responsible for MOR1 phosphorylation. (A) Western blot of HEK293 cells coexpressing HA-MOR1 and myc-TRPV1 treated combinatorially with capsaicin (1 µM) for 5 min before and after 15 min of DAMGO. (B) Western blot of membrane fractions collected from HEK293 cells coexpressing myc-TRPV1, HA-MOR1-WT, and FLAG-GRK5. Before membrane isolation, cells were treated combinatorially with RTX (1 µM) for 5 min, followed by DAMGO (1 µM) with or without EGTA (5 mM) for 15 min. (C) Immunofluorescence of HEK293 cells coexpressing myc-empty vector or myc-TRPV1 with FLAG-GRK5 with or without RTX (1 µM) for 5 min. (D and E) Calmodulin-Sepharose pulldown with lysate from HEK293 cells containing transiently overexpressed FLAG-GRK5 WT or FLAG-EV (D) or truncated FLAG-GRK5 Δ20–39 (E) in the presence (2 mM CaCl2) or absence (5 mM EGTA) of calcium. (F) Western blot of HEK293 cells coexpressing myc-TRPV1, HA-MOR1-WT, and FLAG-GRK5 (WT, 1–540, Δ20–39). Cells treated with capsaicin or ionomycin (1 µM) for 5 min, followed by DAMGO (1 µM) for 15 min.
Ca2+/CaM binds GRK5 and inhibits its enzymatic function (13, 14, 23). Using lysates from HEK293 cells transiently overexpressing FLAG-GRK5, we confirmed robust calcium-dependent binding of GRK5 to calmodulin-conjugated Sepharose beads (Fig. 3D). GRK5 possesses 590 amino acids and binds the plasma membrane at both the N and C termini (13, 14). There are two predicted calmodulin binding domains, one at the N terminus (aa 20–39) and one at the C terminus (aa 541–590). GRK5 with residues 20–39 deleted (GRK5 Δ20–39) (Fig. 3E) and truncated GRK5 1–540 (Fig. S3B) both display reduced calcium-dependent binding to calmodulin compared with WT GRK5. We hypothesized that a Ca2+/CaM-insensitive GRK5 might rescue MOR1 phosphorylation in the presence of capsaicin. When transiently overexpressed, GRK5 Δ20–39, but not GRK5 1–540, partially rescues the phosphorylation of MOR1 following treatment with capsaicin (1 µM) and DAMGO (1 µM) (Fig. 3F). Treatment of cells with ionomycin (1 µM), a calcium ionophore, instead of capsaicin does not inhibit DAMGO-induced phosphorylation of MOR1 (Fig. 3F). In summary, these results show that TRPV1 activation prevents GRK5 from phosphorylating MOR1 by inducing its translocation from the cellular membrane to the nucleus. This process depends on calmodulin binding to the N terminus of GRK5.
Discussion
The principal finding of this study is the striking interaction of TRPV1 with MOR1 and its impact on MOR1 phosphorylation and internalization. MOR1 signaling can be dichotomized into influences on GTP binding contrasted with β-arrestin–dependent effects. We demonstrate that TRPV1 activation has the potential to mitigate opioid side effects by inhibiting MOR1 phosphorylation while leaving intact the analgesic mechanism, G protein signaling (Fig. 4). The TRPV1 interactions do not appear to alter MOR1-dependent GTP binding, but do decrease MOR1 phosphorylation and internalization (Fig. 4D), presumably reflecting down-regulation of the β-arrestin pathway. Conceivably, a two-drug combination to activate both TRPV1 and MOR1 has the potential to improve the pharmacologic profile of existing opiates.
Fig. 4.
TRPV1-mediated calcium influx regulates MOR1 phosphorylation via GRK5. (A) Basal conditions. (B) Opioids alone activate both G protein signaling and induce MOR1 phosphorylation. MOR1 phosphorylation subsequently initiates β-arrestin recruitment, leading to MOR1 internalization. (C) Calcium influx after TRPV1 activation causes Ca2+/CaM binding of GRK5 and translocation to the nucleus. (D) TRPV1 activation before opioid treatment blocks MOR1 phosphorylation. This prevents β-arrestin from internalizing MOR1, but allows G protein signaling.
Physical and/or physiological interactions of GPCRs with TRP channels, including TRPV4 with the angiotensin II receptor (24) and TRPM8 with MOR1 (25), have been reported previously. Previous studies have emphasized the influence of GPCR activation on TRP channel activity (26), degradation (24), and/or localization (25). We find that regulation operates in both directions, with TRPV1 capable of influencing the signaling and localization of MOR1. Using TRPV1 agonists to modulate MOR1 for therapeutic purposes would require the cell-autonomous coexpression of TRPV1. TRPV1 is widely expressed throughout the nervous system, although likely at low levels (20), and has been reported in DRGs, the principal pain relay for the peripheral nervous system. MOR1 is coexpressed in a subset of TRPV1-positive neurons (27).
Diminished MOR1 phosphorylation appears to reflect dissociation of GRK5 from the membrane and its translocation to the nucleus via Ca2+/CaM. Regulation of GRKs by Ca2+/CaM was first demonstrated in vitro (13). The authors found that the ability of purified GRK5 and GRK6 to phosphorylate rhodopsin was inhibited by Ca2+/calmodulin, whereas the effects on GRK1 and GRK2 were minimal. Subsequent work showed that while GRK5 has calmodulin binding domains at both the N and C termini, the C-terminal binding domain is a major regulator of GRK5 kinase activity (14, 23). However, other investigators have shown that the binding of Ca2+/CaM to the N-terminal binding site is necessary for interrupting the interaction of GRK5 with actin (28) and its nuclear translocation (29). In our hands, overexpression of the C-terminal binding domain mutant GRK5 1–540 does not rescue phosphorylation of MOR1, suggesting that Ca2+/CaM does not inhibit the kinase function of GRK5. Overexpression of the N-terminal Ca2+/CaM binding domain mutant partially rescues MOR1 phosphorylation, which agrees with our hypothesis that nuclear translocation, not loss of kinase activity, is most likely the cause of reduced MOR1 phosphorylation.
Our finding that TRPV1 activation can rapidly translocate GRK5 from the plasma membrane has several implications. Previous work on GRK5 nuclear translocation showed that it occurs downstream of G αq signaling and IP3 receptor activation (29, 30). Our results imply that TRPV1 activation involves a different pathway through which calcium may enter the cell, with signaling consequences similar to those of IP3 receptor activation. Not all pathways of calcium entry affect GRK5 equally. For example, in HEp2 cells, GRK5 is normally present in the nucleus and can undergo nuclear export in the presence of ionomycin (23). In our hands, GRK5 is not expressed at high levels in the nucleus at baseline in HEK293 cells, and ionomycin treatment does not block MOR1 phosphorylation. These findings suggest that cell-specific dynamics determine which sources of calcium and which stimuli control GRK5 localization and activity.
In summary, we have demonstrated an interaction among TRPV1, MOR1, and GRK5. Calcium influx through TRPV1 leads to the nuclear translocation of GRK5, which blocks its ability to phosphorylate MOR1. This interaction leaves the G protein-mediated analgesic signaling of MOR1 intact, but inhibits β-arrestin–mediated internalization and desensitization of MOR1. Thus, TRPV1 agonists may have therapeutic potential as regulators of tolerance, respiratory depression, and gastrointestinal side effects of opiates.
Methods
Molecular Biology.
Rat TRPV1 cDNA (a generous gift from Michael Caterina, Johns Hopkins University School of Medicine, Baltimore), human MOR-1 cDNA (a generous gift from Gavril W. Pasternak, Memorial Sloan-Kettering Cancer Center, New York), and GRKs 2–6 (generous gifts from Robert Lefkowitz, Duke University, Durham, NC) were cloned into pCMV-HA (Clontech), pCMV-myc (Clontech), or p3XFLAG-CMV-7 (Sigma-Aldrich) as indicated above using standard methods. Site-directed mutagenesis was performed in accordance with standard methods similar to the Stratagene QuikChange method.
Cell Culture/Transfection Conditions.
HEK293 cells (American Type Culture Collection) were cultured in DMEM supplemented with l-glutamate (Invitrogen), 10% FBS (Gemini Bioproducts), and 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen) at 37 °C with 5% CO. Transient transfections were carried out using Polyfect (Qiagen), following the provider’s protocols for HEK293 cells grown to 65–85% confluency. For drug treatment experiments, cells were transfected at 24 h before drug exposure. Drugs were dissolved in cell culture media at doses indicated above for the indicated times.
Immunoblotting/Immunoprecipitation/Pulldowns.
Cells were lysed in lysis buffer (20 mM Tris⋅HCl pH 7.4, 140 mM NaCl, and 1% IPEGAL 630) supplemented with complete Protease Inhibitor Mixture (Roche) and PhosSTOP Phosphatase Inhibitor Tablets (Roche) on ice. Lysate protein content was determined using Bradford reagent (Sigma-Aldrich). Lysates were boiled in 1× NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen) and subjected to SDS/PAGE using the Novex system (Invitrogen) following the manufacturer’s instructions, transferred to PVDF membranes (EMD Millipore), blocked in 3% BSA (Sigma-Aldrich) in Tris-buffered saline/0.1% Tween-20 for 1 h at room temperature, and then incubated with primary antibodies overnight at 4 °C. In figures containing multiple Western blot panels, triplicate gels were run containing equivalent samples. Each protein of interest was therefore probed separately, with no stripping or reprobing.
The following antibodies were used: GAPDH 6C5 (CB1001, 1:20,000; Calbiochem), Na+/K+ ATPase (a6f, 1:1,000; Developmental Studies Hybridoma Bank), β-tubulin (2146S, 1:1,000; Cell Signaling Technology), FLAG M2 (F3165, 1:20,000; Sigma-Aldrich), phospho-µ-opioid receptor (Ser375) (34515, 1:2,000; Cell Signaling Technology), HA (11867423001, 1:5,000; Roche), myc 9E10 (11667203001, 1:10,000; Roche), HA-HRP (ab1190, 1:5,000; Abcam), Amersham ECL mouse IgG HRP-linked (NA931, 1:10,000; GE Healthcare Life Sciences), and Amersham ECL rabbit IgG HRP-linked (NA934, 1:10,000; GE Healthcare Life Sciences). Coimmunoprecipitation was performed according to the manufacturer’s instructions using EZview Red c-Myc agarose (Sigma-Aldrich). Calmodulin pulldowns were performed using Calmodulin Sepharose 4B (GE Healthcare Life Sciences) according to the manufacturer’s instructions. β-tubulin, GAPDH, or sodium potassium ATPase loading controls were performed in all cases. All Western blot analyses were performed at least in triplicate, with equal loading confirmed for each gel.
Confocal Immunofluorescence.
HEK293 cells were seeded onto 35-mm cell culture dishes with coverslips attached (MatTek). Dishes were coated with 0.01% poly-l-ornithine solution (Sigma-Aldrich) for 2 h at 37 °C and then rinsed twice with warm PBS before cell plating. HEK293 cells were transfected using lipid transfection reagent as indicated above, but with 75–90% of the DNA transfected as empty vector plasmids (e.g., FLAG-Empty, myc-Empty) to reduce signal intensity and ensure that overexpression did not result in localization artifacts. Cells were then fixed with 4% paraformaldehyde in 1× PBS for 15 min, followed by −20 °C methanol for 7 min, then blocked for 30 min (1% BSA/PBS), washed, incubated in primary antibody in 1× PBS/0.5% BSA/0.05% Tween-20 for 1 h at room temperature, washed three times in 1× PBS/0.5% BSA/0.05% Tween-20, and finally incubated in secondary antibody for 1 h at room temperature. Cells were treated with Hoechst 33342 at 2 µg/mL (Thermo Fisher Scientific) for 15 min before a final three washes in 1× PBS/0.5% BSA/0.05% Tween-20. Antibodies used were FLAG M2 (F3165, 1:200; Sigma-Aldrich), HA (11867423001, 1:200; Roche), goat anti-rat IgG Alexa Fluor 488 (A-11006, 1:200; Molecular Probes), goat anti-mouse IgG Alexa Fluor 568 (A-11004, 1:200; Molecular Probes), and goat anti-rabbit IgG Alexa Fluor 488 (A-11008, 1:200; Molecular Probes). Line plot profiles were constructed in Fiji (31), and the list was exported to GraphPad Prism for chart construction. Lines were drawn to transect two membranes of each cell without passing through the nucleus. Intensity was normalized for each image individually. Coverslips were imaged using a Zeiss AxioObserver with a 780-quasar confocal module and FCS or a Zeiss LSM 780 confocal microscope with a 40× LD-LCI C-Apochromat objective (32). The Zeiss microscope used ZEN imaging software. Brightness and contrast were adjusted across the entire image using Imaris (Bitplane) or Fiji to maximize image clarity.
Membrane Fractionation/35S GTPγS Incorporation.
Membrane fractionation and [35S] GTPγS (Perkin-Elmer) incorporation were performed in accordance with our previously published protocol (33). In brief, a crude membrane fraction (34) was isolated as follows. Cells were pelleted and snap-frozen in Buffer 1 (10 mM Hepes pH 7.4, 1 mM EGTA, 1 mM DTT, 10% sucrose, and protease and phosphatase inhibitors; Roche) and then centrifuged at 1,000 × g for 10 min. The supernatants were saved, and the pellet was rehomogenized in Buffer 1 and then centrifuged at 1,000 × g for 10 min at 4 °C. The supernatants were saved and combined, then centrifuged at 11,000 × g for 20 min at 4 °C. The pellet was saved, resuspended in Buffer 2 (10 mM Hepes pH 7.4, 1 mM EGTA, 1 mM DTT, and 1 mM MgCl2), centrifuged at 21,000 × g for 20 min at 4 °C, resuspended in Buffer 2, diluted to 1 mg/mL, and used for [35S] GTPγS incorporation, as described previously (35). For this, 10 μg of membrane protein was incubated in binding buffer (50 mM Hepes pH 7.4, 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% wt/vol BSA, 10 μM GDP, and 0.1 nM [35S] GTPγS) and the appropriate ligand for 2 h at 4 °C with rotation. Background binding was determined using 100 μM unlabeled GTPγS. Equilibration was terminated by rapid filtration on GF/B filters (GE Healthcare Lifesciences) and three washes with wash buffer (50 mM Tris⋅HCl pH 7.4, 5 mM MgCl2, and 50 mM NaCl). Radioactivity bound to the filter was quantified by scintigraphy. Data analysis and graphical presentation were performed using GraphPad Prism. All [35S] GTPγS assays were performed in triplicate or quadruplicate.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717005114/-/DCSupplemental.
References
- 1.Pert CB, Snyder SH. Opiate receptor: Demonstration in nervous tissue. Science. 1973;179:1011–1014. doi: 10.1126/science.179.4077.1011. [DOI] [PubMed] [Google Scholar]
- 2.Pasternak GW, Pan YX. Mu opioids and their receptors: Evolution of a concept. Pharmacol Rev. 2013;65:1257–1317. doi: 10.1124/pr.112.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raehal KM, Schmid CL, Groer CE, Bohn LM. Functional selectivity at the μ-opioid receptor: Implications for understanding opioid analgesia and tolerance. Pharmacol Rev. 2011;63:1001–1019. doi: 10.1124/pr.111.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of β-arrestin- and G protein-biased agonists. Trends Mol Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohn LM, et al. Enhanced morphine analgesia in mice lacking β-arrestin 2. Science. 1999;286:2495–2498. doi: 10.1126/science.286.5449.2495. [DOI] [PubMed] [Google Scholar]
- 6.Matthes HW, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 7.Bohn LM, Gainetdinov RR, Lin FT, Lefkowitz RJ, Caron MG. Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature. 2000;408:720–723. doi: 10.1038/35047086. [DOI] [PubMed] [Google Scholar]
- 8.Raehal KM, Walker JK, Bohn LM. Morphine side effects in beta-arrestin 2 knockout mice. J Pharmacol Exp Ther. 2005;314:1195–1201. doi: 10.1124/jpet.105.087254. [DOI] [PubMed] [Google Scholar]
- 9.Halls ML, et al. Plasma membrane localization of the μ-opioid receptor controls spatiotemporal signaling. Sci Signal. 2016;9:ra16. doi: 10.1126/scisignal.aac9177. [DOI] [PubMed] [Google Scholar]
- 10.Doll C, et al. Deciphering µ-opioid receptor phosphorylation and dephosphorylation in HEK293 cells. Br J Pharmacol. 2012;167:1259–1270. doi: 10.1111/j.1476-5381.2012.02080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glück L, et al. Loss of morphine reward and dependence in mice lacking G protein-coupled receptor kinase 5. Biol Psychiatry. 2014;76:767–774. doi: 10.1016/j.biopsych.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Premont RT, Koch WJ, Inglese J, Lefkowitz RJ. Identification, purification, and characterization of GRK5, a member of the family of G protein-coupled receptor kinases. J Biol Chem. 1994;269:6832–6841. [PubMed] [Google Scholar]
- 13.Pronin AN, Satpaev DK, Slepak VZ, Benovic JL. Regulation of G protein-coupled receptor kinases by calmodulin and localization of the calmodulin binding domain. J Biol Chem. 1997;272:18273–18280. doi: 10.1074/jbc.272.29.18273. [DOI] [PubMed] [Google Scholar]
- 14.Pronin AN, Carman CV, Benovic JL. Structure-function analysis of G protein-coupled receptor kinase-5: Role of the carboxyl terminus in kinase regulation. J Biol Chem. 1998;273:31510–31518. doi: 10.1074/jbc.273.47.31510. [DOI] [PubMed] [Google Scholar]
- 15.Schulz S, et al. Morphine induces terminal micro-opioid receptor desensitization by sustained phosphorylation of serine-375. EMBO J. 2004;23:3282–3289. doi: 10.1038/sj.emboj.7600334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caterina MJ, et al. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 17.Tominaga M, et al. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 1998;21:531–543. doi: 10.1016/s0896-6273(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 18.Caterina MJ, et al. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- 19.Chen S-R, Pan H-L. Loss of TRPV1-expressing sensory neurons reduces spinal μ opioid receptors but paradoxically potentiates opioid analgesia. J Neurophysiol. 2006;95:3086–3096. doi: 10.1152/jn.01343.2005. [DOI] [PubMed] [Google Scholar]
- 20.Roberts JC, Davis JB, Benham CD. [3H]Resiniferatoxin autoradiography in the CNS of wild-type and TRPV1 null mice defines TRPV1 (VR-1) protein distribution. Brain Res. 2004;995:176–183. doi: 10.1016/j.brainres.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Cavanaugh DJ, et al. Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J Neurosci. 2011;31:5067–5077. doi: 10.1523/JNEUROSCI.6451-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu R, et al. Inositol polyphosphate multikinase is a transcriptional coactivator required for immediate early gene induction. Proc Natl Acad Sci USA. 2013;110:16181–16186. doi: 10.1073/pnas.1315551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson LR, Scott MG, Pitcher JA. G protein-coupled receptor kinase 5 contains a DNA-binding nuclear localization sequence. Mol Cell Biol. 2004;24:10169–10179. doi: 10.1128/MCB.24.23.10169-10179.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla AK, et al. Arresting a transient receptor potential (TRP) channel: Beta-arrestin 1 mediates ubiquitination and functional down-regulation of TRPV4. J Biol Chem. 2010;285:30115–30125. doi: 10.1074/jbc.M110.141549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapovalov G, et al. Opiates modulate thermosensation by internalizing cold receptor TRPM8. Cell Rep. 2013;4:504–515. doi: 10.1016/j.celrep.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Shen WL, et al. Function of rhodopsin in temperature discrimination in Drosophila. Science. 2011;331:1333–1336. doi: 10.1126/science.1198904. [DOI] [PubMed] [Google Scholar]
- 27.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 28.Freeman JL, De La Cruz EM, Pollard TD, Lefkowitz RJ, Pitcher JA. Regulation of G protein-coupled receptor kinase 5 (GRK5) by actin. J Biol Chem. 1998;273:20653–20657. doi: 10.1074/jbc.273.32.20653. [DOI] [PubMed] [Google Scholar]
- 29.Gold JI, et al. Nuclear translocation of cardiac G protein-coupled receptor kinase 5 downstream of select Gq-activating hypertrophic ligands is a calmodulin-dependent process. PLoS One. 2013;8:e57324. doi: 10.1371/journal.pone.0057324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martini JS, et al. Uncovering G protein-coupled receptor kinase-5 as a histone deacetylase kinase in the nucleus of cardiomyocytes. Proc Natl Acad Sci USA. 2008;105:12457–12462, and erratum (2008) 105:17206. doi: 10.1073/pnas.0803153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindelin J, et al. Fiji: An open source platform for biological image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewald AJ. Practical considerations for long-term time-lapse imaging of epithelial morphogenesis in three-dimensional organotypic cultures. Cold Spring Harb Protoc. 2013;2013:100–117. doi: 10.1101/pdb.top072884. [DOI] [PubMed] [Google Scholar]
- 33.Vasavda C, Zaccor NW, Scherer PC, Sumner CJ, Snyder SH. Measuring G-protein-coupled receptor signaling via radio-labeled GTP binding. J Vis Exp. 2017;124:e55561. doi: 10.3791/55561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salah-Uddin H, et al. Pharmacological assessment of m1 muscarinic acetylcholine receptor-gq/11 protein coupling in membranes prepared from postmortem human brain tissue. J Pharmacol Exp Ther. 2008;325:869–874. doi: 10.1124/jpet.108.137968. [DOI] [PubMed] [Google Scholar]
- 35.Brillet K, Kieffer BL, Massotte D. Enhanced spontaneous activity of the mu opioid receptor by cysteine mutations: Characterization of a tool for inverse agonist screening. BMC Pharmacol. 2003;3:14. doi: 10.1186/1471-2210-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.