Abstract
The present study aimed at identifying novel molecular cancer drug targets and biomarkers by analyzing the gene expression profiles of high-grade prostate cancer (PC), using a cDNA microarray combined with laser microbeam microdissection. A number of genes were identified that were transactivated in high-grade PC. First, a novel molecular target and diagnostic biomarker, shisa family member 2 (SHISA2), was identified as an overexpressed gene in high-grade PC cells. The reverse transcription-semi-quantitative polymerase chain reaction and immunohistochemical analysis validated the overexpression of SHISA2 (295 amino acids in length), specifically in high-grade PC cells with Gleason scores of between 8 and 10, relative to normal prostate epithelium. Knockdown of SHISA2 expression by short interfering RNA resulted in the marked suppression of PC cell viability. By contrast, exogenous SHISA2 expression in transfected cells promoted PC cell proliferation, indicating its oncogenic effects. Notably, as a result of cDNA microarray analysis, protein Wnt-5a (WNT5A) was focused upon and the expression of WNT5A was identified to be downregulated in SHISA2-knockdown. Western blot analysis validated significant downregulation of WNT5A by SHISA2-knockdown and upregulation of WNT5A by SHISA2 overexpression. The results of the present study indicated that SHISA2 may affect WNT5A synthesis. Furthermore, the secreted SHISA2 protein was determined in the culture medium of PC cells. We hypothesize that SHISA2 is involved in the regulation of WNT5A and in the aggressiveness of PC via the Wnt signaling pathway through WNT5A. Furthermore, SHISA2 may be a molecular target for cancer drugs, and a useful diagnostic biomarker for the prognosis and therapeutic effect in cancer.
Keywords: SHISA2, prostate cancer, protein Wnt-5a, cDNA microarray, laser microbeam microdissection
Introduction
Prostate cancer (PC) has the highest rate of malignancy in males and is recorded as the second leading cause of cancer-associated mortality in Europe and the USA (1). The incidence of PC has markedly increased in the majority of developed countries, which may be due to the prevalence of a Western lifestyle and the age distribution of the populations (1,2). The prostate-specific antigen (PSA) blood test is a widely used screening test for PC. However, the prevalence of PC is markedly higher than has been traditionally believed, and a proposed lowering of the PSA threshold for biopsies may result in the increased over diagnosis and over treatment of PC (3–5). Surgery and radio therapy are effective for the treatment of localized disease, but ~30% of treated patients with PC experience disease relapses (6–8). Therefore, it is desirable to find a biomarker to detect early cancer and to be able to reflect the prognosis for patients and the therapeutic effect.
Clinically high-grade PC with Gleason scores of between 8 and 10 exhibits rapid growth and is more likely to exhibit spread beyond the prostate (9). The majority of patients with advanced disease respond poorly to androgen deprivation therapy and typically acquire a castration-resistant phenotype (6,7,10). There are limited treatment options for these patients, including docetaxel combined with prednisone, which are able to provide a minimal effect (11,12). Castration-resistant PC progresses aggressively and, eventually, patients succumb. Therefore, the development of novel therapies, on the basis of the underlying molecular mechanisms of PC progression, is required.
We previously analyzed gene expression profiles of high-grade PC, using a cDNA microarray combined with laser microbeam microdissection, to enrich populations of cancer cells (13,14). The results of the microarray data are available from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (accession no. GSE6811 and GSE45016).
In the present study, shisa family member 2 (SHISA2) overexpression was identified in clinical high-grade PC with Gleason scores (GSs) between 8 and 10. In addition, SHISA2 was positively involved in cell proliferation and the progression of high-grade PC. The results of the present study may provide novel insights into the underlying molecular mechanisms of PC progression and provide insights to aid the development of novel therapeutic drugs and a potential diagnostic biomarker for PC.
Materials and methods
Patients and tissue samples
A total of 114 frozen or paraffin-embedded tissue samples were obtained from 114 male patients (mean age, 70 years; age range 57–85 years) with PC undergoing open prostatectomy or prostatic needle biopsy at Kochi Medical School Hospital (Nankoku, Japan) with appropriate written informed consent. Patient information was previously described by Satake et al (14). All of the surgical specimens were diagnosed and scored by a single pathologist using the Gleason Grading system as described previously (13,14), and GSs were between 6 and 10. High-grade PCs included in the surgical specimens of PCs were characterized and identified by criteria as previously described (14,15). All protocols were approved by the Ethical Committee of Kochi University.
Cell lines
The LNCaP, 22Rv1, DU145 and PC-3 cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). LNCaP is a human prostate carcinoma epithelial cell line derived from a left supraclavicular lymph node metastasis. 22Rv1 is a human prostate carcinoma epithelial cell line derived from a xenograft that was serially propagated in mice following castration-induced regression. DU145 is a human prostate carcinoma epithelial cell line derived from a brain metastasis. PC-3 is a human prostate carcinoma epithelial cell line derived from a bone metastasis. All cell lines were cultured between 3 and 10 passages, following purchase as monolayers, in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) and 1% antimycotic solution (Thermo Fisher Scientific, Inc.). Cells were maintained in incubators at 37°C in a humidified atmosphere containing 5% CO2.
Reverse transcription-semi-quantitative polymerase chain reaction (RT-sqPCR)
Purification of PC cells and normal prostatic epithelial cells from frozen PC tissues was described previously (13,14). Total RNA from tissues and PC cell lines was extracted using the RNeasy kit (Qiagen, Inc., Valencia, CA, USA) and RNase-Free DNase Set (Qiagen, Inc.), according to the manufacturer's protocol. Subsequently, RNA was reverse transcribed to single-stranded cDNA using an oligo d(T)12-18 primer and Super Script Reverse Transcriptase II (Invitrogen; Thermo Fisher Scientific, Inc.) at 42°C for 50 min. Appropriate dilutions of each single-stranded cDNA was prepared and the cDNA content was normalized to that of β-actin (ACTB). PCR was then performed using single-stranded cDNA as the PCR template. Primer sequences were as follows: SHISA2 forward, 5′-ATTGTTGGCTCCGTGTTTGT-3′ and reverse, 5′-ACTGTTGGTGTGAGGGAAGG-3′; and ACTB forward, 5′-TTGGCTTGACTCAGGATTTA-3′ and reverse, 5′-ATGCTATCACCTCCCCTGTG-3′. PCR conditions were as follows: Initial denaturation at 98°C for 10 sec, followed by a number of cycles (21 cycles for ACTB; 28 cycles for SHISA2) of denaturation at 98°C for 10 sec, annealing at 55°C for 5 sec and elongation at 72°C for 45 sec on a TGradient Thermocycler (Biometra GmbH, Göttingen, Germany). PCR products were analyzed by 2% agarose gel electrophoresis followed by ethidium bromide staining.
Generating SHISA2 antibody
The NH2-terminal peptide (CDNDRQQGAGEPGRA) and COOH-terminal peptide (HTNSEQKMYPAVTV) of the human SHISA2 protein were synthesized and used to immunize rabbits. Immune sera were purified on affinity columns packed with CNBr-activated Sepharose 4B (GE Healthcare, Chicago, IL, USA) conjugating each of the peptide antigens.
Immunohistochemical analysis
A total of 114 frozen or paraffin-embedded tissue samples were obtained from 114 PC patients undergoing prostatectomy or prostatic needle biopsy at Kochi Medical School Hospital (Nankoku, Japan) with appropriate informed consent. Frozen samples were embedded in Tissue-Tek OTC Compound (Sakura Finetek USA, Inc., Torrance, CA, USA) immediately after tissue procurement and stored at −80°C until their use. Frozen sections (4-µm thick) were cut. For paraffin-embedded samples, the tissues were fixed in 10% formalin neutral buffer solution for 1 day at room temperature. The tissue specimens were embedded in paraffin wax (Merck KGaA, Darmstadt, Germany) after fixation. Paraffin sections (2-µm thick) were cut. All of the surgical specimens were diagnosed and scored by a single pathologist. An immunohistochemical study was performed using the Ventana automated immunohistochemical system (Discovery; Ventana Medical Systems, Inc., Tucson, AZ, USA). Serial sections were incubated with a 1:100 diluted solution of purified anti-SHISA2 antibody (generated as described above) and a 1:100 diluted solution of anti-Wnt-5a antibody (catalog no. ab86720; 3D10; Abcam, Cambridge, UK) for 16 min at 37°C. All sections were independently scored in a blinded manner by a clinical pathologist. The intensity of SHISA2 staining was assessed using an immunohistochemical score of the tumor on the basis of strength: Negative, weak, moderate or strong staining. Comparisons between the SHISA2 expression levels in three groups were analyzed and the groups were formulated depending on the GS of the patient (group 1, GS 6; group 2, GS 7; and group 3, GS between 8 and 10). Prostate cancers with a Gleason score of 6 usually have rather good prognoses. By contrast, those with Gleason scores of 8–10 tend to be advanced disease that are unlikely to be cured. Additionally, those with Gleason score 7 are intermediate in clinical aggressiveness.
Construction of the SHISA2 expression vector
Full-length human SHISA2 cDNA (accession no. NM_001007538) was amplified using cDNA derived from human embryonic 293 cells and primers containing hemagglutinin (HA)-tag sequences in the COOH-terminus, and were inserted into the pIRESneo3 vector (Clontech, Mountain view, CA, USA). Primer sequences were as follows: SHISA2 forward, 5′-AGGGTGGTGCCATGTGGG-3′ and reverse, 5′-TTAAGCGTAATCTGGAACATCGTATGGGTATACAGTCACCGCTGG-3′.
Subcellular localization
PC-3 cells (1×105) per well were seeded onto a chamber slide and cultured to 50% confluence. Cells were transfected with the pIRESneo3-SHISA2-HA expression vector using FuGENE6 reagent (Roche Diagnostics, Basal, Switzerland) according to the manufacturer's protocol. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature and incubated with PBS containing 0.1% Triton X-100 for 2 min at room temperature, and subsequently stained with a standard protocol. Primary antibodies used were rat anti-HA monoclonal antibody (mAb) (3F10; catalog no. 12158167001; Roche Diagnostics) and mouse anti-inositol-requiring enzyme 1 (IRE1) mAb, as an endoplasmic reticulum (ER) marker (9F2; catalog no. ab96481; Abcam), and all antibodies were used in a 1:250 dilution and incubated for 60 min at RT. Subsequently, cells were incubated with the following secondary antibodies: Alexa Fluor (AF) 568-conjugated anti-mouse immunoglobulin G (catalog no. A11004; Thermo Fisher Scientific, Inc.) and AF 488-conjugated anti-rat immunoglobulin G (catalog no. A11006; Thermo Fisher Scientific, Inc.), and were used in a 1:250 dilution and incubated for 60 min at RT. Cells were analyzed using a FV500 confocal microscope (Olympus, Tokyo, Japan).
Western blot analysis
PC-3 cells (1×106) were seeded onto a 10-cm dish and cultured to 50% confluence. Cells were transfected with 10 µg of the pIRESneo3 empty vector or pIRESneo3-SHISA2-HA expression vector using FuGENE6 reagent (Roche Diagnostics), according to the manufacturer's protocol. Cells were lysed using IP Lysis Buffer (Thermo Fisher Scientific, Inc.) and protein concentration was determined by the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Cell lysates (30 µ (30 es and culture medium (DMEM without FBS) were separated on SDS-PAGE (12% gels), transferred to polyvinylidene difluoride (PVDF) membranes, and subsequently blocked with 5% milk for 30 min at RT. Membranes were then incubated with anti-HA mAb or anti-SHISA2 pAb for 60 min at RT. Signals were visualized using the Enhanced Chemiluminescence Plus Western Blotting Detection system (GE Healthcare). The anti-ACTB antibody (dilution, 1:5,000; clone AC-15; Sigma-Aldrich; Merck KGaA) served as a loading control. ACTB was used as a loading control (cell lysate). Silver stain was used as a loading control (medium).
Construction of short hairpin RNA (shRNA) expression vectors
Plasmids designed to express shRNA were prepared by cloning double-stranded oligonucleotides into the pBAsi-hU6 Neo vector (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's protocol. Target sequences for SHISA2 were as follows: Sense strand sequence SHISA2-si1, 5′-CCGTGTTTGTCGCCTTTAT-3′; SHISA2-si2, 5′-CCAGATTGTGCCACATCAA-3′; and negative control short interfering (si)RNA (siCONTROL), 5′-GCGCGCTTTGTAGGATTCG-3′, as described previously (12–14).
Cell viability assay
PC-3 cells (2×105), that highly expressed SHISA2, were seeded onto a 6-well plate and cultured to 50% confluence. Cells were transfected with shRNA expression vectors (SHISA2-si1, SHISA2-si2 and siCONTROL) using FuGENE6 (Roche Diagnostics), according to the manufacturer's protocol and cultured in DMEM containing 800 µg/ml geneticin (Thermo Fisher Scientific, Inc.) for 14 days. Cells were fixed with 100% methanol and stained with 0.1% crystal violet-H2O for the colony formation assay. Cell viability was determined using Cell Counting Kit-8 (Dojindo Molecular Technologies, Kumamoto, Japan) 10 days after transfection according to the manufacturer's protocol. Absorbance was determined at 450 nm as a reference with a Microplate Reader iMark (Bio-Rad Laboratories, Inc.). These experiments were performed in triplicate. Preliminarily, the knockdown effects of the shRNA-expression vectors on endogenous SHISA2 expression were validated 7 days after transfection by western blot analysis as aforementioned.
Generation of SHISA2-overexpressing cells and cell proliferation assay
PC-3 cells (1×106) were seeded onto a 10-cm dish and cultured to 50% confluence. Cells were transfected with 4 µg of the pIRESneo3 empty vector or pIRESneo3-SHISA2-HA expression vector using FuGENE6 reagent (Roche Diagnostics), according to the manufacturer's protocol and selected for by culturing in an DMEM containing 400 µg/ml geneticin (Thermo Fisher Scientific, Inc.) for 16 days, and subsequently discrete colonies were collected. Stable clones were maintained in DMEM, as aforementioned, and assayed for exogenous SHISA2 expression by western blot analysis as aforementioned. The proliferation of PC-3 cells that stably expressed SHISA2 (PC-3-SHISA2 clone mixture) and those transfected with pIRESneo3 empty vector (PC-3-Mock clone mixture) were examined using Cell-Counting Kit-8 (Dojindo Molecular Technologies, Inc.). PC-3-SHISA2 and PC-3-Mock cells were seeded at a concentration of 3×103 cells/well onto 48-well plates. The assay was performed every 24 h for 5 days, according to the manufacturer's protocol. In addition, 22Rv1-stable transformants were generated in the same way.
cDNA microarray analysis and data acquisition. PC-3 cells (1×106) were seeded onto a 10-cm dish and cultured to 50% confluence. Cells were transfected with pBA-siSHISA2 or pBA-siCONTROL using FuGENE6 (Roche Diagnostics), according to the manufacturer's protocol. At 7 days after transfection, total RNA was extracted using the RNeasy Mini kit (Qiagen, Inc.) according to the manufacturer's protocol. GeneChip array data were compared using the Kurabo custom analysis service (Kurabo Industries Ltd., Osaka, Japan) as described previously (14). Briefly, total RNA was reverse transcribed to cDNA using a T7 oligo d(T) primer (Affymetrix; Thermo Fisher Scientific, Inc.). The cDNA synthesis product was used in an in vitro transcription (IVT) reaction involving T7 RNA polymerase (Thermo Fisher Scientific, Inc.). A biotin-labeled ribonucleotide mix (Thermo Fisher Scientific, Inc.) was used in the first cycle of IVT amplification. Labeled aRNA products were fragmented, loaded onto the GeneChip Human Genome U133 Plus 2.0 array (Thermo Fisher Scientific, Inc.) and hybridized according to the manufacturer's protocol. Streptavidin-phycoerythrin (Thermo Fisher Scientific, Inc.) was used as a fluorescent conjugate to detect hybridized target sequences. Raw intensity data from the GeneChip array were analyzed using the GeneChip Operating Software 1.4 (Thermo Fisher Scientific, Inc.).
Statistical analysis
Results are expressed as the mean ± standard deviation or frequency. Data analysis was performed using the statistical software package JMP (version 7; SAS Institute Inc., Cary, NC, USA). Spearman's univariate correlation analysis was conducted between SHISA2 levels and GSs for non-parametric variables. A comparison of three groups (siCONTROL, SHISA2-si1 and SHISA2-si2) was analyzed statistically using One-way ANOVA and Tukey's test. Cell proliferation assay was performed as triplicate and Statistical significance was calculated using Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
SHISA2 overexpression in high-grade PC cells
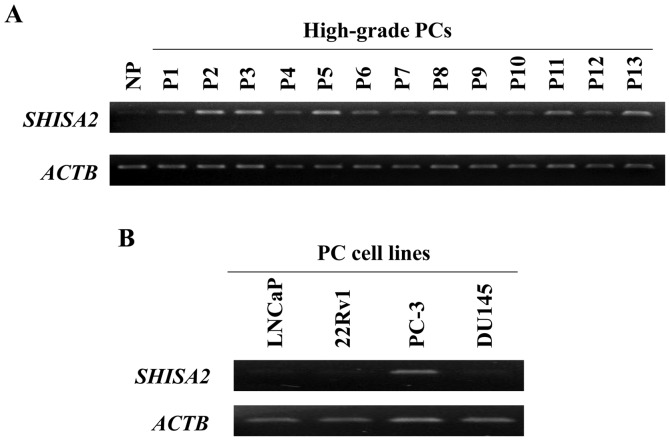
Gene expression profiles of high-grade PC cells, purified from clinical PC tissues with high GSs (between 8 and 10), were analyzed (NCBI Gene Expression Omnibus; accession no. GSE45016) (14). Among the genes that were transactivated in high-grade PC cells, compared with normal prostate epithelial cells (NP), SHISA2 was focused on in the present study. RT-sqPCR validated the fact that SHISA2 expression was increased in 13 clinical high-grade PC cell samples compared with that in NP cells (Fig. 1). Additionally, SHISA2 was expressed only in PC-3 cells (androgen-independent PC cell line) among 4 human PC cell lines.
Figure 1.
SHISA2 overexpression in high-grade PC cells. (A) RT-sqPCR analysis validated that SHISA2 was overexpressed in micro dissected high-grade PC cells, relative to NP micro dissected epithelial cells. ACTB was used to quantify cDNA contents. (B) RT-sqPCR analysis of SHISA2 in 4 human PC cell lines. ACTB was used to quantify cDNA contents. SHISA2, shisa family member 2; PC, prostate cancer; RT-qPCR, reverse transcription-semi-quantitative polymerase chain reaction; NP, normal prostate; ACTB, β-actin.
Expression and subcellular localization of SHISA2 protein in PC cells
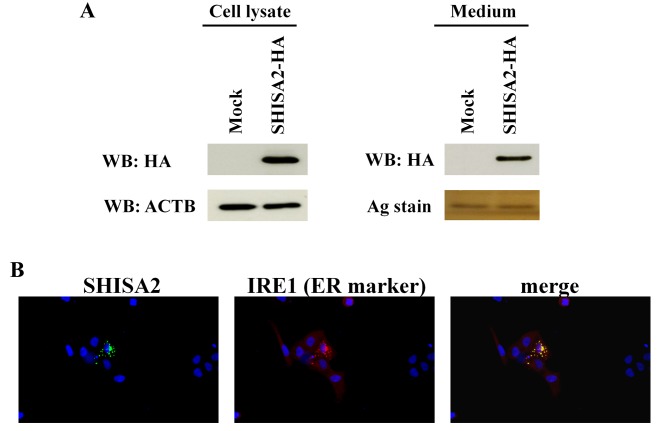
To determine the expression of SHISA2 protein in PC cells, the pIRESneo3-SHISA2-HA expression vector was transfected into PC-3 cells. SHISA2 protein was detected in the cell lysate and culture medium using western blot analysis (Fig. 2A). Subcellular localization of SHISA2 protein was validated in the ER of PC-3 cells using double-fluorescent immunostaining with anti-HA antibody and anti-IRE1 antibody (ER marker) (Fig. 2B).
Figure 2.
Expression of SHISA2 protein in PC cells. Western blot analysis with anti-HA-tag antibody validated transient expression of exogenous SHISA2 in the cell lysate and medium. ACTB was used as a loading control (cell lysate). Silver stain was used as a loading control (medium). (B) Double fluorescent immunostaining using anti-HA antibody and anti-IRE1 antibody (ER marker). SHISA2, shisa family member 2; PC, prostate cancer; ACTB, β-actin; WB, western blotting; IRE1, inositol-requiring enzyme 1; ER, endoplasmic reticulum; HA, hemagglutinin.
Immunohistochemical analysis of SHISA2 in clinical PC tissues
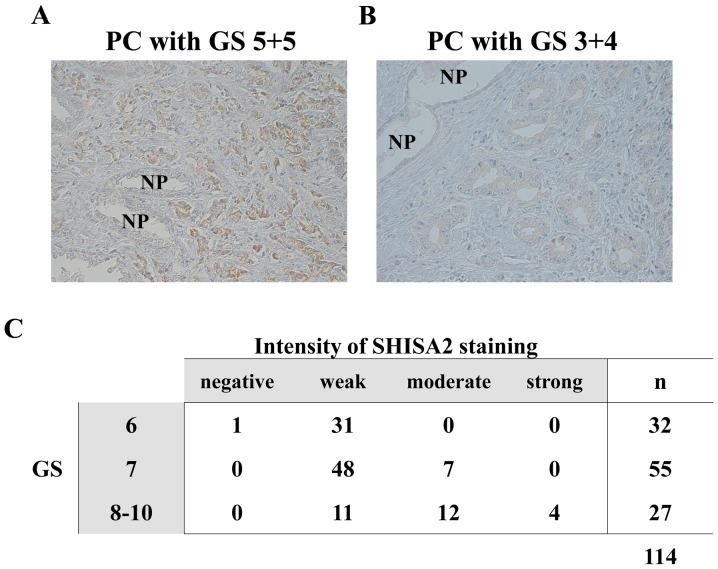
To validate the overexpression of SHISA2 protein in clinical PC cells, immunohistochemical analysis was performed on 114 clinical PC tissues with various GSs, using a polyclonal antibody specific to human SHISA2. As presented in Fig. 3A, a strong immunohistochemical signal for SHISA2 was determined predominantly in the cytoplasm of PC cells with GS 5+5=10, whereas PC cells with GS 3+4=7 revealed weak immune reactivity to anti-SHISA2 antibody (Fig. 3B). Adjacent NP epithelium in the same patient revealed no or very weak signal for SHISA2. Overall, the results demonstrated that SHISA2 was expressed in high-grade PCs.
Figure 3.
Immunohistochemical analysis of SHISA2 in clinical PC tissues. Immunoreactivity with anti-SHISA2 antibody was observed in high-grade PCs with high GS, exhibiting strong positive immunostaining in the cytoplasm of PC cells. (A) Representative image of PC cells with GS 5+5 (magnification, ×200). (B) Representative image of PC cells with GS 3+4 (magnification, ×200). Adjacent NP epithelium revealed a weak or no signal for SHISA2. (C) Association between SHISA2 expression and GS in PC. A significant association was determined between the expression level of SHISA2 and GS, using univariate Spearman's rank analysis (r=0.5287; P<0.0001). SHISA2, shisa family member 2; PC, prostate cancer; GS, Gleason score; NP, normal prostate.
To investigate the clinical and pathological significance of SHISA2 expression in PC tissues, the association between SHISA2 expression and GS in 114 PC tissues with various GSs was analyzed. As presented in Fig. 3C, a significant association was observed between SHISA2 expression levels and a high GS, as determined using the univariate Spearman's rank correlation analysis (r=0.5287; P<0.0001).
Knockdown of SHISA2 expression by shRNA decreases PC cell numbers
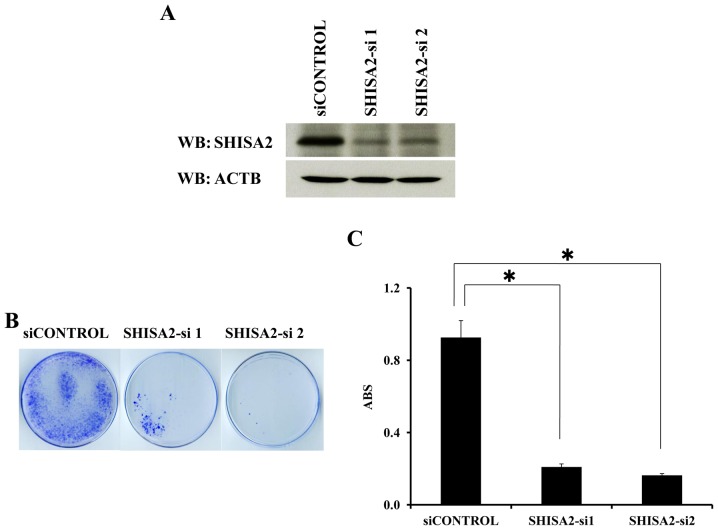
To examine the biological roles of SHISA2 overexpressed in PC cells, shRNA expression vectors (SHISA2-si1, SHISA2-si2 and siCONTROL) were constructed and transfected into PC-3 cells expressing endogenous SHISA2 (Fig. 1B). SHISA2-si1 and SHISA2-si2 exhibited a marked knockdown effect on endogenous SHISA2 protein, as determined using western blot analysis (Fig. 4A). Furthermore, transfection resulted in a decreased number of PC colonies and viable PC cells, as determined using a cell viability assay (P<0.01, Tukey's test). By contrast, transfection of the negative control (siCONTROL) exhibited a limited or no effect on SHISA2 expression and did not affect the viability of PC-3 cells (Fig. 4B and C).
Figure 4.
Knockdown of SHISA2 expression by siRNA decreases PC cell numbers. (A) siRNA-knockdown effect of SHISA2 expression in PC-3 cells. Western blot analysis was performed using cells that had been transfected with shRNA-expressing vectors specific to SHISA2 and a negative control vector. ACTB was used as a loading control. (B) Colony formation assay of PC-3 cells transfected with the indicated shRNA-expressing vectors specific to SHISA2 and a negative control vector. Cells were visualized with 0.1% crystal violet staining after 14 days incubation with geneticin. (C) Cell viability assays of PC-3 cells transfected with the indicated shRNA-expressing vectors specific to SHISA2 and a negative control vector. Each mean value is plotted with error bars indicating the standard deviation. ACTB was used as a loading control. Experiments were performed in triplicate. *P<0.01, Tukey's test. SHISA2, shisa family member 2; siRNA, short interfering RNA; PC, prostate cancer; ACTB, β-actin; WB, western blotting; ABS, absorbance at 450 nm.
SHISA2 overexpression promotes PC cell proliferation
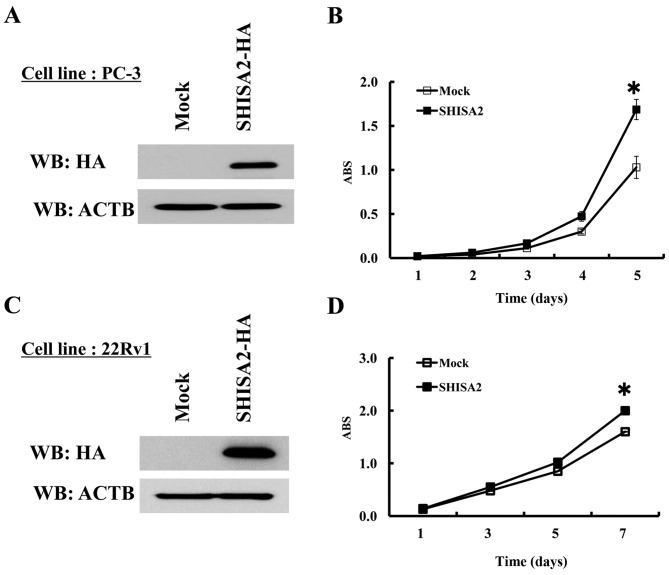
To identify the potential oncogenic function of SHISA2, a stable transformant was established from PC-3 cells, which constitutively expressed exogenous SHISA2 (Fig. 5A). In addition, control PC-3 cells were prepared, which were transfected with the empty vector (Mock), and the proliferation was compared between the two. Western blot analysis determined an increased level of exogenous SHISA2 expression in the stable clone. A cell proliferation assay revealed that PC-3-SHISA2-overexpressing clones grew more rapidly compared with PC-3-mock clones (P<0.01, Student's t-test), indicating that SHISA2 overexpression promoted PC cell proliferation (Fig. 5B). Additionally, a stable transformant was established from 22Rv1 cells, in which SHISA2 is not expressed endogenously (Fig. 5C). As with PC-3, a cell proliferation assay revealed that 22Rv1-SHISA2-overexpressing clones grew more rapidly compared with 22Rv1-mock clones (P<0.01, Student's t-test) (Fig. 5D).
Figure 5.
SHISA2 overexpression promoted cancer cell proliferation. (A) Western blot analysis determined constitutive expression of exogenous SHISA2-HA in stable PC-3 transformants. (B) Cell proliferation assay revealed that the SHISA2-HA-overexpressing clones grew more rapidly, compared with the mock clones. (C) Western blot analysis determined constitutive expression of exogenous SHISA2-HA in stable 22Rv1 transformants. (D) Cell proliferation assay revealed that the SHISA2-HA-overexpressing clones grew more rapidly than the mock clones. ACTB was used as a loading control. *P<0.01, analyzed using the Student's t-test. SHISA2, shisa family member 2; WB, western blotting; ABS, absorbance measured at 450 nm; ACTB, β-actin; HA, hemagglutinin.
SHISA2 affects WNT5A production
The physiological and pathological functions of SHISA in humans and human cancer types remains unknown. To understand the association between SHISA2 overexpression and the aggressiveness of clinical high-grade PCs, gene expression analysis was performed using a cDNA microarray. Gene expression patterns of PC-3 cells transfected with the two shRNA expression vectors (siSHISA2 and siCONTROL) were compared. The microarray data are available from the NCBI Gene Expression Omnibus (accession no. GSE48833).
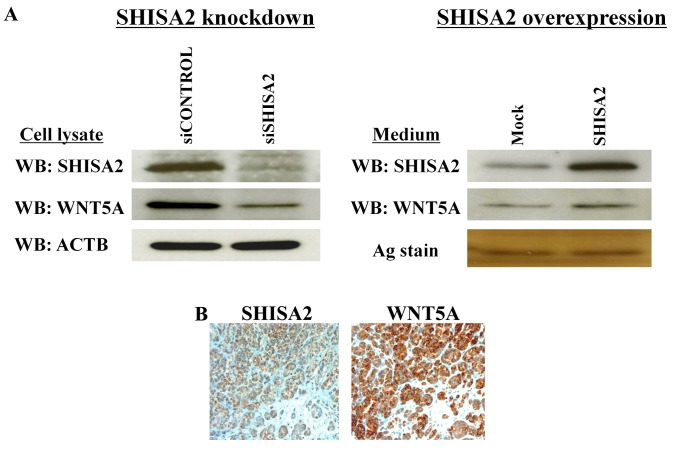
The results of the microarray analysis, with a focus on SHISA2-knockdown in PC-3 cells, identified that WNTs and FGFs were downregulated (Table I). Western blot analysis identified marked downregulation of WNT5A by SHISA2-knockdown, and upregulation of WNT5A by SHISA2-overexpression (Fig. 6A). Furthermore, it was validated that SHISA2 was co-expressed with WNT5A in clinical PC tissues using immunohistochemical analysis (Fig. 6B).
Table I.
Downregulation of WNTs and FGFs by SHISA2-knockdown.
| Gene symbol | Gene name | Log ratio |
|---|---|---|
| WNT5A | Wingless-type MMTV integration site family, member 5A | −3.6 |
| FGF20 | Fibroblast growth factor 20 | −3.6 |
| WNT5A | Wingless-type MMTV integration site family, member 5A | −3 |
| WNT6 | Wingless-type MMTV integration site family, member 6 | −2.2 |
| FGF18 | Fibroblast growth factor 18 | −1.8 |
| FGF18 | Fibroblast growth factor 18 | −0.9 |
| WNT10B | Wingless-type MMTV integration site family, member 10B | −0.8 |
| FGF18 | Fibroblast growth factor 18 | −0.8 |
| FGF11 | Fibroblast growth factor 11 | −0.5 |
| FGF21 | Fibroblast growth factor 21 | −0.4 |
| FGF3 | Fibroblast growth factor 3 | −0.2 |
SHISA2, shisa family member 2.
Figure 6.
Association of SHISA2 with WNT5A in PC cells. (A) Western blot analysis demonstrated that expression levels of WNT5A were downregulated by SHISA2-knockdown and upregulated in SHISA2 overexpression. ACTB was used as a loading control (cell lysate). Silver stain was used as a loading control (medium). (B) Representative picture of PC (magnification, ×200). Immunohistological analysis revealed the co-expression of SHISA2 and WNT5A in serial sectionsof clinical PC tissues. ACTB was used as a loading control. SHISA2, shisa family member 2; PC, prostate cancer; WB, western blotting; WNT5A, protein Wnt-5a; ACTB, β-actin; si, short interfering.
Discussion
To the best of our knowledge, the present study is the first to identify a novel molecular target and diagnostic biomarker, SHISA2, as an overexpressed gene in high-grade PC cells. To characterize the molecular features of clinical high-grade PCs, gene expression profiles were analyzed using genome-wide cDNA microarrays, combined with micro dissection, and a number of trans-activated genes in clinical high-grade PCs were identified (14,15). In the present study, in order to obtain molecular cancer drug targets and diagnostic biomarkers highly specific to PC, candidate genes were selected on the basis of the following criteria: i) Genes that were transactivated in PC cells relative to NP epithelial cells (to identify cancer-specific molecular targets and biomarkers); ii) genes that encode secreted protein (to identify serum biomarkers); and iii) androgen-independent genes (to identify biomarkers distinct from the androgen receptor signaling pathway, including PSA). SHISA2 was focused on as it met the aforementioned conditions.
Shisa was initially identified by Yamamoto et al (16) as a novel antagonist against Wnt and FGF signaling for head formation in Xenopus. ‘Shisa’ was termed on the basis of a type of sculpture that is common to southern Japan (Okinawa) and exhibits a large head, similar to the Egyptian sphinx. Shisa is a transcription factor-type molecule that physically interacts with immature forms of the Wnt receptor Frizzled and the FGF receptor within the ER, and inhibits posttranslational maturation and trafficking to the cell surface (16). In vertebrates, the Shisa family has nine known members. Human SHISA2 is an orthologue of the Xenopus SHISA2. The latter serves an essential role in segmental patterning during Xenopus somitogenesis (17). However, the physiological and pathological functions of SHISA2 in humans and human cancer types remains unknown.
In the present study, it was identified that clinically high-grade PC cellsoverexpressed SHISA2. Knockdown of SHISA2 expression by siRNA resulted in the marked suppression of PC cell viability. By contrast, exogenous SHISA2 expression in transfected cells promoted PC cell proliferation. The results of the present study suggested that SHISA2 was involved in the viability and proliferation of PC cells. Furthermore, secreted SHISA2 protein in the culture medium was detected. It was hypothesized that, if secreted SHISA2 protein promotes the viability and proliferation of PC cells in an autocrine or paracrine manner, SHISA2 may be detected in the serum of patients with PC as a useful diagnostic biomarker in predicting PC progression. In addition, neutralization of SHISA2 by highly specific antibodies may be used in a therapeutic strategy to treat PC.
Notably, as a result of the cDNA microarray analysis in the present study, the expression of WNT5A was identified to be downregulated by SHISA2-knockdown. Previous studies have demonstrated that WNT5A is highly upregulated in PC tissue from patients with advanced PC, and WNT5A overexpression was associated with the aggressiveness and poor prognosis of PC (18,19). Additionally, it has been identified that WNT5A contributes to gastric cancer progression by increasing metastatic potential and that WNT5A upregulates laminin subunit γ2 to mediate cancer cell aggressiveness (20,21). Furthermore, western blot analysis validated a significant downregulation of WNT5A by SHISA2-knockdown and the upregulation of WNT5A by SHISA2 overexpression. Immunohistochemical analysis revealed the co-expression of SHISA2 and WNT5A in serial sectionsof clinical PC tissues, which suggested that SHISA2 affects WNT5A production.
The WNT signaling pathway through which WNT5A was regulated by SHISA2 may be essential in the cell viability and proliferation of PC, although the underlying molecular mechanisms remain unknown and further investigations are required. The detection of SHISA2 and its inhibition may provide a novel target to develop a diagnostic biomarker and/or a molecular target in the treatment of PC.
Acknowledgements
The present study was supported by the Suzuki Foundation for Urological Medicine, by a Grant-in-Aid for Scientific Research (grant no. 21791508) and by the Japanese Society for the Promotion of Science (grant no. 23791762).
References
- 1.Grönberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–864. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 2.Hsing AW, Devesa SS. Trends and patterns of prostate cancer: What do they suggest? Epidemiol Rev. 2001;23:3–13. doi: 10.1093/oxfordjournals.epirev.a000792. [DOI] [PubMed] [Google Scholar]
- 3.Roddam AW, Hamdy FC, Allen NE, Price CP. UK Prostate Cancer Risk Management Programme: The impact of reducing the prostate-specific antigen threshold and including isoform reflex tests on the performance characteristics of a prostate-cancer detection programme. BJU Int. 2007;100:514–517. doi: 10.1111/j.1464-410X.2007.07000.x. [DOI] [PubMed] [Google Scholar]
- 4.Müntener M, Kunz U, Eichler K, Puhan M, Schmid DM, Sulser T, Strebel RT. Lowering the PSAthreshold for prostate biopsy from 4 to 2.5 ng/ml: Influence on cancer characteristics and number of men needed to biopt. Urol Int. 2010;84:141–146. doi: 10.1159/000277589. [DOI] [PubMed] [Google Scholar]
- 5.Shao YH, Albertsen PC, Roberts CB, Lin Y, Mehta AR, Stein MN, DiPaola RS, Lu-Yao GL. Risk profiles and treatment patterns among men diagnosed as having prostate cancer and a prostate-specific antigen level below 4.0 ng/ml. Arch Intern Med. 2010;170:1256–1261. doi: 10.1001/archinternmed.2010.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 7.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: Directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 8.Han M, Partin AW, Piantadosi S, Epstein JI, Walsh PC. Era specific biochemical recurrence-free survival following radical prostatectomy for clinically localized prostate cancer. J Urol. 2001;166:416–419. doi: 10.1097/00005392-200108000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Hurwitz MD, Schultz D, Richie JP, Wein AJ, Whittington R, Malkowicz SB, D'Amico AV. Radical prostatectomy for high-gradeprostate cancer. Urology. 2006;68:367–370. doi: 10.1016/j.urology.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Grossmann ME, Huang H, Tindall DJ. Androgen receptor signaling in androgen refractory prostate cancer. J Natl Cancer Inst. 2001;93:1687–1697. doi: 10.1093/jnci/93.22.1687. [DOI] [PubMed] [Google Scholar]
- 11.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Theodore C, James ND, Turesson I, et al. Docetaxel plus predonisone or mitoxantrone plus predonisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 12.Petrylak DP, Tangen CM, Hussain MH, Lara PN, Jr, Jones JA, Taplin ME, Burch PA, Berry D, Moinpour C, Kohli M, et al. Docetaxel and estramustine compared with mitoxantrone and predonisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Furihata M, Tsunoda T, Ashida S, Takata R, Obara W, Yoshioka H, Daigo Y, Nasu Y, Kumon H, et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. doi: 10.1158/0008-5472.CAN-06-4040. [DOI] [PubMed] [Google Scholar]
- 14.Satake H, Tamura K, Furihata M, Anchi T, Sakoda H, Kawada C, Iiyama T, Ashida S, Shuin T. The ubiquitin-like molecule interferon-stimulated gene 15 is overexpressed in human prostate cancer. Oncol Rep. 2010;23:11–16. [PubMed] [Google Scholar]
- 15.Anchi T, Tamura K, Furihata M, Satake H, Sakoda H, Kawada C, Kamei M, Shimamoto T, Fukuhara H, Fukata S, et al. SNRPE is involved in cell proliferation and progression of high grade prostate cancer through the regulation of androgen receptor expression. Oncol Lett. 2012;3:264–268. doi: 10.3892/ol.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto A, Nagano T, Takehara S, Hibi M, Aizawa S. Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors, Wnt and FGF. Cell. 2005;120:223–235. doi: 10.1016/j.cell.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 17.Nagano T, Takehara S, Takahashi M, Aizawa S, Yamamoto A. Shisa2 promotes the maturation of somatic precursors and transition to the segmental fate in Xenopus embryos. Development. 2006;133:4643–4654. doi: 10.1242/dev.02657. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto H, Oue N, Sato A, Hasegawa Y, Yamamoto H, Matsubara A, Yasui W, Kikuchi A. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene. 2010;29:2036–2046. doi: 10.1038/onc.2009.496. [DOI] [PubMed] [Google Scholar]
- 19.Thiele S, Rauner M, Goettsch C, Rachner TD, Benad P, Fuessel S, Erdmann K, Hamann C, Baretton GB, Wirth MP, et al. Expression profile of WNT molecules in prostate cancer and its regulation by aminobisphosphonates. J Cell Biochem. 2011;112:1593–1600. doi: 10.1002/jcb.23070. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto H, Kitadai Y, Yamamoto H, Oue N, Ohdan H, Yasui W, Kikuchi A. Laminin gamma2 mediates Wnt5a-induced invasion of gastric cancer cells. Gastroenterology. 2009;137:242–252. doi: 10.1053/j.gastro.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Hanaki H, Yamamoto H, Sakane H, Matsumoto S, Ohdan H, Sato A, Kikuchi A. An anti-Wnt5a antibody suppresses metastasis of gastric cancer cells in vivo by inhibiting receptor-mediated endocytosis. Mol Cancer Ther. 2012;11:298–307. doi: 10.1158/1535-7163.MCT-11-0682. [DOI] [PubMed] [Google Scholar]