Abstract
Hepatocellular carcinoma (HCC) remains one of the most common types of malignancy with high mortality and morbidity rates. Previous studies have suggested that microRNAs (miRs) serve pivotal functions in various types of tumor. The aim of the present study was to assess the association between miR-34a expression and HCC cell migration and invasion, and the potential underlying mechanisms. The miR-34a overexpression vector or scramble control was transfected into human Hep3B and Huh7 cell lines. Transwell assays, and Matrigel and wound healing assays were used to detect the effects of miR-34a expression on HCC cell invasion and migration, respectively. The expression of miR-34a and the mRNA expression of other associated proteins were detected using quantitative reverse transcription polymerase chain reaction, and protein levels were measured using western blot analysis. Compared with the control, miR-34a expression was significantly downregulated in Hep3B and Huh7 cells, but this was reversed by the transfection with exogenous miR-34a (P<0.01). The number of migrated or invaded cells was significantly reduced by the overexpression of miR-34a in Hep3B or Huh7 cells (P<0.01). The expression of sirtuin 1 was upregulated, while the level of acetylate-p53 was downregulated by overexpression of miR-34a. Taken together, the results of the present study suggested that the overexpression of miR-34a may have suppressed HCC metastasis via inhibited cell migration and invasion.
Keywords: hepatocellular carcinoma, microRNA-34a, cell migration, cell invasion, sirtuin 1
Introduction
Hepatocellular carcinoma (HCC) remains one of the most common malignancies, and is a leading cause of cancer-associated mortality due to its high mortality rate (1). Although previous studies have demonstrated that surgery is the first choice for HCC treatment, the diagnostic methods for HCC remain restricted due to hidden lesions and an increased metastatic rate (2,3). The 5-year survival rate of patients with HCC was 5–9% in the United States in 2009 (4). Therefore, it is important to evaluate biomarkers for HCC treatment.
microRNAs (miRNAs/miRs) are endogenous, highly conserved non-coding RNAs, 20–22 nucleotides in length, that function in a variety of biological processes at the transcriptional or post-transcriptional level by targeting the 3′-untranslated regions of genes (5). Previous evidence has suggested that various miRNAs are involved in the progression and underlying biology of HCC (6,7). For example, Tsai et al (8) indicated that the tumor suppressor miR-122 regulates HCC metastasis, and Meng et al (9) suggested that miR-21 expression is abnormal in HCC tissue and is correlated with HCC growth via regulation of cytochrome b6-f complex subunit 8, chloroplastic expression. Additionally, miR-34a is considered to be a tumor suppressor in various types of cancer, including colon and breast cancer, and to be involved in a variety of biological processes (9,10). Previous studies have revealed a significant correlation between miR-34a expression and cancer metastasis, including in HCC (11,12). However, few studies refer to the involvement of miR-34a in HCC metastasis. Li et al (13) indicated that miR-34a functions as an inhibitor for HCC cell migration and invasion via the downregulation of c-Met expression. Although several studies have suggested the involvement of miR-34a in HCC cell migration or invasion, the basic mechanism remains unknown.
In the present study, the potential effects of miR-34a expression on HCC cell migration and invasion were investigated using Hep3B and Huh7 cells. Comprehensive experimental methods were used to analyze the potential underlying mechanism.
Materials and methods
Cell lines and cell culture
The human hepatocellular carcinoma cell lines Hep3B and Huh7 and the normal hepatocyte cell line THLE-2 were obtained from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2.
Cell transfection
Cells were plated onto a 60-mm dish, and following incubation for 24 h, the overexpression vector for miR-34a (sense, 5′-UGGCAGUGUCUUAGCUGGUUGU-3′ and antisense, 5′-AACCAGCUAAGACACUGCCAUU-3′) (Sangon Biotech Co., Ltd., Shanghai, China) or the scramble control miRNA (sense, 5′-UGUCAGCUUUGGAGCUGGUUGU-3′ and antisense, 5′-AACCUAAGAUGCCACCAGCAUU-3′) was transfected into the Hep3B and Huh7 cells using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Inc.). Following transfection for 48 h at 37°C, cells were prepared for additional analysis.
Cell invasion assay
For cell invasion assay, the Transwell with Matrigel method was performed as previously described (14). Briefly, following transfection for 48 h, hepatocellular cells (5×105/ml) were re-suspended in serum-free DMEM containing 0.01% bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for an additional 24 h. The upper chamber of the Transwell plate was covered with serum-free DMEM supplemented with 50 mg/l Matrigel, and then air-dried at 4°C for 15 min. Then, 50 µl fresh serum-free DMEM medium containing 10 g/l BSA was added, and cultured for 30 min at 37°C. Following removal of this medium, 200 µl cell suspension was added into the upper chamber of the Transwell plate, and 600 µl complete DMEM culture supplemented with 10% FBS (Sigma-Aldrich; Merck KGaA). After 48 h of incubation at 37°C in 5% CO2, the Transwell plate was washed with PBS buffer to remove the cells on the upper side of the microporous membrane, followed with fixation in ice-cold alcohol for 30 min. Non-migratory cells were removed from the upper surface of the filter with a cotton swab. Finally, the cells were stained with 0.1% crystal violet for 30 min at room temperature, and then decolorized with 33% acetic acid. The absorbance of eluents was observed at an optical density of 570 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Cells transfected with the control vector served as the negative control.
Cell migration assay
For the cell migration assay, the wound healing asssay was performed as previously described (15). Hep3B and Huh7 cells (1×105) transfected with miR-34a overexpression or scramble controls were seeded in 24-well plates and grown overnight to achieve confluence. The monolayer cells were scratched using a 20 µl pipette tip to create the wound. The floating cells were removed by washing twice with PBS (Sigma-Aldrich; Merck KGaA). Subsequently, serum-free DMEM was added to permit wound healing. The rate of wound closure was assessed using images captured after 24 h. Image analysis was performed using Image-Pro Plus software version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA from the Hep3B or Huh7 cells was collected 48 h after transfection and isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., USA) as previously described (16). The samples were treated with RNase-free DNase I (Promega Corporation, Madison, WI, USA). Consequently, the concentration and purity for the isolated RNA were measured with SMA 400 UV-VIS (Merinton Instrument, Ltd., Beijing, China). Purified RNA at a density of 0.5 µg/µl with nuclease-free water was used for cDNA synthesis using the PrimerScript 1st Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.), with the following temperature protocol: 30°C for 10 min, 42°C for 50 min, and 95°C for 5 min. The expressions of targets in the cells were detected in an Eppendorf Mastercycler (Brinkmann Instruments, Westbury, NY, USA) using the SYBR ExScript RT-qPCR kit (Takara Bio, Inc., Otsu, Japan). The total reaction system of 20 µl volume was as follows: 1 µl cDNA from the above PCR, 10 µl SYBR Premix EX Taq, 1 µl each of the primers (10 µM), and 7 µl ddH2O. The PCR thermocycler conditions were as follows: Denaturation at 50°C for 2 min; 95°C for 10 min; followed by 45 cycles of 95°C for 10 sec and 60°C for 1 min. Melting curve analysis of amplification products was performed at the end of each PCR to confirm that only one product was amplified and detected. These data were quantified using the 2−ΔΔCq method (17). GAPDH was used as the internal control. Primers used for the amplification of the targets are listed in Table I.
Table I.
Primers used for target amplification in the present study.
| Name | Primer | Sequence (5′-3′) |
|---|---|---|
| GAPDH | Sense | GGGTGGAGCCAAACGGGTC |
| Antisense | GGAGTTGCTGTTGAAGTCGCA | |
| SIRT1 | Sense | CAGAGCAT CACACGCAAGC |
| Antisense | CAGGAAACAG AAACCCCAGC | |
| p53 | Sense | TTCCTCTTCCTGCAGTACTC |
| Antisense | ACCCTGGGCAACCAGCCCTGT |
SIRT, sirtuin; p53, tumor protein 53.
Western blot analysis
Cells cultured for 48 h were lysed with radioimmunoprecipitation buffer (Sangon Biotech Co., Ltd.) containing phenylmethanesulfonyl fluoride (Sigma-Aldrich; Merck KGaA), and the lysates were then centrifuged at 12,000 × g for 10 min at 4°C. The supernatants were collected, and protein concentrations were determined using a bicinchoninic protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). The proteins (30 µg/lane) were separated using SDS-PAGE on a 10% gel, as previously described (18) followed by transfer onto a polyvinylidene fluoride membrane (Merck KGaA). The membranes were blocked using Tris-buffered saline with 0.05% Tween-20 (TBST) containing 5% non-fat milk for 1 h at room temperature, and then incubated with rabbit anti-human antibodies [(purchased from Abcam (Cambridge, MA, USA)] against sirtuin 1 (SIRT1; cat. no. ab12193), tumor protein 53 (p53; cat. no. ab131442), acetylate-p53 (Ac-p53; cat. no. ab75754) and GAPDH (cat. no. ab37168; all 1:100) overnight at 4°C. Subsequently, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (cat. no. ab205718; 1:5,000; Abcam) for 1 h at room temperature. Finally, the polyvinylidene fluoride membranes were washed 3 times with 1X TBST buffer for 10 min each time. The signals were detected following incubation of the membranes with a chromogenic substrate using the SuperSignal® West Pico chemiluminescent western blotting substrate (Pierce; Thermo Fisher Scientific, Inc.). Analysis was performed using the Image Guage 4.0 program (Fuji, Tokyo, Japan). GAPDH served as the internal control.
Statistical analysis
All experiments were conducted independently 3 times. All data are presented as the mean ± standard deviation. The significant difference for the data was calculated using SPSS v19.0 statistical software. P-values were calculated using one-way analysis of variance followed by Duncan's multiple-range test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-34a in Hep3B and Huh7 cells
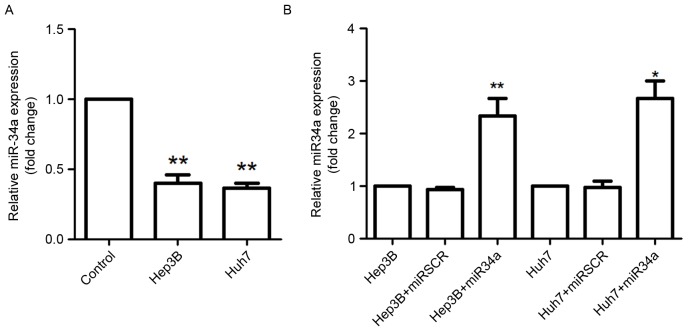
The relative expression levels of miR-34a in THLE-2, Hep3B and Huh7 cells were assessed using RT-qPCR analysis (Fig. 1). The results demonstrated that miR-34a expression was significantly decreased in Hep3B and Huh7 cells compared with that in the THLE-2 cells (P<0.01; Fig. 1A). The present study further analyzed the expression of miR-34a in Hep3B and Huh7 cells following transfection with miR-34a overexpression vectors, and the results suggested that miR-34a was highly expressed in these cells by miR-34a overexpression transfection compared with miR-34a expression in scramble control cells (Huh7 cells, P<0.05; Hep3B cells, P<0.01; Fig. 1B).
Figure 1.
Expression of miR-34a in the two types of hepatocellular cells. (A) miR-34a expression was downregulated in Hep3B and Huh7 cells. (B) Relative expression of miR-34a was significantly upregulated in Hep3B and Huh7 cells following the transfection of miR-34a overexpression vectors. *P<0.05, **P<0.01 vs. control group. miR, microRNA.
miR-34a overexpression suppresses hepatocellular cell migration
When cells were transfected with miR-34a overexpression vectors, the number of migrated cells was significantly decreased compared with that for the scramble control group (P<0.01; Fig. 2). There was no significant difference between the number of migrated cells for the negative control group and that for the scramble treated cells.
Figure 2.
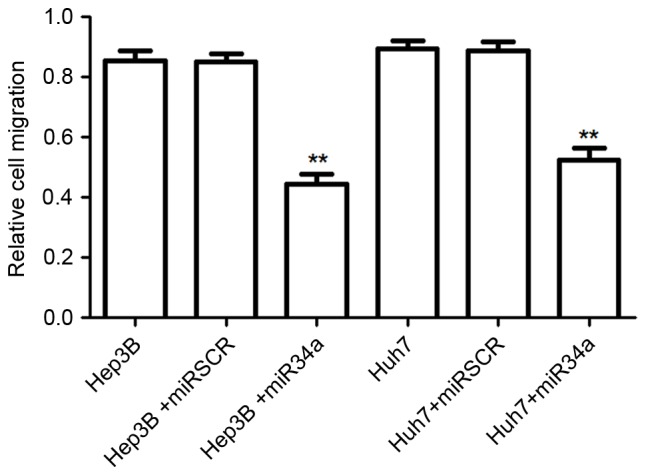
Effects of miR-34a on hepatocellular cell migration. Compared with the control cells, overexpression of miR-34a significantly decreased the number of migrated Hep3B and Huh7 cells. **P<0.01 vs. control cells. miR, microRNA; miRSCR, scramble miR-34a.
miR-34a overexpression suppresses hepatocellular cell invasion
The effect of miR-34a expression on hepatocellular cell invasion was also detected. The results suggested that the number of invasive cells was significantly decreased by the overexpression of miR-34a in Hep3B and Huh7 cells compared with that in the control group (P<0.01; Fig. 3). However, there was no significant difference in the number of invasive cells between control cells and the scramble treated cells.
Figure 3.
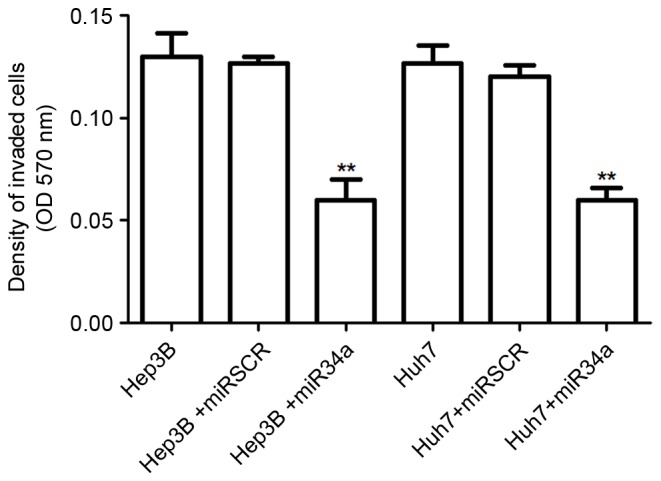
Effects of miR-34a on hepatocellular cell invasion. Compared with the control cells, the overexpression of miR-34a significantly decreased the number of invaded Hep3B and Huh7 cells. **P<0.01 vs. control cells. miR, microRNA; miRSCR, scramble miR-34a; OD, optical density.
Effects of miR-34a expression on cell metastasis-associated protein expressions
To further investigate the potential mechanism underlying the effect of miR-34a on hepatocellular cell migration and invasion, the expression of proteins including SIRT1, p53 and Ac-p53 were analyzed (Fig. 4). The results demonstrated that the SIRT1 mRNA and protein levels were significantly decreased by the overexpression of miR-34a (P<0.01; Fig. 4A and B). The Ac-p53 protein level was significantly increased by the overexpression of miR-34a in Hep3B and Huh7 cells, compared with that in control cells (P<0.05; Fig. 4A and B).
Figure 4.
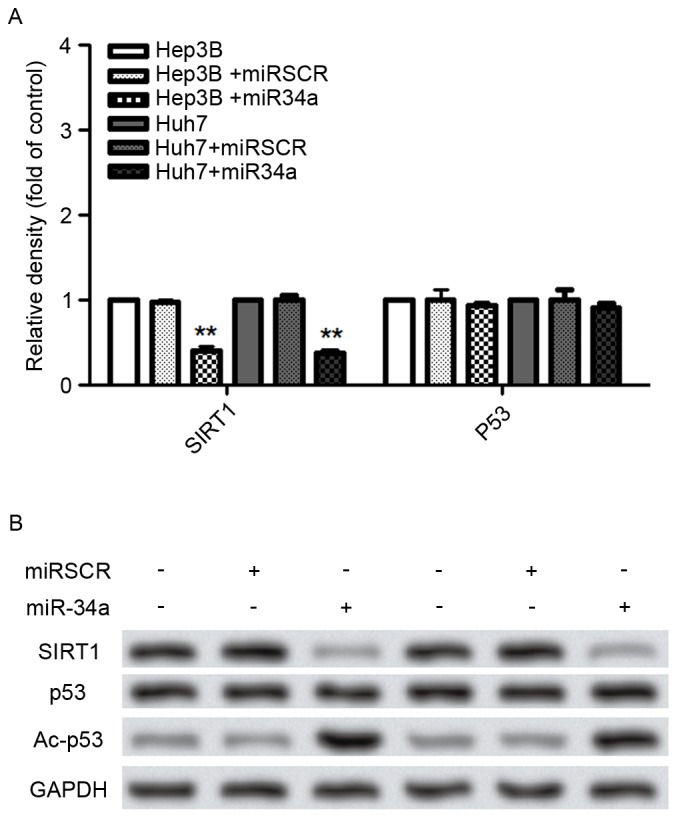
Effects of miR-34a expression on cell metastasis-associated protein expressions in hepatocellular cells. (A) Overexpression of miR-34a significantly decreased the mRNA levels of SIRT1. (B) Overexpression of miR-34a decreased the protein expression of SIRT1, but increased the expression of Ac-p53. **P<0.01 vs. control cells. miR, microRNA; miRSCR, scramble miR-34a; SIRT, sirtuin; p53, tumor protein 53, Ac-p53, acetylated p53.
Discussion
Previous studies have demonstrated that miRNAs serve crucial functions in tumor biology, and miR-34a has been suggested to be associated with HCC cell migration and invasion (6,19). The present study analysed the function of miR-34a expression in HCC cell migration and invasion, and its potential underlying mechanism. In agreement with previous data (20), the results indicated that miR-34a was downregulated in the two types of HCC cells. Following transfection with the miR-34a overexpression vector, the numbers of migrated and invaded cells were significantly decreased by the overexpression of miR-34a in Hep3B and Huh7 cells. Additionally, the protein and mRNA levels of SIRT1 were suppressed, while the protein levels for Ac-p53 were increased by overexpression of miR-34a.
These results demonstrated that the number of the migrated and invaded Hep3B or Huh7 cells was significantly reduced by the overexpression of miR-34a, suggesting that miR-34a expression was associated with HCC cell migration and invasion. Cell migration and invasion are important biological processes associated with tumor metastasis (21). Previous studies have demonstrated that miR-34a functions as a tumor suppressor in various tumors, including colon cancer and neuroblastoma (9,22). miR-34a has been suggested to be involved in the venous metastasis of Hepatitis B virus-positive HCC (23). Except for the study conducted by Li et al (13), the involvement of miR-34a expression in HCC cell migration and invasion has not been fully discussed. Based on the results of the present study, it was hypothesized that the overexpression of miR-34a may inhibit HCC metastasis through suppressing cell migration and invasion.
Concurrently, SIRT1 is an nicotine adenine dinucleotide-dependent deacetylase that is involved in multiple biological processes, including DNA damage, apoptosis and proliferation (24). A previous study demonstrated that SIRT1 expression is correlated with breast cancer invasion and metastasis (25), and Hao et al (26) suggested that the overexpression of SIRT1 promotes HCC metastasis through the epithelial mesenchymal transition. In the present study, SIRT1 expression was downregulated by the overexpression of miR-34a, indicating that miR-34a may suppress HCC metastasis via the downregulation of SIRT1. Conversely, Lewis et al (27) demonstrated that the absence of p53 promotes HCC metastasis in a mouse model. It has been revealed that deacetylated p53 is associated with cell growth (28). Ac-p53 is associated with breast cancer cell migration and invasion through the targeting of the SMAR1 gene (29). The results of the present study demonstrated that the overexpression of miR-34a increased Ac-p53 expression, suggesting that miR-34a may suppress HCC cell migration and invasion by increasing Ac-p53 expression.
Taken together, the results of the present study revealed that the overexpression of miR-34a may be a suppressor of HCC metastasis. The overexpression of miR-34a inhibits Hep3B and Huh7 cell migration and invasion, and downregulates SIRT1 expression while increasing Ac-p53 expression. The present study may provide a theoretical basis for studies investigating the mechanisms of HCC pathogenesis and metastasis, and for the potential application of miR-34a in HCC metastasis diagnosis. However, additional studies are needed to explore the basic underlying mechanisms.
References
- 1.Utsunomiya T, Shimada M, Kudo M, Ichida T, Matsui O, Izumi N, Matsuyama Y, Sakamoto M, Nakashima O, Ku Y, et al. Nationwide study of 4741 patients with non-B non-C hepatocellular carcinoma with special reference to the therapeutic impact. Ann Surg. 2014;259:336–345. doi: 10.1097/SLA.0b013e31829291e9. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Sherman M. American Association for the Study of Liver Diseases: Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 4.Martin RC, Scoggins CR, McMasters KM. Safety and efficacy of microwave ablation of hepatic tumors: A prospective review of a 5-year experience. Ann Surg Oncol. 2010;17:171–178. doi: 10.1245/s10434-009-0686-z. [DOI] [PubMed] [Google Scholar]
- 5.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 6.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 7.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 9.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 14.Adini A, Fainaru O, Udagawa T, Connor KM, Folkman J, D'Amato RJ. Matrigel cytometry: A novel method for quantifying angiogenesis in vivo. J Immunol Methods. 2009;342:78–81. doi: 10.1016/j.jim.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Ding C, Chen C, Zhang Z, Xiao H, Xie F, Lei L, Chen Y, Mao B, Jiang M, et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29:835–842. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- 16.Hummon AB, Lim SR, Difilippantonio MJ, Ried T. Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques. 2007;42(467–470):472. doi: 10.2144/000112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ish-Shalom S, Lichter A. Analysis of fungal gene expression by real time quantitative PCR. Methods Mol Biol. 2010;638:103–114. doi: 10.1007/978-1-60761-611-5_7. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Kuramitsu Y, Takashima M, Yokoyama Y, Iizuka N, Tamesa T, Sakaida I, Oka M, Nakamura K. Identification of four isoforms of aldolase B down-regulated in hepatocellular carcinoma tissues by means of two-dimensional Western blotting. In Vivo. 2011;25:881–886. [PubMed] [Google Scholar]
- 19.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 20.Pogribny IP, Starlard-Davenport A, Tryndyak VP, Han T, Ross SA, Rusyn I, Beland FA. Difference in expression of hepatic microRNAs miR-29c, miR34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest. 2010;90:1437–1446. doi: 10.1038/labinvest.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Condeelis J, Pollard JW. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 23.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng L, Yuan Z, Li Y, Ling H, Izumi V, Fang B, Fukasawa K, Koomen J, Chen J, Seto E. Ubiquitinated sirtuin 1 (SIRT1) function is modulated during DNA damage-induced cell death and survival. J Biol Chem. 2015;290:8904–8912. doi: 10.1074/jbc.M114.612796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung YR, Kim H, Park SY, Park IA, Jang JJ, Choe JY, Jung YY, Im SA, Moon HG, Lee KH, et al. Distinctive role of SIRT1 expression on tumor invasion and metastasis in breast cancer by molecular subtype. Hum Pathol. 2015;46:1027–1035. doi: 10.1016/j.humpath.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Hao C, Zhu PX, Yang X, Han ZP, Jiang JH, Zong C, Zhang XG, Liu WT, Zhao QD, Fan TT, et al. Overexpression of SIRT1 promotes metastasis through epithelial-mesenchymal transition in hepatocellular carcinoma. BMC Cancer. 2014;14:978. doi: 10.1186/1471-2407-14-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine AJ, Bargonetti J, Bond GL, Hoh J, Onel K, Overholtzer M, Stoffel AK, Walsh CA, Jin S. The p53 network. Protein Rev. 2005;2:1–23. doi: 10.1007/0-387-30127-5_1. [DOI] [Google Scholar]
- 28.Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107:815–818. doi: 10.1016/S0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- 29.Singh K, Mogare D, Giridharagopalan RO, Gogiraju R, Pande G, Chattopadhyay S. p53 target gene SMAR1 is dysregulated in breast cancer: Its role in cancer cell migration and invasion. PLoS One. 2007;2:e660. doi: 10.1371/journal.pone.0000660. [DOI] [PMC free article] [PubMed] [Google Scholar]