Abstract
Although emerging evidence has indicated that single nucleotide polymorphisms (SNPs) in microRNAs (miRNAs) are associated with susceptibility to gastric cancer, a limited number of studies have revealed the underlying molecular mechanisms. In the present study, the results suggested that miR-1269a rs73239138 has a role in decreasing the risk of gastric cancer. The level of miR-1269a variant expression was significantly downregulated compared with the wild-type miR-1269a in the gastric cells (Fig. 1). Furthermore, overexpression of miR-1269a inhibited apoptosis of gastric cancer cells. Expression of the miR-1269a variant inhibited the function of miR-1269a by increasing the apoptotic rate and the expression of Bik, Bim and Bak was upregulated consistently. In addition, zinc-finger protein 70 (ZNF70) was identified to be a target gene of miR-1269a, which was downregulated by miR-1269a and upregulated by miR-1269a variant. ZNF70 was indicated to exert a role as a tumor suppressor in gastric cancer. To the best our knowledge, the present study for the first time highlights a critical role of miR-1269a variant rs73239138 in decreasing the susceptibility to gastric cancer by downregulating its expression and targeting ZNF70, which promotes apoptosis of gastric cancer cells. This SNP is indicated to serve as a potential biomarker and therapeutic target for gastric cancer.
Keywords: miR-1269a, rs73239138, gastric cancer, zinc-finger protein 70, apoptosis
Introduction
There are ~738,000 mortalities per year caused by gastric cancer, which has been recognized as a major cause of cancer-associated mortality in the world (1). Over 70% of new cases and mortalities are occurring in developing countries, most of which are in advanced stage when first diagnosed due to the lack of sensitive and early diagnosis markers (2). Additionally, recurrence and metastasis of advanced gastric cancers are commonly observed following surgery and chemical therapy (3,4). Therefore, it is urgently required to identify novel and efficient methods for early diagnosis and offer positive treatments.
microRNAs (miRNAs) are a type of small non-coding single-strand RNAs that regulate gene expression post-transcriptionally. It is reported that 30–60% of human genes are regulated by miRNAs, primarily by binding to target sites on messenger RNAs (mRNAs) (5–7). miRNAs have a pivotal role in regulating biological processes in the development of gastric cancer (5). Specifically, miRNAs contribute to carcinogenesis by altering the expression of oncogenes and tumor suppressors to affect proliferation, apoptosis motility and invasion (8). Numerous studies have revealed that small changes in miRNAs expression may cause significant changes in target mRNA translation and protein synthesis and therefore affect the occurrence and development of gastric cancer (9,10).
Single nucleotide polymorphisms (SNPs) are the third generation of genetic markers for the study of complex genetic diseases, pharmacogenomics and human evolution (11,12). SNPs in microRNA may reconstruct the secondary structure of pre-miRNAs, which are named pri-miRNAs, or interfere with the maturation or target selection of miRNA (13), leading to the alteration of cancer formation. Therefore, miRNA SNPs may be regarded as biomarkers for cancer diagnosis and prognosis. However, a limited number of studies have investigated the molecular mechanisms of microRNA SNPs in gastric cancer.
miR-1269a, which is located at human chromosome 4, is associated with the occurrence and development of various tumors including primary hepatocellular carcinoma (14), colorectal cancer (15) and breast cancer (16). miR-1269a has been reported to be upregulated in hepatocellular carcinoma, as one of the independent cancer predictors via suppressing Forkhead box protein O1 expression (14). However, little has been reported in the literature regarding the role of miR-1269a in gastric cancer or the involvement of its SNP, rs73239138.
In the present study, to the best of our knowledge, for the first time an association was identified between rs73239138 in microRNA-1269a and the susceptibility to gastric cancer. The present study further revealed the molecular role of this variation. The findings of the present study may assist to clarify the roles of SNPs in miRNAs in the development and occurrence of gastric cancer and lead to proposals of novel methods for early prediction, detection, diagnosis, treatment and prognosis.
Materials and methods
Study population
The 373 gastric cancer patients were recruited from the Huashan Hospital and Huadong Hospital Affiliated to Fudan University (Shanghai, China), Renji Hospital and Tongren Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Shanghai, China) and PLA 175 Hospital (Xiamen, China), between January 2011 and December 2014. The mean age was 62.13 years and the age distribution was 27–90 years. The patients were diagnosed by pathology or imaging by enhanced computed tomography (CT) scan or positron emission tomography-CT scan. A total of 402 cancer-free controls were collected from the Taizhou longitudinal study without a history of other types of cancer or restriction of age and sex (17). All subjects were unrelated Han Chinese. All materials from patients, including peripheral blood samples and epidemiological investigations, were obtained with informed consent, and the whole study was approved by the Ethic Review Committees of Huashan Hospital and School of Life Science, Fudan University (Shanghai, China).
The prediction of microRNA target gene
The target genes of the miRNA were obtained according to the miRNA target gene database (TargetScan 6.2 http://www.targetscan.org/vert_62/).
Gene ontology (GO) analysis
GO analysis was applied in order to organize genes into hierarchical categories and uncover the target genes of miRNA on the basis of biological process (18). In detail, two-sided Fisher's exact test and χ2 test were used to classify the GO category, and the false discovery rate (FDR) (19) was calculated to correct the P-value. The present study selected only GOs that had a P-value of <0.01 and a FDR of <0.05.
Pathway analysis
Based on KEGG, pathway analysis was performed using Fisher's exact test and χ2 test, and the threshold of significance was defined by P-values and FDRs. The screening criteria were P<0.05 (20–22).
DNA extraction and genotyping
Genomic (g)DNA of each subject was isolated by using the AxyPrep™ Blood Genomic DNA Miniprep kit (Axygen Biosciences; Corning Incorporated, Corning, NY, USA). Concentration of gDNA was determined using NanoDrop (C723 ND-1000 UV/Vis; NanoDrop Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA). DNA samples were detected by agarose gel electrophoresis to ensure the DNA quality and a concentration >20 ng/µl were diluted into a working dilution of 10 ng/µl and added to 96-well plates for subsequent genotyping.
The SNPs were genotyped using matrix-assisted laser desorption/ionization-time of flight-based assay (MALDI-TOF) (23). The primers were designed using MassARRAY Assay Design software (Table I) and synthetized by Shanghai Generay Biotech Co., Ltd (Shanghai, China). The primer sequences are listed in Table I. Genotyping was performed using the MassARRAY Compact system (Sequenom Inc., CA, USA), and the data were analyzed using TyperAnalyzer software version 4.0 (Sequenom Inc.).
Table I.
Primer sequences for miR-1269a variant rs73239138.
| Primer sequences (5′-3′) | |||||||
|---|---|---|---|---|---|---|---|
| SNP IDa | Substitution | SNP location | miRNA | Location | Amplification primer 1 | Amplification primer 2 | Extension primer |
| rs73239138 | G/A | 67142620 | hsa-mir-1269a | Mat | ACGTTGGATGA | ACGTTGGATGA | CAGGGAAGC |
| AGTCTCATGAT | CCTGAGGAATG | CAGTAGCA | |||||
| AGGCCATC | CCTGGAC | ||||||
SNP ID from NCBI dbSNP database; miRNA, microRNA; mat, mature region of the miRNA.
Cell lines and cell culture
A total of two human gastric cell lines (HGC-27 and MGC-803) used in the present study, and the cell lines were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China), which were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc,) with 10% FBS (Biological Industries Israel, Kibbutz Beit Haemek, Israel). The cells were all maintained in a humidified incubator containing 5% CO2 at 37°C.
Reverse transcription-quantitative PCR analysis
miRNA was quantified using the miRcute miRNA qPCR Detection kit (SYBR green; Tiangen Biotech Co., Ltd., Shanghai, China) and analyzed using the 7500 Fast Real-Time Sequence detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used for stem-loop reverse-transcription PCR for miR-1269a and 5S were purchased from Tiangen Biotech Co., Ltd. The relative expression levels of the miRNA were normalized to the expression of small nuclear RNA 5S and calculated using 2−[(Cq of miRNA) - (Cq of 5S)] (24).
Cell transfection
The wild-type and variant miR-1269a expression plasmids were generated by cloning the wild-type and variant pre-miR-1269a sequence into the retroviral transfer plasmid CMV-MCS-SV40-neomycin (Shanghai GeneChem Co., Ltd., Shanghai, China), respectively. The concentration of DNA plasmids transfected was 1 µg/ml in DMEM with Penicillin-Streptomycin (Gibco; Thermo Fisher Scientific, Inc.). The miRNA-1269a mimics and miRNA-1269a inhibitor of the wild-type and variant type as well as the miR-negative control (NC) were purchased from GenePharma (Shanghai GenePharma Co., Ltd., Shanghai, China). Zinc-finger protein 70 (ZNF70) small interfering (si)RNA and the siRNA NC were purchased from Biotend (Shanghai, China). Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Cell apoptosis assay
HGC-27 and MGC-803 cell lines were seeded in 6-well plate at a suitable density (60–70%) and after 24, 48 and 72 h of transfection. The cells were washed in PBS, harvested by trypsin, and subsequently treated with 100 µl Muse™ Annexin V Dead Cell reagent (EMD Millipore; Billerica, MA, USA) at room temperature in the dark for 20 min. This was followed by detection of apoptosis using Muse™ Cell Analyzer (EMD Millipore).
Cell proliferation assay
Proliferation was measured using the cell-counting kit-8 (CCK-8 kit; Dojindo Molecular Technologies, Inc., Shanghai, China). According to the manufacturer's instructions, 2×103 cells were seeded into 96-well culture plates and allowed to adhere for 24 h at 37°C. The cells were subsequently transfected with 50 nM negative control or siRNA and incubated at 37°C for 24, 48, 72 or 96 h. At the endpoint, 20 µl CCK-8 (5 g/l) was added for a further 4 h at 37°C. A Multiscan GO spectrophotometer (Thermo Fisher Scientific, Inc.) was used to measure the absorbance at 450 nm.
Western blot analysis
Treated cells were washed twice with ice-cold PBS and then treated with RIPA lysis buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (both from Beyotime Institute of Biotechnology, Haimen, China). Following centrifugation at 10,000 × g for 15 min at 4°C, concentration of the proteins was measured using BCA protein assay kit (Beyotime Institute of Biotechnology). Cell protein lysates were separated on 12% SDS denatured polyacrylamide gel and electro-transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% non-fat milk and were incubated with primary antibodies against ZNF70 (zinc-finger protein 70; cat. no. ab49339; 5:1,000; Abcam, Cambridge, UK), GAPDH (cat. no. 5174S, Cell Signaling Technology Inc., Danvers, MA, USA), Bik (cat. no. 4592S, Cell Signaling Technology Inc., Danvers, MA, USA), Bim (cat. no. 2933S, Cell Signaling Technology Inc., Danvers, MA, USA) and Bak (cat. no. 12105S, Cell Signaling Technology Inc., Danvers, MA, USA) at 4°C overnight, which were all diluted to 1:1,000. The membranes were then washed 3 times with TBST (Bioscience Shanghai, Inc., China) and incubated with anti-rabbit immunoglobulin G at room temperature ~1 h, horseradish peroxidase-linked antibody (cat. no. 7074; 1:2,000; Cell Signaling Technology Inc.), according to the manufacturer's instructions. Finally, the images were captured using a gel imaging analysis system (Tanon 4100; Tanon Science and Technology Co., Ltd., Shanghai, China).
Statistical analysis
All statistical tests were performed using the software SPSS (version 19. 0; IBM Corp., Armonk, NY, USA) and Excel (Microsoft Corporation, Redmond, WA, USA). Using Pearson's chi-squared test of goodness of fit, genotype frequencies were evaluated for Hardy-Weinberg equilibrium. Binary logistic regression was applied to analyze the association between the genotypes of miRNA SNP and the susceptibility to gastric cancer following adjustments for age, sex, smoking status and drinking status. Odds ratios and 95% confidence intervals (CI) were also calculated. Pearson's chi-squared test of independence was then used to investigate the association between the SNPs and qualitative clinical indexes, including age, sex, smoking and alcohol consumption status, and tumor size in gastric cancer patients. A two-sided P<0.05 was considered to indicate a statistically significant difference.
The data from cellular experiments are expressed as the mean ± standard deviation for three independent experiments. Subsequently, ANOVA and multiple comparisons test were used to evaluate the significant difference between three groups of data, and the student's t-test was used to evaluate the significant difference between two groups of data. Statistics was performed with the use of GraphPad Prism (version 6.0; GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-1269a rs73239138 decreases the risk of gastric cancer
The present study included 373 patients with gastric cancer and 402 healthy subjects, and the demographic characteristics were summarized in Table II. It was indicated that there were significant differences between the patients with gastric cancer and the healthy controls in terms of age, sex and smoking status. However, there were no differences between the two groups for alcohol consumption.
Table II.
General characteristics of gastric cancer patients and unaffected controlsa.
| Characteristic | Cases (n=373) | Controls (n=402) | P-value |
|---|---|---|---|
| Age | 62.13±12.10 | 62.37±8.76 | <0.001b |
| Sex, n (%) | <0.001b | ||
| Total | 367 | 402 | |
| Male | 267 (72.8) | 175 (43.5) | |
| Female | 100 (27.2) | 227 (56.5) | |
| Smoking, n (%) | 0.012b | ||
| Total | 357 | 402 | |
| Neverc | 273 (76.5) | 274 (68.2) | |
| Everc | 84 (23.5) | 128 (31.8) | |
| Consumption of alcohol, n (%) | 0.099 | ||
| Total | 357 | 402 | |
| Neverc | 315 (88.2) | 338 (84.1) | |
| Everc | 42 (11.8) | 64 (15.9) | |
| Size of tumor foci (n=209), mean ± SD | 4.28±2.70 | ||
| Tumor sites, n (%) | |||
| Total | 303 | ||
| Non-cardia cancer | 230 (75.9) | ||
| Cardia cancer | 73 (24.1) | ||
| Organ metastasis, n (%) | 321 | ||
| Negative, M0 | 253 (78.8) | ||
| Positive, M1 | 68 (21.2) | ||
| Lymph-node metastasis, n (%) | |||
| Total | 340 | ||
| Negative (N0) | 158 (46.5) | ||
| Positive (N1-N3) | 182 (53.5) |
Certain patients were excluded from sex, smoking, the consumption of alcohol analysis, tumor sites analysis, organ metastasis and lymph-node metastasis analysis due to missing data (n=6, n=16, n=16, n=70, n=52 and n=33, respectively).
P<0.05.
Never means that patients never smoke or drink alcohol, and ever means that patients have smoked or consumed alcohol.
As shown in Table III, compared with the wild-type GG, the AA genotype was associated with a significant decreased risk of gastric cancer (P=0.045; OR, 0.610; 95% CI, 0.376–0.990). However, there was no statistically significant association between the GA genotype and gastric cancer risk (P=0.567; OR, 1.106; 95% CI, 1.106 (0.783–1.564). A similar association was identified in the recessive model (AA vs. GG + AG; P=0.014; OR, 0.576; 95% CI, 0.371–0.896). Collectively, the findings indicated that miR-1269a rs73239138 may have a role in decreasing the risk of gastric cancer.
Table III.
Association between genotypes/alleles of miR-1269a rs73239138 and the risk of gastric cancer.
| Gastric cancer patients | ||||
|---|---|---|---|---|
| Genotype | Control, n (%) | Number, n (%) | OR (95% CI)a | P-valueb |
| rs7329138 | ||||
| GG | 144 (36.1) | 131 (35.1) | ||
| GA | 180 (45.1) | 193 (51.7) | 1.106 (0.783–1.564) | 0.567 |
| AA | 75 (18.8) | 49 (13.1) | 0.610 (0.376–0.990) | 0.045b |
| Dominant model | ||||
| AA + GA vs. GG | 0.956 (0.689–1.326) | 0.788 | ||
| Recessive model | ||||
| AA vs. GG + GA | 0.576 (0.371–0.896) | 0.014b | ||
| G | 468 (58.6) | 455 (61.0) | ||
| A | 330 (41.4) | 291 (39.0) | 0.841 (0.669–1.056) | 0.135 |
ORs and P-values were all obtained following adjustment for age, sex, smoking status and alcohol consumption status.
P<0.05. CI, confidence interval; OR, odds ratio.
Decreased expression of miR-1269a variant in human gastric cancer cell lines
Subsequently, the expression levels of miR-1269a in HGC-27 and MGC-803 gastric cancer cell lines transfected with pre-miR1269a and per-miR1269a variant plasmid were analyzed by RT-qPCR. As shown in Fig. 1, the expression level of miR-1269a was significantly downregulated in cells that were transfected with pre-miR1269a-variant compared with pre-miR1269a wild-type-transfected cells (P<0.0001), indicating that the decreased expression of miR-1269a by the SNP may have contributed to the development of gastric cancer.
Figure 1.
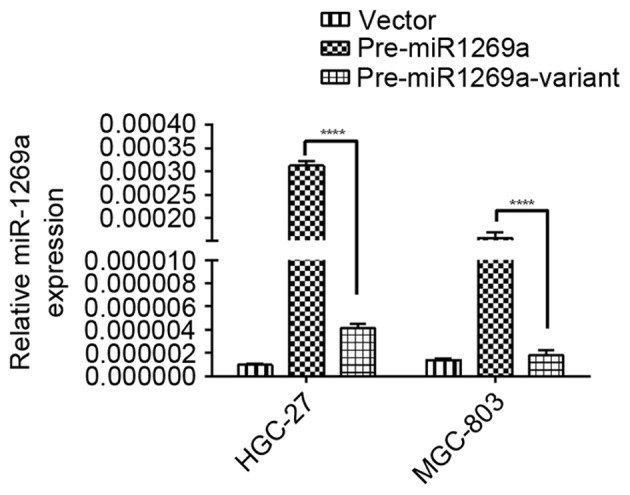
Decreased miR-1269a variant-type expression in human gastric cancer cell lines compared with miR-1269a wild-type. Relative expression of miR-1269a in HGC-27 and MGC-803 cells transfected with vector, pre-miR1269a and per-miR1269a-variant plasmid for 48 h. Data represent the mean ± standard deviation from three independent experiments. ****P<0.0001. miRNA, microRNA.
Increased apoptosis in cells transfected with miR-1269a variant compared with cells transfected with wild-type miR-1269a
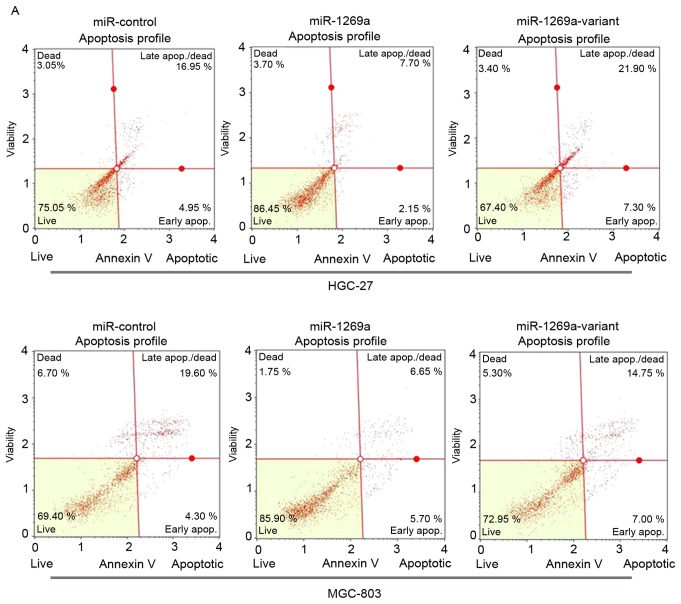
To determine the functional alteration of miR-1269a variant in gastric cancer progression, the authors of the present study examined the rate of apoptosis of MGC-803 and HGC-27 cells transfected with wild-type miR-1269a and mimics of miR-1269a variant by flow cytometric (FCM) analysis. As shown in Fig. 2A and B, the proportion of apoptotic cells was significantly decreased in MGC-803 or HGC-27 cells transfected with wild-type miR-1269a mimics after 72 h compared with control miR mimics. However, the apoptotic rate of cells transfected with miR-1269a variant type was significantly increased compared with cells transfected with miR-1269a wild-type (miR-NC vs. miR-1269a vs. miR-1269a variant, 19.10 vs. 11.62 vs. 22.95% in HGC-27; 21.17 vs. 13.97 vs. 20.73% in MGC-803). These results suggested that wild-type miR-1269a may have promoted tumor genesis in gastric cancer through prohibiting the apoptosis of gastric cancer cells.
Figure 2.
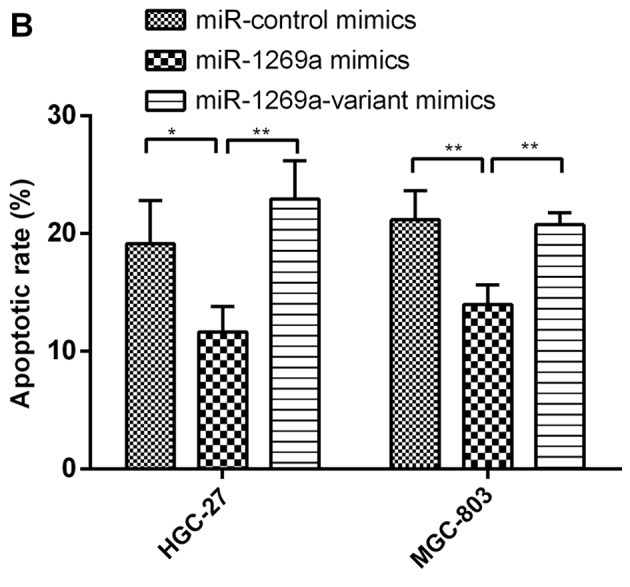
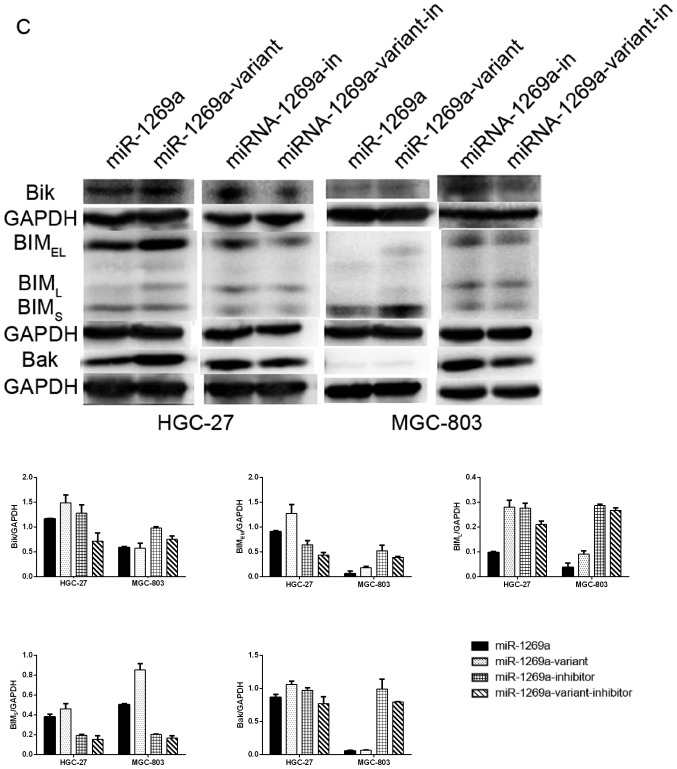
Overexpression of miR-1269a variant increases the apoptosis of gastric cancer cells. (A) A representative FCM analysis chart. The effect of miR1269a and miR-1269a variant on apoptosis was examined by FCM analysis. HGC-27 and MGC-803 cells were transfected with miR-control, miR1269a mimics and miR-1269a variant mimics and their inhibitors. Subsequently, the cells were analyzed for apoptotic rate following staining with Muse™ Annexin V Dead Cell reagent. (B) Quantitative data from FCM analysis. Data represent the mean ± standard deviation from three independent experiments. *P<0.05; **P<0.01. (C) Western blot analysis of apoptosis proteins (Bik, Bim and Bak) in HGC-27 and MGC-803 cells, which were transfected with miR1269a mimics and miR-1269a variant mimics. GAPDH served as the loading control. FCM, flow cytometry. Bik, Bcl-2-interacting killer; miRNA, microRNA; Bak, Bcl-2 homologous antagonist/killer.
The authors further detected the expression levels of proteins involved in apoptosis (Bik, Bim and Bak) between cells transfected with wild-type and miR-1269a variant by western blotting. As expected, compared with cells transfected with miR-1269a variant, the expression of Bik, Bim and Bak was downregulated in gastric cancer cells transfected with wild-type miR-1269a. However, the expression of these proteins was upregulated in cells transfected with the wild-type inhibitor compared with cells transfected with inhibitor of miR-1269a variant (Fig. 2C). Collectively, these results indicated that wild-type miR-1269a may have a role in the proliferation of gastric cancer cells by inhibiting apoptosis. However, expression of miR-1269a variant increased the apoptosis of gastric cancer cells and reduced the tumor-promoting effect of miR-1269a.
ZNF70 is a candidate target gene of miR-1269a and serves as a tumor suppressor gene in gastric cancer cells
It is important to investigate potential mRNAs, which are regulated by miR-1269a, which may further evaluate the function of miR-1269a. The potential target genes were obtained using the miRNA target gene database (TargetScan 6.2). Then, the authors used gene ontology (GO) analysis and pathway analysis to exclude any unlikely targets. The authors obtained a total of 23 predicted target genes of hsa-miR-1269a (Table IV), and excluded those by overexpressing miR-1269a in order to investigate which target did not respond (data not shown). As presented in Fig. 3A, a binding site in ZNF70 mRNA 3′-untranslated region (UTR) was identified. Using western blotting, it was demonstrated that ZNF70 expression was downregulated in MGC-803 and HGC-27 cells overexpressing miR-1269a compared with cells transfected with miR-control. However, this effect was reversed in cells overexpressing miR-1269a variant. Similarly, ZNF70 expression was upregulated in cells transfected with miR-1269a inhibitor and reversed in cells transfected with miR-1269a variant inhibitor compared with cells transfected with miR-inhibitor control (Fig. 3B). Therefore, these results suggest that miR-1269a targets and inhibits ZNF70 in gastric cancer cells.
Table IV.
The predicted target genes of miRNA-1269a.
| No. | Gene symbol | Transcript ID | Gene full name | Total context+score |
|---|---|---|---|---|
| 1 | DACT1 | NM_001079520 | Dapper, antagonist of β-catenin, homolog 1 (Xenopus laevis) | −0.47 |
| 2 | INTS6 | NM_001039938 | Integrator complex subunit 6 | −0.47 |
| 3 | RBMS3 | NM_001003792 | RNA binding motif, single stranded interacting protein 3 | −0.41 |
| 4 | ZNF70 | NM_021916 | Zinc finger protein 70 | −0.37 |
| 5 | DAZ2 | NM_001005785 | Deleted in azoospermia 2 | −0.35 |
| 6 | VPS13B | NM_017890 | Vacuolar protein sorting 13 homolog B (yeast) | −0.33 |
| 7 | DDX5 | NM_004396 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | −0.27 |
| 8 | AFAP1 | NM_001134647 | Actin filament associated protein 1 | −0.23 |
| 9 | APPBP2 | NM_006380 | Amyloid β precursor protein (cytoplasmic tail) binding protein 2 | −0.22 |
| 10 | NLN | NM_020726 | Neurolysin (metallopeptidase M3 family) | −0.21 |
| 11 | ONECUT1 | NM_004498 | One cut homeobox 1 | −0.19 |
| 12 | NFX1 | NM_002504 | Nuclear transcription factor, X-box binding 1 | −0.19 |
| 13 | USP9Y | NM_004654 | Ubiquitin specific peptidase 9, Y-linked | −0.18 |
| 14 | RAB3GAP2 | NM_012414 | RAB3 GTPase activating protein subunit 2 (non-catalytic) | −0.18 |
| 15 | CACNA1E | NM_000721 | Calcium channel, voltage-dependent, R type, α 1E subunit | −0.16 |
| 16 | WNT10A | NM_025216 | Wingless-type MMTV integration site family, member 10A | −0.12 |
| 17 | CUX2 | NM_015267 | Cut-like homeobox 2 | −0.11 |
| 18 | SPTB | NM_001024858 | Spectrin, β, erythrocytic | −0.1 |
| 19 | KLF3 | NM_016531 | Kruppel-like factor 3 (basic) | −0.1 |
| 20 | FAM55C | NM_001134456 | Family with sequence similarity 55, member C | −0.07 |
| 21 | SLC24A2 | NM_001193288 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | −0.04 |
| 22 | MTDH | NM_178812 | Metadherin | −0.04 |
| 23 | LRP6 | NM_002336 | Low density lipoprotein receptor-related protein 6 | >-0.02 |
miRNA, microRNA.
Figure 3.
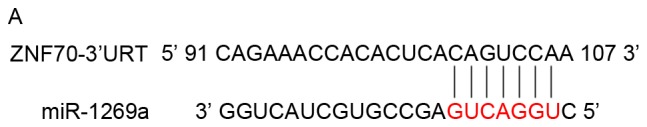
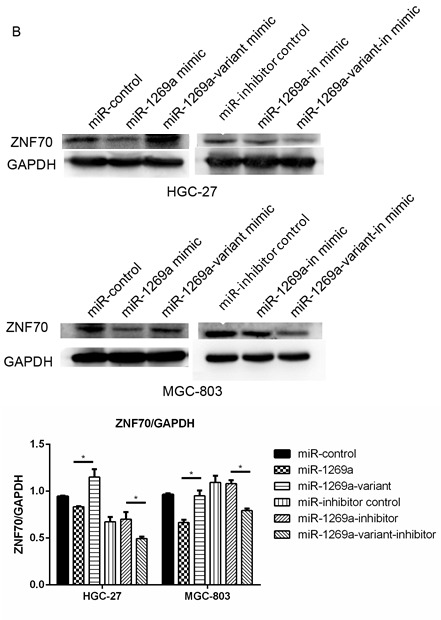
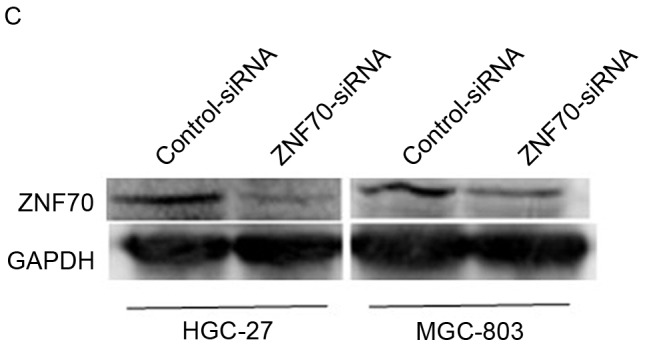
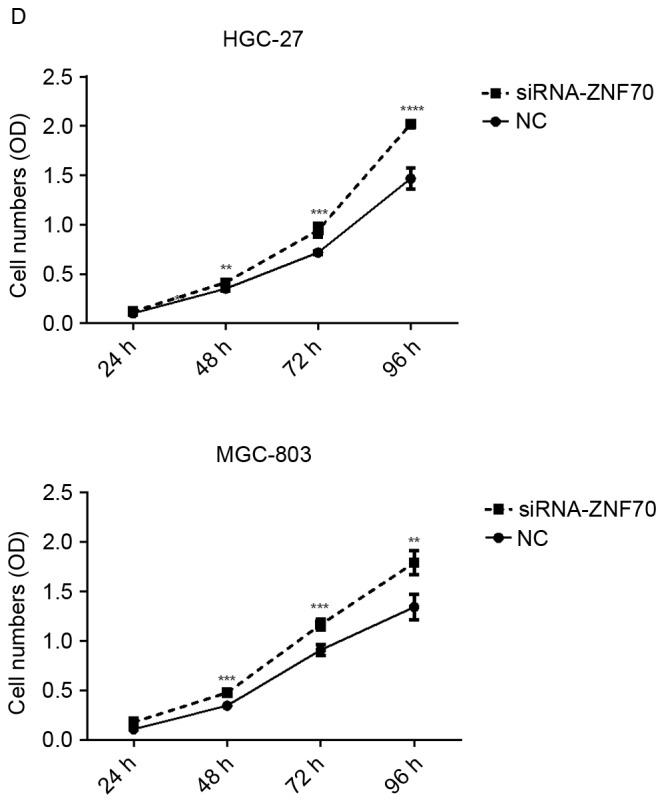
ZNF70 is a candidate target gene of miR-1269a and serves as a tumor suppressor in gastric cancer cells. (A) Sequence alignment of miR-1269a and putative ZNF70-3′UTR. (B) Western blot analysis of expression levels of ZNF70 in HGC-27 and MGC-803 cells that were transfected with miR-control, miR1269a mimics and miR-1269a-variant mimics and their inhibitors. GAPDH served as the loading control. (C) Western blot analysis of ZNF70 in ZNF70-siRNA-transfected cells. GAPDH served as the loading control. (D) The effects of ZNF70 on miR-1269a-mediated HCC proliferation as analyzed by cell-counting kit-8 assay. *P<0.05, **P<0.01 and ***P<0.001. ZNF70, zinc-finger protein 70; siRNA, small-interfering RNA; miRNA, microRNA; OD, optical density; UTR, untranslated region.
To further confirm ZNF70, as a target of miR-1269a, is involved in the pathology of gastric cancer, the authors of the present study have used siRNA to inhibit the endogenous ZNF70, and the siRNA interference efficiency is presented as Fig. 3C. As shown in Fig. 3D, the CCK-8 analysis revealed that when ZNF70 was suppressed, the growth rates of gastric cancer cells were increased significantly compared with the negative control. This result suggested that ZNF70 may be a tumor suppressor gene and the suppression of ZNF70 may have an important role in miR-1269a-mediated gastric cancer progression.
Discussion
Numerous studies have reported that miRNAs are upregulated or downregulated in gastric cancer, indicating that overexpressed oncogenic miRNAs (oncomiRs) or downregulated tumor suppressor miRNAs may affect apoptosis and proliferation in gastric cancer by inhibiting target genes (10). For example, it was reported that the overexpression of miR-181a, an oncomiR, in a gastric cancer cell line led to increased proliferation and inhibition of apoptosis via repression of tumor suppressor kruppel like factor 6 (25). Furthermore, a number of recent studies have focused on SNPs in miRNAs and have identified a number of SNPs that contribute to gastric cancer risk, including rs3746444 (in miR-499), rs2296616 (in miR-107) and rs2910164 (in miR-146a) (26–28). However, the specific functions of SNPs in miRNA remain unknown.
Apoptosis is an important cell death process that occurs in physiological and pathological conditions. An imbalance of the delicate relationship between physiological and pathological conditions can lead to the development of cancer. Previously, little is known about the effect of miR-1269a on apoptosis in cancer. To the best of our knowledge, the present study describes the first report of the observation that overexpression of miR-1269a may significantly inhibit the apoptosis of gastric cancer cells in vitro.
Additionally, it was indicated that miR-1269a inhibited the expression of apoptosis proteins, including Bik, Bim and Bak. Meanwhile, the low expression of miR-1269a variant had a reverse effect of wild-type, which suggested that miR-1269a variant suppressed the gastric cancer development by promoting the apoptosis of gastric cancer cells.
In the present study, three databases, TargetScan 6.2, gene ontology (GO) analysis and pathway analysis, were used to determine the target genes of miR-1269a. A total of 23 potential target genes were obtained, and the expression levels of the target genes were analyzed by RT-qPCR following transfection with miR-1269a mimics in HGC-27 and MGC-803 cells (data not shown). It was observed that there was a decreased level of ZNF70 expression in cells transfected with miR-1269a mimics in HGC-27 and MGC-803 cells compared with cells transfected with miR-control (Fig. 3B). Additionally, the effect on ZNF70 expression was reversed in gastric cells overexpressing miR-1269a variant mimics. Therefore, the findings indicated that ZNF70 is a target gene of miR-1269a and ZNF70 is involved in mediating the inhibitory effect of miR-1269a variant on gastric cancer cells.
Zinc finger proteins are a type of abundant proteins present in eukaryotic genomes. The functions of zinc finger proteins are extraordinarily diverse, including DNA recognition, RNA packaging, transcriptional activation, regulation of apoptosis, protein folding and assembly and lipid binding (29). For example, ZNF217 is an oncogenic protein that is involved in various types of human cancer and has a role in regulating transcription, and inducing positive transcriptional regulation of target gene (30). ZNF70, located at human chromosome 22, was reported to be a part of the transcription factor complex, which determines cell fate and participates in gene regulation, tissue differentiation and mammalian evolution (31). However, the specific function of ZNF70 in cancer remains unknown. In the present study, the findings indicated that ZNF70 is a target gene of miR-1269a. Furthermore, the results suggest that ZNF70 may serve as a tumor-suppressor gene in gastric cancer. Therefore, these results indicate that ZNF70 is involved in epigenetic regulation via miRNAs. The regulation of ZNF70 by miR-1269a is important in tumor development and progression and its downstream pathway would be of interest for future studies.
In conclusion, microRNA-1269a rs73239138 has a protective role in the susceptibility of gastric cancer. The primary mechanism may be that a decreased expression of miR-1269a variant increases the expression of the target tumor suppressor gene ZNF70 and decreases the risk of gastric cancer by upregulating apoptosis. To the best of our knowledge, the present study has, for the first time, revealed an important association between a miR-1269a SNP and gastric cancer progression. A potential molecular mechanism was elucidated, and ZNF70 was identified as a target gene of target gene of miR-1269a, which is also a tumor suppressor gene. The present study indicates an important role in cancer development for miRNAs and SNPs that are present in miRNAs. Furthermore, the present study indicates that rs73239138 is a potential predictive marker and therapeutic target for gastric cancer.
Acknowledgements
The present study was supported by grants from the Shanghai Municipal Commission of Health and Family Planning (grant no. 20134132), the Ministry of Science and Technology of China (grant no. 2013CB945401), the National Natural Science Foundation of China (grant no. 81300327) and the Ministry of Education of China (grant no. 20130071110048), the Key Discipline Construction Project of Pudong Health Bureau of Shanghai (grant no. PWZx2014-07) and the Shanghai Municipal Science and Technology Commission (grant no. 13ZR1437200).
Glossary
Abbreviations
- miRNA
microRNA
- FCM
flow cytometry
- mRNA
messenger RNA
- SNP
single nucleotide polymorphism
- oncomiR
oncogenic miRNA
- GO
gene ontology
- ZNF217
zinc-finger protein 217
- ZNF70
zinc-finger protein 70
- NC
negative control
- OR
odds ratio
- CI
confidence interval
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- siRNA
small-interfering RNA
References
- 1.Nagini S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4:156–169. doi: 10.4251/wjgo.v4.i7.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.PDQ Screening and Prevention Editorial Board: Stomach (Gastric) Cancer Prevention (PDQ®): Health Professional Version, corp-author. PDQ Cancer Information Summaries. National Cancer Institute (US); Bethesda (MD): 2002. [Google Scholar]
- 3.Li J, Zhang S, Liu J, Shao M, Chen L. Review of clinical investigation on recurrence of gastric cancer following curative resection. Chin Med J. 2012;125:1479–1495. [PubMed] [Google Scholar]
- 4.Roukos DH, Kappas AM. Limitations in controlling risk for recurrence after curative surgery for advanced gastric cancer are now well-explained by molecular-based mechanisms. Ann Surg Oncol. 2001;8:620–621. doi: 10.1007/s10434-001-0620-5. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R, Su B. Small but influential: The role of microRNAs on gene regulatory network and 3′UTR evolution. J Genet Genomics. 2009;36:1–6. doi: 10.1016/S1673-8527(09)60001-1. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143(35–47):e2. doi: 10.1053/j.gastro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Mizuno K, Seki N, Mataki H, Matsushita R, Kamikawaji K, Kumamoto T, Takagi K, Goto Y, Nishikawa R, Kato M, et al. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int J Oncol. 2016;48:450–460. doi: 10.3892/ijo.2015.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song S, Ajani JA. The role of microRNAs in cancers of the upper gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2013;10:109–118. doi: 10.1038/nrgastro.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- 12.Zheng H, Song F, Zhang L, Yang D, Ji P, Wang Y, Almeida M, Calin GA, Hao X, Wei Q, et al. Genetic variants at the miR-124 binding site on the cytoskeleton-organizing IQGAP1 gene confer differential predisposition to breast cancer. Int J Oncol. 2011;38:1153–1161. doi: 10.3892/ijo.2011.940. [DOI] [PubMed] [Google Scholar]
- 13.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XW, Shen GZ, Cao LQ, Jiang XF, Peng HP, Shen G, Chen D, Xue P. MicroRNA-1269 promotes proliferation in human hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer. 2014;14:909. doi: 10.1186/1471-2407-14-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Kristine Varanko A, Xu Y, et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nat Commun. 2015;6:6879. doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, Loman N, Jonsson G, Naya H, Hoglund M, et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71:78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y, Wang R, Zhang J, Li W, Gao C, Liu J, Wang J. Identification of miR-423 and miR-499 polymorphisms on affecting the risk of hepatocellular carcinoma in a large-scale population. Genet Test Mol Biomarkers. 2014;18:516–524. doi: 10.1089/gtmb.2013.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: A comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie L, Shen Y, Franke D, Sebastian V, Bawendi MG, Jensen KF. Characterization of indium phosphide quantum dot growth intermediates using MALDI-TOF mass spectrometry. J Am Chem Soc. 2016 doi: 10.1021/jacs.6b06468. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Tang N, Hui T, Wang S, Zeng X, Li H, Ma J. Identification of endogenous reference genes for RT-qPCR analysis of plasma microRNAs levels in rats with acetaminophen-induced hepatotoxicity. J Appl Toxicol. 2013;33:1330–1336. doi: 10.1002/jat.2864. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Nie Y, Du Y, Cao J, Shen B, Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour Biol. 2012;33:1589–1597. doi: 10.1007/s13277-012-0414-3. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Lv C, Jin H, Xu M, Kang M, Chu H, Tong N, Wu D, Zhu H, Gong W, et al. A common genetic variation in the promoter of miR-107 is associated with gastric adenocarcinoma susceptibility and survival. Mutat Res. 2014;769:35–41. doi: 10.1016/j.mrfmmm.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Fu B, Song P, Lu M, Wang B, Zhao Q. The association between miR-146a gene rs2910164 polymorphism and gastric cancer risk: A meta-analysis. Biomed Pharmacother. 2014;68:923–928. doi: 10.1016/j.biopha.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Cai M, Zhang Y, Ma Y, Li W, Min P, Qiu J, Xu W, Zhang M, Li M, Li L, et al. Association between microRNA-499 polymorphism and gastric cancer risk in Chinese population. Bull Cancer. 2015;102:973–978. doi: 10.1016/j.bulcan.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 29.Laity JH, Lee BM, Wright PE. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 30.Cohen PA, Donini CF, Nguyen NT, Lincet H, Vendrell JA. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget. 2015;6:41566–41581. doi: 10.18632/oncotarget.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]