Abstract
The present study aimed to identify the molecular mechanisms of microRNA-124 (miRNA-124/miR-124) in hepatoblastoma. The GSE6207 microarray dataset, obtained from the Gene Expression Omnibus database, included samples extracted from HepG2 cells transfected with miR-124 duplex (the experimental group) or negative control (the control group) at 4, 8, 16, 24, 32, 72 and 120 h after transfection. Differentially expressed genes (DEGs) were screened between the two groups. miR-124 activity was inferred based on the expression of its target genes. The mRNAs targeted by miR-124 were predicted and a miR-124-target mRNA network was constructed. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed for the target genes. The number of DEGs was highest at 72 h. The experimental group had higher miR-124 activity than that of the control group at 4, 8, 16, 24 and 120 h. Small GTPase-mediated signal transduction and Ras protein signal transduction were significant GO terms enriched with syndecan binding protein (SDCBP), Ras homolog family member G (RHOG) and Rho-GDP dissociation inhibitor-α (ARHGDIA). Regulation of actin cytoskeleton, D-glutamine and D-glutamate metabolism, and axon guidance were significant pathways. Axon guidance pathway was associated with neuropilin (NRP1), MET proto-oncogene, receptor tyrosine kinase (MET) and semaphorin 7A, GPI membrane anchor (SEMA7A). Small GTPase-mediated signal transduction, Ras protein signal transduction, regulation of actin cytoskeleton pathway, D-glutamine and D-glutamate metabolism pathway, axon guidance pathway, SDCBP, RHOG, ARHGDIA, NRP1, SEMA7A, and MET may be implicated in the underlying mechanisms of miR-124 overexpression in hepatoblastoma.
Keywords: HepG2 cells, microRNA-124, Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, differentially expressed genes
Introduction
Liver cancer is the sixth-leading cause of cancer and the second-leading cause of cancer-associated mortality worldwide (1), leading to 746,000 mortalities in 2012 (1). Hepatoblastoma, an embryonal malignancy originated in hepatocytes, is the most common primary liver tumor in children, and is usually diagnosed in the first 3 years of life (2). The incidence of hepatoblastoma in children has gradually increased, and the overall 5-year survival rate in children is ~70% (3,4). The most common manifestation of hepatoblastoma is abdominal distension or presence of a mass (2). Hepatoblastoma can generally be cured via tumor resection if the tumor is single, smaller and non-metastasic, but a number of children cannot be cured due to a late diagnosis, multiple subtypes and metastases, which cause a poor prognosis (5). Therefore, it is necessary to investigate the molecular mechanisms of hepatoblastoma and lay the basis for refining the current diagnosis or treatment interventions. The HepG2 cell line is originated from hepatoblastoma (6); thus, HepG2 cells were selected in the present study to investigate the molecular mechanisms of hepatoblastoma.
MicroRNAs (miRNAs/miRs) are small non-coding RNA molecules involved in diverse cell activities, including regulation of cell proliferation, differentiation, apoptosis and carcinogenesis (5,6). miR-124 is the most abundant miRNA in the adult brain and serves a tumor-suppressive role in diverse types of cancer, including those of the breast and prostate (7,8). miR-124 is potentially involved in liver cancer: miR-124 suppresses aggressive hepatocellular carcinoma by repressing Rho-associated, coiled-coil containing protein kinase 2 and enhancer of zeste 2 polycomb repressive complex 2 subunit (9). miR-124 overexpression appears to repress the migration and invasion of intrahepatic cholangiocarcinoma cells in vitro (10). This evidence indicates that miR-124 may serve a complicated tumor-suppressive role in hepatoblastoma.; however, its underlying molecular mechanism remains elusive.
The activity of miRNAs is negatively associated with the expression of their target mRNAs (11–13). On this basis, miR-124 transfection has been used to identify its downregulated target mRNAs by microarray time-course experiments (14). These target genes may be implicated in various biological functions and signaling pathways, thereby contributing to the progression of hepatoblastoma. To further investigate the underlying mechanisms of miR-124 overexpression in hepatoblastoma, the present study applied a range of bioinformatics approaches to a microarray dataset (GSE6207). Differentially expressed genes (DEGs) between human hepatoblastoma HepG2 cells transfected with a miR-124 duplex or a negative control RNA duplex at different time points (4, 8, 16, 24, 32, 72 and 120 h) were screened, followed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for the screened DEGs. Changes in miR-124 activity over the time course were also analyzed. miR-124-target mRNA pairs were then selected to construct miR-124-target mRNA networks for each time point. Gene ontology (GO) function and KEGG pathway enrichment analysis were performed for the targets genes of miR-124. The present study therefore provides additional data concerning the molecular mechanisms of miR-124 in hepatoblastoma.
Materials and methods
Microarray data and preprocessing
The present study used the GSE6207 microarray dataset, downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) repository based on the Affymetrix Human Genome U133 Plus 2.0 Array platform (14). It included RNA samples extracted from HepG2 cells transfected with a miRNA-124 duplex or a negative control, with one sample for each time point (4, 8, 16, 24, 32, 72 and 120 h). The gene expression data were sequentially preprocessed with background correction, quantile normalization and probe summarization using robust multiarray analysis (RMA) of the affy package in R (15). The expression values of several probes that correspond to a certain gene were averaged, and the averaged expression value was defined as the expression value of the gene.
DEG screening
DEGs at each time point were screened in the HepG2 cells transfected with the miRNA-124 duplex and those transfected with the negative control RNA duplex. As there was only one sample for each experimental condition at each time point, |log2 fold change| ≥0.58 was set as the strict cutoff (16). The expression values of DEGs at different time points were hierarchically clustered using Cluster software (version 3.0; http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm), which was originally developed by Michael Eisen while at Stanford University (17), and displayed in a heat map.
KEGG pathway enrichment analysis
To unravel the pathways involving the DEGs at each time point, KEGG pathway enrichment analysis was performed for the DEGs using Database for Annotation, Visualization and Integration Discovery (DAVID), with P<0.05 as the cutoff value (18).
Inferred activity of miR-124
The regulatory activity of miRNAs is negatively associated with the expression of their target mRNAs, which could be measured by microarray. When the expression of the target mRNA is downregulated, the activity of the miRNAs is enhanced, and when the target mRNA expression is upregulated, the activity of the corresponding miRNA is inhibited (12). On this basis, the miR-124 activity at each time point in the study was inferred by combing microarray expression data with miRNA target predictions, as described in a previous study [the number of permutations for binding vectors (k)=1,000] (12).
Construction of miRNA-target mRNA networks
The target mRNAs of miRNA-124 at each time point were predicted using five repositories: miRanda (http://www.microrna.org/microrna/home.do), MirTarget2 (http://nar.oxfordjournals.org/cgi/content/abstract/34/5/1646), PicTar (http://pictar.mdc-berlin.de/), PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) and TargetScan (http://www.targetscan.org/vert_71/) (19–23). These repositories provide information on the integration of miRNA target prediction with expression files. Only the miR-124-target mRNA pairs that were validated in at least three out of the five databases were selected and included in the miR-124-target gene network. Expression values of miRNA-124-target genes at different time points were hierarchically clustered using Cluster software. Furthermore, GO function (24) and KEGG pathway enrichment analysis were performed for functional annotation of the obtained target genes in the network, with P<0.05 as the strict threshold.
Results
Screening of DEGs at different time points
DEGs in HepG2 cells transfected with the miRNA-124 duplex and negative control were screened at 4, 8, 16, 24, 32, 72 and 120 h. As shown in Fig. 1, the total count of DEGs increased following transfection, reaching its peak at 72 h and then decreasing. Of the DEGs, there were more downregulated genes than upregulated genes at 4, 8, 16, 24, 32 and 72 h after transfection. The exception was 120 h, when there were more upregulated genes than downregulated genes. The expression data of all DEGs at seven time points is displayed in Fig. 2.
Figure 1.
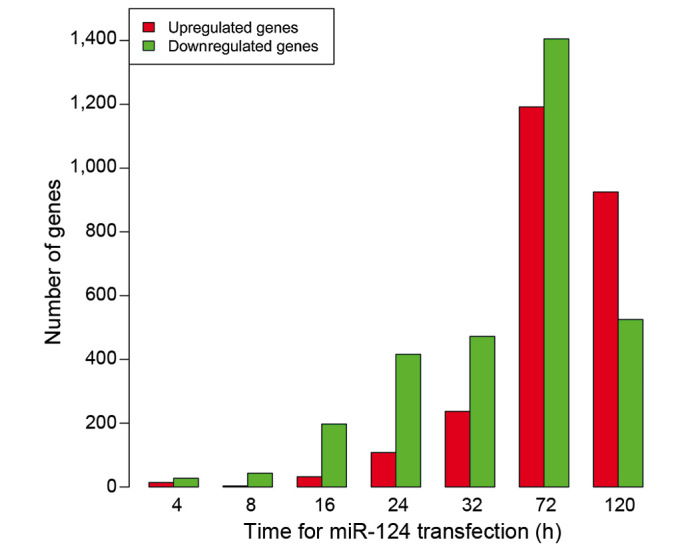
Number of DEGs at different time points. The total number of DEGs increased following transfection, peaking at 72 h and then decreasing. DEG, differently expressed gene; miR-124, microRNA-124.
Figure 2.
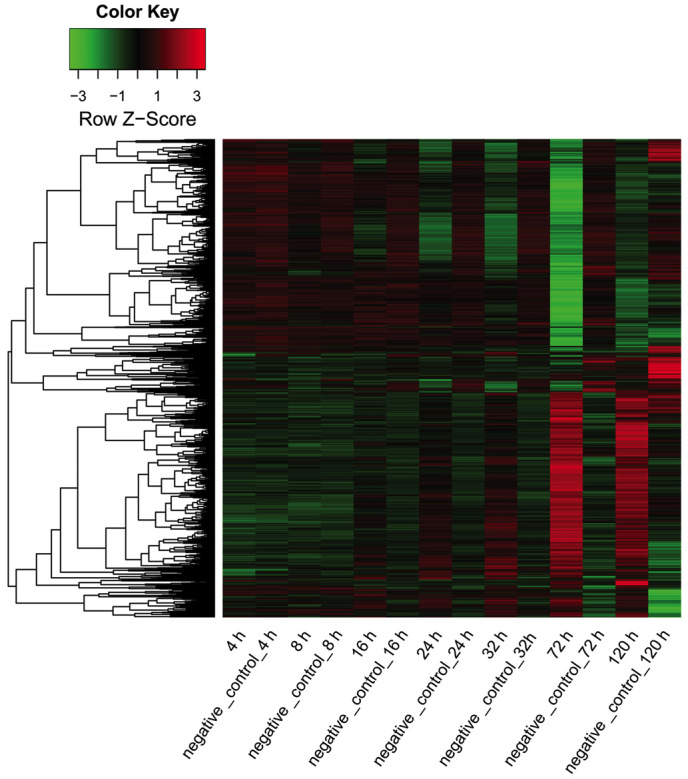
Heat map showing the expression of all differently expressed genes in the microRNA-124 transfection and negative control groups at various time points. Red area, upregulated genes; black area, genes with unaltered expression; green area, downregulated genes.
KEGG pathway enrichment analysis for DEGs
Pathways significantly enriched for upregulated DEGs at different time points are shown in Table I. No significant pathways were detected at 4, 8 or 16 h; genes involved in the tumor protein p53 signaling and retinol metabolism pathways were significantly associated with upregulated genes at 24 h after transfection. Expression of genes involved in the p53, systemic lupus erythematosus and mitogen-activated protein kinase (MAPK) signaling pathways were significantly upregulated at 32 h; those involved in axon guidance and adherens junction pathways were significantly upregulated at 72 h; those involved in glycerolipid metabolism and p53 signaling pathways were significantly upregulated at 120 h. With respect to downregulated DEGs (Table II), no significant pathway was detected at 4 or 16 h; those involved in the ribosome pathway (hsa03010: ribosome) were significantly downregulated at 8 h; those involved in small cell lung cancer, sphingolipid metabolism and the cell cycle were significantly downregulated at 24 h; those involved in small cell lung cancer, the cell cycle and pathways in cancer were significantly downregulated at 32 h; those involved in the cell cycle and DNA replication pathways were significantly associated with downregulated genes at 72 h; and those involved in the complement and coagulation cascades, cell cycle, and valine, leucine and isoleucine degradation pathways were significantly downregulated at 120 h.
Table I.
KEGG pathways enriched with upregulated differentially expressed genes at different time points.
| Time after transfection, h | KEGG pathway identity | Enriched genes, n | P-value |
|---|---|---|---|
| 24 | hsa04115:p53 signaling pathway | 5 | 1.00×10−3 |
| hsa00830:Retinol metabolism | 3 | 5.00×10−2 | |
| 32 | hsa04115:p53 signaling pathway | 5 | 1.02×10−2 |
| hsa05322:Systemic lupus erythematosus | 5 | 3.53×10−2 | |
| hsa04010:MAPK signaling pathway | 8 | 4.90×10−2 | |
| 72 | hsa04360:Axon guidance | 17 | 4.41×10−3 |
| hsa04520:Adherens junction | 12 | 6.03×10−3 | |
| hsa04144:Endocytosis | 20 | 1.44×10−2 | |
| hsa00051:Fructose and mannose metabolism | 7 | 1.48×10−2 | |
| hsa05130:Pathogenic Escherichia coli infection | 9 | 2.00×10−2 | |
| hsa04115:p53 signaling pathway | 10 | 2.00×10−2 | |
| hsa05410:Hypertrophic cardiomyopathy (HCM) | 11 | 3.09×10−2 | |
| hsa05412:Arrhythmogenic right ventricular cardiomyopathy | 10 | 3.81×10−2 | |
| hsa00983:Drug metabolism | 7 | 4.26×10−2 | |
| hsa05414:Dilated cardiomyopathy | 11 | 4.93×10−2 | |
| 120 | hsa00561:Glycerolipid metabolism | 8 | 4.35×10−3 |
| hsa04115:p53 signaling pathway | 9 | 1.30×10−2 | |
| hsa00140:Steroid hormone biosynthesis | 7 | 1.89×10−2 | |
| hsa04070:Phosphatidylinositol signaling system | 9 | 2.09×10−2 | |
| hsa05120:Epithelial cell signaling in Helicobacter pylori infection | 8 | 3.77×10−2 | |
| hsa00564:Glycerophospholipid metabolism | 8 | 3.77×10−2 | |
| hsa05211:Renal cell carcinoma | 8 | 4.32×10−2 | |
| hsa04650:Natural killer cell-mediated cytotoxicity | 12 | 4.49×10−2 | |
| hsa00983:Drug metabolism | 6 | 4.80×10−2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; hsa, Homo sapiens.
Table II.
KEGG pathways enriched with downregulated differentially expressed genes at different time points.
| Time after transfection, h | Term | Enriched genes, n | P-value |
|---|---|---|---|
| 8 | hsa03010:Ribosome | 3 | 2.30×10−2 |
| 24 | hsa05222:Small cell lung cancer | 9 | 7.02×10−4 |
| hsa00600:Sphingolipid metabolism | 5 | 1.25×10−2 | |
| hsa04110:Cell cycle | 8 | 2.65×10−2 | |
| hsa00565:Ether lipid metabolism | 4 | 4.75×10−2 | |
| 32 | hsa05222:Small cell lung cancer | 11 | 6.88×10−5 |
| hsa04110:Cell cycle | 10 | 5.95×10−3 | |
| hsa05212:Pancreatic cancer | 7 | 1.21×10−2 | |
| hsa05200:Pathways in cancer | 16 | 2.79×10−2 | |
| hsa00480:Glutathione metabolism | 5 | 4.37×10−2 | |
| 72 | hsa04110:Cell cycle | 46 | 7.63×10−20 |
| hsa03030:DNA replication | 20 | 1.43×10−12 | |
| hsa04914:Progesterone-mediated oocyte maturation | 19 | 8.17×10−5 | |
| hsa03040:Spliceosome | 24 | 8.48×10−5 | |
| hsa00240:Pyrimidine metabolism | 20 | 9.97×10−5 | |
| hsa03430:Mismatch repair | 9 | 2.18×10−4 | |
| 120 | hsa04610:Complement and coagulation cascades | 16 | 1.39×10−9 |
| hsa04110:Cell cycle | 11 | 4.75×10−3 | |
| hsa00280:Valine, leucine and isoleucine degradation | 6 | 1.05×10−2 | |
| hsa00260:Glycine, serine and threonine metabolism | 5 | 1.41×10−2 | |
| hsa05020:Prion diseases | 5 | 2.13×10−2 | |
| hsa00910:Nitrogen metabolism | 4 | 3.20×10−2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens.
Activity changes of miRNA-124
The changes in activity of miR-124 following transfection are shown in Fig. 3. The activity of miR-124 varied with time after transfection: It was highest at 4 h after transfection. The activity of miR-124 was higher in the experimental group than that in the control group at 4, 8, 16 and 24 h after transfection. By contrast, the experimental group exhibited decreased miR-124 activity compared with that of the control group at 72 h after transfection. Notably, at 32 h, the activity of miR-124 in the experimental group was similar to that in the control group.
Figure 3.
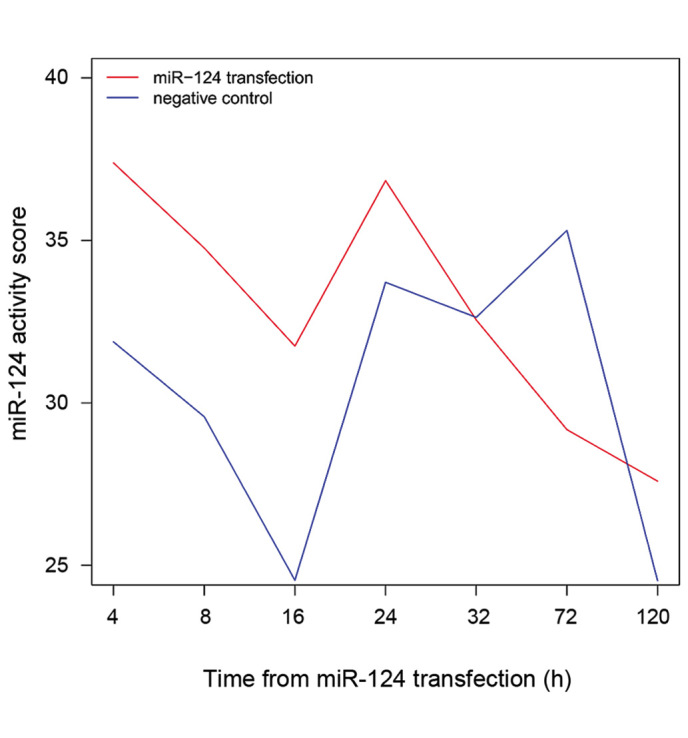
Changes in miR-124 activity across the time course following miR-124 transfection. The activity score of miR-124 in the miR-124 transfection group was highest at 4 h after transfection. miR-124, microRNA-124.
miR-124-target gene network analysis
The target mRNAs of miRNA-124 at each time point were predicted based on the aforementioned five repositories (miRanda, MirTarget2, PicTar, PITA and TargetScan). To investigate the association between miR-124 and its target genes, the miR-124-target gene pairs validated in at least three out of these five databases were used to construct miR-124-target gene networks for each time point. The expression values of all the predicted target genes at different time points are shown in a heat map (Fig. 4).
Figure 4.
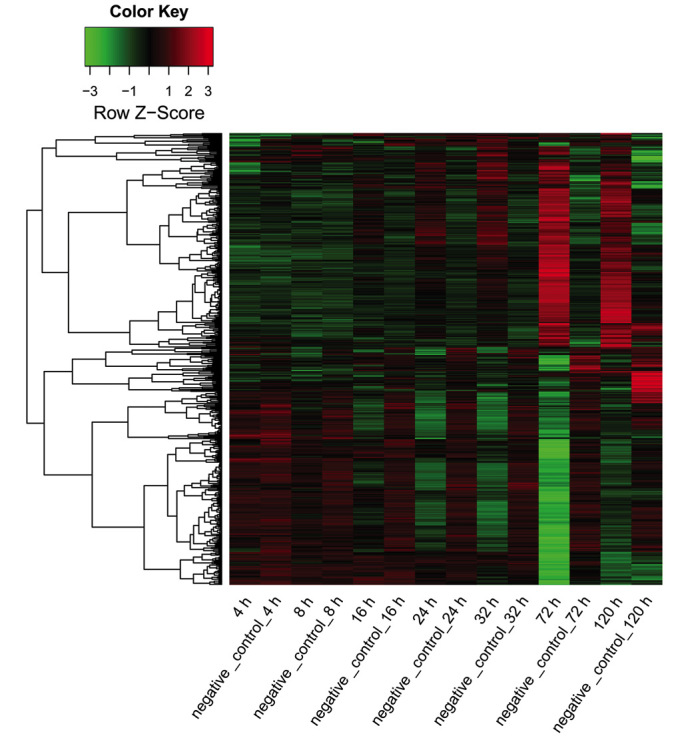
Heat map for the expression of the target genes of miR-124 in the miR-124 transfection and negative control groups at various time points. Red area, upregulated genes; black area, genes with unaltered expression; green area, downregulated genes. miR-124, microRNA-124.
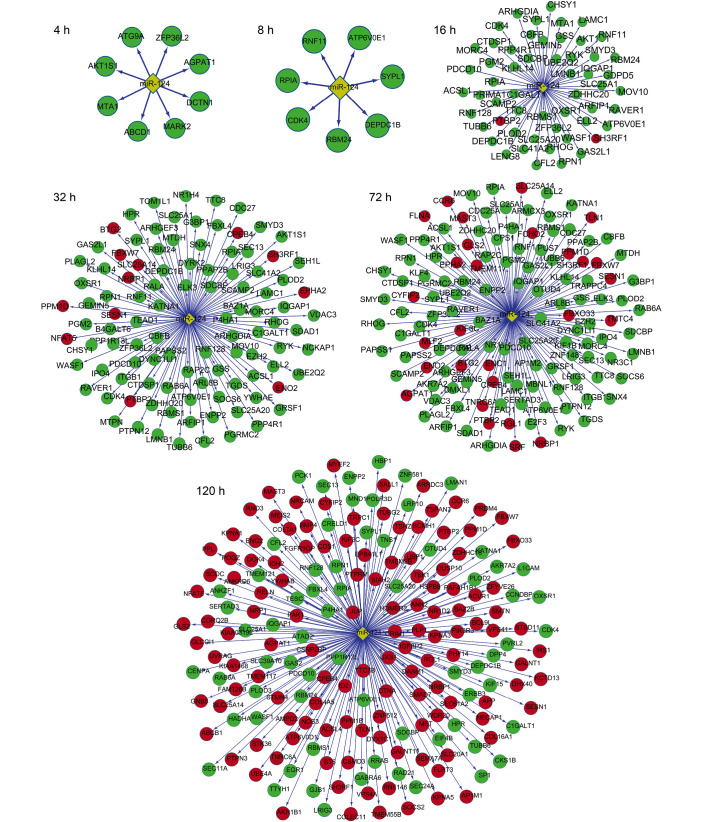
The miR-124-target gene networks are shown in Fig. 5. At 4 and 8 h after transfection, all miR-124-target genes were downregulated, indicating high miR-124 activity. At 16, 24 and 32 h, a small proportion of the target genes were upregulated, whereas the majority of the target genes were downregulated. By contrast, more upregulated genes were observed at 72 and 120 h when compared to that at 16, 24 and 32 h, indicating a reduction in miR-124 activity.
Figure 5.
miR-124-target gene networks at a range of different time points. Red nodes, upregulated genes; green nodes, downregulated genes; yellow diamond nodes, miR-124. miR-124, microRNA-124.
Functional annotation for target genes of miR-124
GO functional enrichment analysis was performed for miR-124-target genes at different time points. No significant GO term was enriched at 4 or 8 h. As shown in Table III, Ras protein signal transduction was a significant GO term at 16 h and was enriched with syndecan binding protein (SDCBP), Ras homolog family member G (RHOG) and Rho GDP dissociation inhibitor-α (ARHGDIA). At 24 h, small GTPase-mediated signal transduction and Ras protein signal transduction were primary GO terms, and the two were significantly enriched with SDCBP, RHOG and ARHGDIA, Rho-guanine nucleotide exchange factor 3 (ARHGEF3), GTPase activating protein (SH3 domain) binding protein 1 (G3BP1) and V-Ral simian leukemia viral oncogene homolog A (RALA). Similarly, at 32 h, the two GO terms were also enriched with the above six genes. Cell cycle and intracellular transport were significant GO terms at 72 h. A total of 44 genes were significantly associated with the cell cycle, such as kinesin family member 23 (KIF23), KIF15, cell division cycle 27 (CDC27) and CDC25A. A total of 38 genes were significantly involved in intracellular transport, such as adaptor-related protein complex 1, µ-1 subunit (AP1M1), adaptor-related protein complex 1, µ-2 subunit (AP1M2), KDEL endoplasmic reticulum protein retention receptor 1 (KDELR1) and KDELR2. At 120 h, cell motion was significantly associated with 17 genes, such as talin 1 (TLN1) and protein tyrosine phosphatase, receptor type, M (PTPRM), while intracellular transport was significantly associated with 18 genes, such as karyopherin-α (KPNA)5, KPNA3 and KPNA1.
Table III.
GO terms enriched with target genes of microRNA-124 at different time points.
| Time after transfection, h | Term | Enriched genes, n | P-value | Genes |
|---|---|---|---|---|
| 16 | GO:0007265~Ras protein signal transduction | 3 | 4.38×10−2 | SDCBP, RHOG, ARHGDIA |
| 24 | GO:0007264~small GTPase mediated-signal transduction | 10 | 1.47×10−4 | ARHGEF3, RAP2C, G3BP1, RALA, SDCBP, RAB6A, ARL8B, IQGAP1, RHOG, ARHGDIA |
| GO:0007265~Ras protein signal transduction | 6 | 5.71×10−4 | ARHGEF3, G3BP1, RALA, SDCBP, RHOG, ARHGDIA | |
| GO:0046907~intracellular transport | 11 | 9.37×10−3 | SLC25A20, SLC25A14, NRBP1, SCAMP2, TOM1L1, IPO4, SDCBP, SEC13, SLC25A1, ARFIP1, YWHAE | |
| GO:0006928~cell motion | 9 | 1.13×10−2 | ENPP2, WASF1, KATNA1, SDCBP, LAMC1, PPAP2B, ITGB1, YWHAE, ARHGDIA | |
| GO:0015031~protein transport | 11 | 2.41×10−2 | SDAD1, SCAMP2, TOM1L1, SEH1L, IPO4, SDCBP, SEC13, SNX4, RAB6A, ARFIP1, YWHAE | |
| 32 | GO:0007264~small GTPase mediated-signal transduction | 11 | 1.61×10−4 | ARHGEF3, RAP2C, G3BP1, RALA, SDCBP, RAB6A, ARL8B, IQGAP1, RHOG, ARHGDIA, RGL1 |
| GO:0007265~Ras protein signal transduction | 6 | 1.52×10−3 | ARHGEF3, G3BP1, RALA, SDCBP, RHOG, ARHGDIA | |
| GO:0006928~cell motion | 11 | 4.64×10−3 | TLN1, CCR6, ENPP2, WASF1, KATNA1, SDCBP, LAMC1, SRF, PPAP2B, ITGB1, ARHGDIA | |
| GO:0046907~intracellular transport | 13 | 6.11×10−3 | SCAMP2, NRBP1, AP1M2, ARFIP1, FLNA, SLC25A20, SLC25A14, KIF1B, IPO4, SDCBP, SEC13, TRAPPC4, SLC25A1 | |
| GO:0051329~interphase of mitotic cell cycle | 5 | 9.28×10−3 | PPM1D, KATNA1, CDK4, ITGB1, CDC25A | |
| 72 | GO:0007049~cell cycle | 44 | 3.69×10−7 | KIF23, CKS1B, E2F3, TUBB2B, CDC16, ITGB1, SESN1, CTNNB1, CCNE2, APP, NDE1, RAD21, DUSP13, SEH1L, CENPA, KATNA1, RB1CC1, TFDP2, VPS4A, PPP3CA, HELLS, CSRP2BP, NASP, KIF15, SUV39H1, SMAD3, MND1, ILF3, GAS2, MCM2, CDK4, CDC27, CDC25A, SMC4, MCM6, PPM1G, PA2G4, PPM1D, FANCD2, PLK1, GAS2L1, SETD8, SIAH2, ACVR1 |
| GO:0046907~intracellular transport | 38 | 1.79×10−6 | NUP98, AP1M1, AP1M2, SEC24A, NRBP1, EIF5A, BCL2L1, KLC2, LMAN1, SLC25A20, AP1S1, APP, AP2B1, NDE1, CSE1L, ZFYVE9, VPS4A, SLC25A1, PPP3CA, SAR1B, KDELR1, XPOT, GABARAPL2, KDELR2, SCAMP2, VPS41, ARFIP1, SLC25A12, SLC25A14, TOM1L1, IPO4, TRAPPC4, SEC13, SDCBP, KPNA5, KPNA3, TRAPPC1, SSR2 | |
| GO:0022402~cell cycle process | 33 | 8.30×10−6 | KIF23, TUBB2B, CDC16, ITGB1, SESN1, CTNNB1, APP, NDE1, RAD21, DUSP13, SEH1L, CENPA, KATNA1, PPP3CA, HELLS, CSRP2BP, KIF15, SMAD3, MND1, ILF3, GAS2, CDK4, CDC27, CDC25A, SMC4, PPM1G, PA2G4, PPM1D, FANCD2, PLK1, GAS2L1, SETD8, ACVR1 | |
| GO:0048193~Golgi vesicle transport | 13 | 8.58×10−5 | GABARAPL2, AP1M1, SCAMP2, NRBP1, SEC24A, AP1M2, VPS41, LMAN1, AP1S1, SEC13, TRAPPC4, TRAPPC1, SAR1B | |
| GO:0070727~cellular macromolecule localization | 24 | 2.12×10−4 | KDELR2, AP1M1, NUP98, SEC24A, AP1M2, EIF5A, VPS41, ARFIP1, CASC3, CTNNB1, AP1S1, AP2B1, CSE1L, TOM1L1, IPO4, ZFYVE9, SEC13, SDCBP, KPNA5, PPP3CA, SAR1B, KPNA3, KDELR1, SSR2 | |
| 120 | GO:0006928~cell motion | 17 | 1.86×10−4 | TLN1, NRP1, PTPRM, ENPP2, WASF1, MET, L1CAM, NRCAM, APP, CCR6, TNS1, KATNA1, SDCBP, NOS3, PAFAH1B1, RELN, ACVR1 |
| GO:0046907~intracellular transport | 18 | 2.27×10−3 | AP1M1, SEC24A, NRBP1, YWHAB, VPS41, LMAN1, SLC25A20, APP, SLC25A14, VPS4A, SDCBP, SEC13, SLC25A1, PAFAH1B1, KPNA5, KPNA3, KPNA1, SEC61A2 | |
| GO:0032355~response to estradiol stimulus | 5 | 3.94×10−3 | BMP4, SOCS2, ENO2, NOS3, IGFBP2 | |
| GO:0016477~cell migration | 10 | 6.03×10−3 | NRCAM, TNS1, NRP1, KATNA1, MET, SDCBP, NOS3, PAFAH1B1, RELN, ACVR1 | |
| GO:0006796~phosphate metabolic process | 22 | 6.12×10−3 | ATP6V0E1, NRBP1, PTPRM, PTPN3, SLC20A1, ENPP2, ERBB3, SMAD7, TBK1, STK36, MET, DUSP10, OXSR1, PPM1B, CDK4, MAST3, PPM1D, APP, RELN, PAK1, ATP6V0D1, ACVR1 |
GO, gene ontology.
The potential signaling pathways and target genes that may be involved in these signaling pathways were also analyzed (Table IV). No significant pathways were enriched with target genes at 4, 8 or 16 h. Regulation of the actin cytoskeleton pathway was a significant pathway at 24 h, and was enriched with several target genes, including cofilin 2 (CFL2) and WAS protein family, member 1 (WASF1). The target genes were predominantly associated with small cell lung cancer and ether lipid metabolism pathways at 32 h. At 72 h, the D-glutamine and D-glutamate metabolism pathway was significantly enriched with glutamate dehydrogenase 1 (GLUD1) and GLUD2. At 120 h, axon guidance, adherens junction and the transforming growth factor β signaling pathway were enriched. The axon guidance pathway was significant at 72 and 120 h, and was enriched with a number of target genes, such as neuropilin 1 (NRP1), MET proto-oncogene, receptor tyrosine kinase (MET) and semaphorin 7A, GPI membrane anchor (SEMA7A).
Table IV.
Kyoto Encyclopedia of Genes and Genomes pathways enriched with target genes of microRNA-124 at different time points.
| Time after transfection, h | Term | Enriched genes, n | P-value | Genes |
|---|---|---|---|---|
| 24 | hsa04810:Regulation of actin cytoskeleton | 5 | 4.87×10−2 | CFL2, WASF1, ITGB1, IQGAP1, NCKAP1 |
| 32 | hsa05222:Small cell lung cancer | 4 | 3.14×10−2 | E2F3, LAMC1, CDK4, ITGB1 |
| hsa00565:Ether lipid metabolism | 3 | 3.34×10−2 | ENPP2, PPAP2B, AGPAT1 | |
| 72 | hsa04360:Axon guidance | 15 | 6.34×10−6 | NRP1, MET, DPYSL2, ITGB1, EPHA2, SEMA6A, ROBO1, SEMA7A, SEMA3F, CFL2, NFAT5, PAK1, PPP3CA, NFATC3, RASA1 |
| hsa04110:Cell cycle | 14 | 2.20×10−5 | E2F3, E2F5, SMAD3, MCM2, CDC16, CDK4, CDC27, CDC25A, MCM5, MCM6, CCNE2, RAD21, PLK1, TFDP2 | |
| hsa05212:Pancreatic cancer | 9 | 5.74×10−4 | E2F3, RALBP1, RELA, ARAF, PIK3CD, SMAD3, RALA, BCL2L1, CDK4 | |
| hsa05222:Small cell lung cancer | 9 | 1.60×10−3 | CCNE2, CKS1B, E2F3, RELA, PIK3CD, LAMC1, BCL2L1, CDK4, ITGB1 | |
| hsa00471:D-Glutamine and D-glutamate metabolism | 3 | 4.05×10−3 | GLS2, GLUD2, GLUD1 | |
| 120 | hsa04360:Axon guidance | 7 | 1.29×10−2 | NRP1, CFL2, SEMA7A, MET, NFAT5, L1CAM, PAK1 |
| hsa04520:Adherens junction | 5 | 2.86×10−2 | PTPRM, WASF1, MET, PVRL2, IQGAP1 | |
| hsa04350:TGF-β signaling pathway | 5 | 4.21×10−2 | BMP4, ID2, SP1, SMAD7, ACVR1 |
hsa, Homo sapiens; TGF, transforming growth factor.
Discussion
miRNAs participate in a variety of cell activities via suppression of the expression of their target genes (25). The molecular mechanism by which miR-124 results in suppression of hepatoblastoma is not entirely clear. The present study revealed that the total number of DEGs in hepatoblastoma cells increased with time following miR-124 transfection, reached a peak at 72 h and then decreased, compared with the findings in cells treated with a negative control. miR-124 overexpression led to more downregulated genes than upregulated genes at 4, 8, 16, 24, 32 and 72 h, which was partly consistent with the fact that miR-124 inhibits the expression of its target genes (26). However, a proportion of DEGs were upregulated following miR-124 transfection. An explanation for this observation is that the upregulated DEGs may be the result of downregulation of DEGs by miR-124 overexpression. miRNAs may regulate the expression of other miRNAs (12,27), which could therefore strengthen or inhibit the activity of other endogenous miRNAs, thus contributing to differential expression of their target genes. These inhibited miRNAs may cause upregulation of their target genes.
Small GTPases are intracellular G proteins and hydrolase enzymes that hydrolyze guanosine triphosphate. They serve a key role in diverse cell activities, including cell growth, differentiation, movement and lipid vesicle transport (28,29). Ras protein family members belong to the small GTPase family. Ras is an intracellular anti-apoptotic protein that is closely associated with the phosphoinositide 3-kinase/AKT and MAPK signaling pathways, regulating various activities, including cell apoptosis and growth (30,31). Several lines of evidence have suggested that small GTPases have a regulatory role in hepatoblastoma (32,33). Consistent with this, the present study revealed that small GTPase-mediated signal transduction and Ras protein signal transduction were significant GO categories, enriched with SDCBP, RHOG and ARHGDIA in hepatoblastoma at 24 and 32 h. SDCBP encodes syntenin-1, which is a tandem PDZ domain-containing protein involved in trafficking, signaling and cancer metastasis (34). Syntenin-1 is implicated in regulating colon cancer cell migration (35). However, the role of SDCBP in hepatoblastoma has not been previously reported. RHOG encodes the Ras homology growth-related (RhoG) protein, which is a small GTP-binding protein and a member of the Rac subgroup of the Rho family (36). The Rac signaling pathways serve an essential role in the motility of hepatocellular carcinoma cells (37). ARHGDIA encodes Rho GDP-dissociation inhibitor 1 protein, which participates in regulating Rho activity. The absence of ARHGDIA has been reported to enhance the invasion and metastasis of hepatocellular carcinoma cells (37). However, the role of these genes in hepatoblastoma has not been reported thus far. Thus, the present study speculated that miR-124 overexpression may lead to the inhibition of small GTPase-mediated signal transduction and Ras protein signal transduction by regulating SDCBP, RHOG and ARHGDIA, thereby repressing hepatoblastoma development.
The present study unraveled that regulation of the actin cytoskeleton pathway was significantly enriched with several target genes such as CFL2 at 24 h after transfection. Regulation of the actin cytoskeleton pathway is involved in cancer cell migration, tumor invasion and metastasis (38). CFL2 is an intracellular actin-modulating protein that regulates actin-filament dynamics (39); it indicates that miR-124 overexpression may inhibit cancer cell migration, tumor invasion and metastasis by regulating actin cytoskeleton pathway genes such as CFL2. Furthermore, the present study revealed that the D-glutamine and D-glutamate metabolism pathways were enriched (GLUD1 and GLUD2) at 72 h. The D-glutamine and D-glutamate metabolic pathways are critical components of protein synthesis and cellular energy production (40,41). GLUD1 and GLUD2 are two mitochondrial matrix enzymes that serve a key role in glutamate metabolism and energy homeostasis (42). The findings of the present study revealed that miR-124 transfection may affect glutamate metabolism and energy homeostasis.
Notably, the expression of NRP1, SEMA7A and MET, which are involved in the axon guidance signaling pathway, was significantly increased at 72 and 120 h. An aberrant axon guidance pathway is involved in pancreatic carcinogenesis (43). Nevertheless, the role of the axon guidance pathway in hepatoblastoma has not been clearly characterized. Neuropilin-1, encoded by NRP1, is a co-receptor member for members of the semaphorin family, and is involved in several cellular processes, including angiogenesis and cell survival (44). A previous study revealed that the expression of NRP1 is higher in HepG2 cell lines than that in a human normal hepatic cell line, indicating the effect of NRP1 on hepatoblastoma (45). SEMA7A, which is encoded by SEMA7A, is a membrane-bound semaphorin and promotes axon outgrowth (46). MET encodes hepatocyte growth factor receptor (HGFR), which has tyrosine kinase activity and binds to HGF. It has been reported that invasion of hepatocellular carcinoma cells is dependent on HGF (47). Furthermore MET/HGF inhibitors have been increasingly recognized as anticancer therapies for various types of cancer, including hepatoma (48). These observations indicate that axon guidance pathway genes may be abnormally regulated due to miR-124 overexpression, thereby suppressing hepatoblastoma cell growth and invasion. The present study indicates the presence of an as-yet-undescribed role of the axon guidance pathway in hepatoblastoma.
The current study demonstrated that miR-124 overexpression may perturb small GTPase-mediated signal transduction and Ras protein signal transduction, and abnormally affect the regulation of the actin cytoskeleton, D-glutamine and D-glutamate metabolic and axon guidance pathways in hepatoblastoma. SDCBP, RHOG, ARHGDIA, NRP1, SEMA7A and MET may be promising molecular targets for therapies against hepatoblastoma. These pathways are not specific to hepatoblastoma, but may also be involved in other types of cancer. These findings provide useful information concerning the molecular mechanisms that underlie the tumor-suppressive role of miR-124 in hepatoblastoma. Further experimental studies are warranted to validate the findings of the present study.
References
- 1.Stewart BW, Wild CP. World cancer report 2014. World Health Organization. 2014 [Google Scholar]
- 2.Ringe B, Pichlmayr R, Wittekind C, Tusch G. Surgical treatment of hepatocellular carcinoma: Experience with liver resection and transplantation in 198 patients. World J Surg. 1991;15:270–285. doi: 10.1007/BF01659064. [DOI] [PubMed] [Google Scholar]
- 3.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Sherman M. American Association for the Study of Liver Diseases: Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyers RL, Tiao G, de Ville de Goyet J, Superina R, Aronson DC. Hepatoblastoma state of the art: Pre-treatment extent of disease, surgical resection guidelines and the role of liver transplantation. Curr Opin Pediatr. 2014;26:29–36. doi: 10.1097/MOP.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 6.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 7.Shi XB, Xue L, Ma AH, Tepper CG, Gandour-Edwards R, Kung HJ, deVere White RW. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2013;32:4130–4138. doi: 10.1038/onc.2012.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng H, Song F, Zhang L, Yang D, Ji P, Wang Y, Almeida M, Calin GA, Hao X, Wei Q, et al. Genetic variants at the miR-124 binding site on the cytoskeleton-organizing IQGAP1 gene confer differential predisposition to breast cancer. Int J Oncol. 2011;38:1153–1161. doi: 10.3892/ijo.2011.940. [DOI] [PubMed] [Google Scholar]
- 9.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 10.Zeng B, Li Z, Chen R, Guo N, Zhou J, Zhou Q, Lin Q, Cheng D, Liao Q, Zheng L, Gong Y. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012;586:3271–3278. doi: 10.1016/j.febslet.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 11.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 12.Cheng C, Li LM. Inferring microRNA activities by combining gene expression with microRNA target prediction. PLoS One. 2008;3:e1989. doi: 10.1371/journal.pone.0001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Bernhardt V, Hotchkiss MT, Garcia-Reyero N, Escalon BL, Denslow N, Davenport PW. Tracheal occlusion conditioning in conscious rats modulates gene expression profile of medial thalamus. Front Physiol. 2011;2:24. doi: 10.3389/fphys.2011.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon W, Culverhouse R, Duncan J. Analyzing microarray data using cluster analysis. Pharmacogenomics. 2003;4:41–52. doi: 10.1517/phgs.4.1.41.22581. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 20.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 21.Lall S, Grün D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 22.Wang X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weis SM, Cheresh DA. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat Med. 2011;17:1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 26.Pillai RS. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 29.Jaffe AB, Hall A. Rho GTPases: Biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimura T, Arimura N, Kawano Y, Kawabata S, Wang S, Kaibuchi K. Ras regulates neuronal polarity via the PI3-kinase/Akt/GSK-3beta/CRMP-2 pathway. Biochem Biophys Res Commun. 2006;340:62–68. doi: 10.1016/j.bbrc.2005.11.147. [DOI] [PubMed] [Google Scholar]
- 31.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 32.Wong CM, Yam JW, Ching YP, Yau TO, Leung TH, Jin DY, Ng IO. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- 33.Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beekman JM, Coffer PJ. The ins and outs of syntenin, a multifunctional intracellular adaptor protein. J Cell Sci. 2008;121:1349–1355. doi: 10.1242/jcs.026401. [DOI] [PubMed] [Google Scholar]
- 35.Lee H, Kim Y, Choi Y, Choi S, Hong E, Oh ES. Syndecan-2 cytoplasmic domain regulates colon cancer cell migration via interaction with syntenin-1. Biochem Biophys Res Commun. 2011;409:148–153. doi: 10.1016/j.cbpb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: Regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang L, Li Q, Huang LY, Li DW, Wang YW, Li XX, Cai SJ. Loss of ARHGDIA expression is associated with poor prognosis in HCC and promotes invasion and metastasis of HCC cells. Int J Oncol. 2014;45:659–666. doi: 10.3892/ijo.2014.2451. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maciver SK, Hussey PJ. The ADF/cofilin family: Actin-remodeling proteins. Genome Biol. 2002;3:reviews3007. doi: 10.1186/gb-2002-3-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aledo JC. Glutamine breakdown in rapidly dividing cells: Waste or investment? Bioessays. 2004;26:778–785. doi: 10.1002/bies.20063. [DOI] [PubMed] [Google Scholar]
- 41.Le Bacquer O, Laboisse C, Darmaun D. Glutamine preserves protein synthesis and paracellular permeability in Caco-2 cells submitted to ‘luminal fasting’. Am J Physiol Gastrointest Liver Physiol. 2003;285:G128–G136. doi: 10.1152/ajpgi.00459.2002. [DOI] [PubMed] [Google Scholar]
- 42.Spanaki C, Kotzamani D, Plaitakis A. Widening spectrum of cellular and subcellular expression of human Glud1 and Glud2 glutamate dehydrogenases suggests novel functions. Neurochem Res. 2017;42:92–107. doi: 10.1007/s11064-016-1986-x. [DOI] [PubMed] [Google Scholar]
- 43.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu C, Rodriguez ER, Reimert DV, Shu T, Fritzsch B, Richards LJ, Kolodkin AL, Ginty DD. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev Cell. 2003;5:45–57. doi: 10.1016/S1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Xia J. NRP-1 silencing suppresses hepatocellular carcinoma cell growth in vitro and in vivo. Exp Ther Med. 2013;5:150–154. doi: 10.3892/etm.2012.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasterkamp RJ, Peschon JJ, Spriggs MK, Kolodkin AL. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature. 2003;424:398–405. doi: 10.1038/nature01790. [DOI] [PubMed] [Google Scholar]
- 47.Monvoisin A, Neaud V, Lédinghen V, Dubuisson L, Balabaud C, Bioulac-Sage P, Desmoulière A, Rosenbaum J. Direct evidence that hepatocyte growth factor-induced invasion of hepatocellular carcinoma cells is mediated by urokinase. J Hepatol. 1999;30:511–518. doi: 10.1016/S0168-8278(99)80113-5. [DOI] [PubMed] [Google Scholar]
- 48.Scagliotti GV, Novello S, von Pawel J. The emerging role of MET/HGF inhibitors in oncology. Cancer Treat Rev. 2013;39:793–801. doi: 10.1016/j.ctrv.2013.02.001. [DOI] [PubMed] [Google Scholar]