Abstract
It has been reported that LMO1 gene was associated with progression, metastasis and apoptosis of leukemia, colorectal cancer and lung cancer. However, the association of LMO1 and gastric cancer remains unclear. The aim of this study is to analyze the relation between LMO1 expression and apoptosis of gastric cancer cells and explore the clinical implications of LMO1 in gastric cancer tissues. The results demonstrated that expression levels of LMO1 and Bcl-2 proteins in gastric cancer tissues were higher than those in adjacent tissues, whereas the opposite was detected for Bax expression (P<0.05). LMO1 protein was associated with TNM staging and lymph node metastasis in gastric cancer (P<0.05). The survival rate of the patients with positive LMO1 gastric carcinoma was lower than that with negative LOM1 expression, and LMO1 was as an independent prognostic factor in COX survival analysis (P<0.05). LMO1-siRNA transfected MKN45 cells had a significant decrease in LMO1 expression and the cell viability, despite of an increase in the apoptotic rate (P<0.05). Following LMO1-siRNA transfection, Bcl-2 expression decreased, while the expression of Bax increased (P<0.05). It's concluded that overexpressed LMO1 in gastric cancer could be as one of new markers of poor prognosis.
Keywords: gastric cancer, LMO1 gene, clinicopathological features, RNA interference, apoptosis
Introduction
Gastric cancer is the second most common malignancy in China with highest mortality (1). Despite advanced techniques introduced in surgery, medications and targeted therapy, the overall survival rate and prognosis remains unsatisfactory in patients with gastric cancer (2,3). This may be caused by anti-apoptosis of cancer cells (4–7). In order to enhance prognosis and improve the outcome of treatment, effective suppression of the anti-apoptotic cells in gastric carcinoma may be needed. Therefore, it can be meaningful to identify new genes involved in apoptosis in gastric cancer. LMO1 gene has been reported to have a close relation with lelakemogenesis (8) with characteristics of proto-oncogene. However, little research has been done to explore LMO1 expression and also the mechanism of LMO1 in gastric cancer apoptosis remains ambiguous. In this study, in order to investigate the clinical value and the mechanism of LOM1 in gastric carcinoma, we examined the LOM1 expression level in gastric cancer tissues and cell lines, also the relation between LMO1 expression and clinicopathological features in patients with gastric cancer was investigated and the correlation between LOM1 gene and genes associated with apoptosis in terms of Bcl-2 and Bax was analyzed. Moreover, variations of cell viability, apoptosis rate, and Bcl-2, Bax genes were measured to determine the molecular mechanism of LMO1 in gastric cancer after LMO1-siRNA was synthesized and transfected to MKN45 gastric cancer cell line.
Materials and methods
Patients and tissue samples
Gastric cancer cases were enrolled from Hebei General Hospital (Shijiazhuang, China) between January 2011 and December 2011, diagnosed of gastric cancer and underwent surgery. A total of 96 patients were recruited consisting of 66 males and 30 females, with median age of 58 years (range, 32–81 years). Patients were considered eligible for the present study if they i) had a primary gastric cancer and underwent a tumorectomy; ii) had gastric cancer with no distant metastasis; and iii) had not be treatment of radiation/chemotherapy or targeted therapy before the tumorectomy. Patients were excluded if they met one of these criteria: i) had recurrent gastric cancer; ii) had gastric cancer with other malignancies; or iii) dissented to their participation. All paticipants had complete clinicopathological data and follow-ups for 60 months after surgery. The patient follow-ups ended in December 2015. Samples were obtained from tumor tissues and adjacent normal tissues (3 cm distance from the tumor margin without traces of cancerous cells under microscope). Paraffin-embedded samples were prepared for immunohistochemistry (IHC) after fixed with 10% polyoxymethylene. Fresh cancer and adjacent normal tissues of 20 gastric cancer patients, including 14 males and 6 females (median age of 56.5 years; range, 41–72 years), were collected from February 2017 to May 2017. Informed consent was obtained from participants, and approval from the Institutional Review Board at our hospital was attained.
Cell lines and reagents
Human gastric cancer cell lines including: Hixed gastric adenocarcinoma cells MKN28 (which are a derivative of MKN74 cells) (9), MKN45, SGC-7901, BGC823, MGC803 and human normal gastric epithelial cell lines GES-1 were purchased from Academy of Military Medical Science (Beijing, China). Rabbit anti-human LMO1, Bcl-2, Bax and GAPDH polyclonal antibody were the products of Santa Cruz Co. (Dallas, TX, USA). Horseradish peroxidase-labeled IgG antibodies and the BCA Protein Assay kit were purchased from Zhong Shan-Golden Bridge Biological Technology (Beijing, China). Fetal bovine serum (FBS), PRMI1640 medium, Lipofectamine™ 2000, TRIzol and Real-time PCR kits were supplied by Invitrogen (Carlsbad, CA, USA), while LMO1-siRNA, negative control siRNA and PCR primers were synthesized by GenePharma Co. (Shanghai, China). Cell Counting kit-8 (CCK8) was purchased from Boster Biological Technology (Wuhan, China).
IHC staining for detection of LMO1, Bcl-2 and Bax proteins in gastric cancer tissues and adjacent tissues
Paraffin-embedded tissue sections were dewaxed and rehydrated followed by incubation in 3% H2O2, washing and antigen-retrieval methods. Then IHC assay was applied according to the manufacturer's instruction which includes processes such as blocking, primary antibody incubating, washing and secondary antibody incubating followed by addition of horseradish enzyme labeled chain enzyme working solutions, and finally blotting, re-staining, dehydrating and mounting. Results were evaluated by pathological professionals. Five arbitrary visual fields under magnification, ×400 were selected with 100 cells counted for each field. IHC reactivity for LMO1, Bcl-2 and Bax was read by a scoring system based on both the quantity and quality of brown pigmentation in the plasma of cells. The presence of cells with yellow or brownish-yellow staining was identified as positive. If the positive rate was: <10%=0; 0–30%=1; 30–50%=2; >50%=3. The quality of staining of IHC was also scored if it was: clear=0; light yellow=1; yellow=2; brownish-yellow=3. By multiplication of two scores for each, the final score was calculated, of which if 0–1=(−); 2–3=(+); 4–6=(++); >6=(+++).
Cell culture
All cell lines were cultured in RPMI 1640 media supplemented with 10% FBS, 1% penicillin streptomycin combination in a 5% CO2 at 37°C. Media were changed every second day. Cells growing exponentially were selected for experiment. Experimental samples were divided into three groups according to the transfection status. The three groups were LMO1-siRNA transfection (LMO1-siRNA group), NS-siRNA transfection (control-siRNA group), and Lipofectamine™ 2000 (blank group). Each treatment repeated 3 triple.
LMO1-siRNA transfection and grouping
The sequence of LMO1-siRNA was synthesized as below: 5′-GGGCCCGAGACAAUGUGUAUtt-3′. Meanwhile, one fluorescent-labeled siRNA (NS-siRNA) was used as the negative control, and the sequence was as follows: 5′-UUCUCCGAACGUGUCACGUtt-3′. The MKN45 cells undergoing exponential growth phase were transplanted in 6-well plates with density of 2×105 cells/ml. Transfection was performed using the Lipofectamine™ 2000. Efficiency of transfection was tested under fluorescence microscope in 24 h after transfection. After 48 h, the cells were collected for experiment.
Cell viability of each group (CCK8 cell proliferation assay)
Cells (1×105 cell/well) were planted in 96-well plates and divided into 3 groups which were transfected with LMO1-siRNA, NS-siRNA and Lipofectamine™ 2000 reagent, respectively after 24-hour incubation. 48 h after the interference, 10 µl of CCK8 solution was added to each well of the plates. Bubbles should not to be introduced to the wells. The absorbance (optical density, OD) was measured at 450 nm with microplate reader. The assay was repeated 3 times.
Fluorescence quantitative real-time PCR (qRT-PCR) for the detection of targeted gene expression
Single-step RNA isolation method with TRIzol was applied to extract total RNA from cell or tissue samples. The RNA samples were used for synthesis of the complementary DNA (cDNA) template for qRT-PCR with 2 µg reverse transcription product. Gene expression was detected using SYBR-Green PCR mix (Toyoko) and 1 µg of the template. Reverse transcription PCR (RT-PCR) was performed with 2 µg reverse transcription product for the quantification of target mRNA levels, while GAPDH was employed as an internal standard reference. PCR was performed in a total volume of 20 µl containing 2 µl reverse transcription product, 10 µl of SYBR-Green Mix (Applied Biosystems, Foster City, CA) and 0.5 µl of sense an anti-sense primer each (10 µmol/l). PCR thermal cycling condition was optimized as following: 5 min initial denaturation at 95°C, followed by 45 cycles of 30 sec denaturation at 94°C, 30 sec annealing at 60°C and 30 sec extension at 70°C. The PCR primers used for experiment were designed using Primer 5.0 software with interpretation of the BLAST result to improve the specificity of the designed primers. The template sequences were as follows: LMO1 forward, 5′-TCTACACCAAGGCCAACCTC-3′ and reverse, 5′-CTGCCCTTCCTCATAGTCCA-3′; Bcl-2 forward, 5′-TGTGTGGAGAGCGTCAACC-3′ and reverse, 5′-TGGATCCAGGTGTGCAGGT-3′; Bax forward, 5′-TTTCTGACGGCAACTTCAAC-3′ and reverse, 5′-AGTCCAATGTCCAGCCCAT-3′; GAPDH forward, 5′-GACCCCTTCATTGACCTCAAC-3′ and reverse, 5′-CGCTCCTGGAAGATGGTGAT-3′. The RT-PCR result was examined adopting 1.5% agarose gel electrophoresis, while the result of real-time fluorescent quantitative PCR was analyzed using the 2−ΔΔCt method with GAPDH as internal reference of standard.
Western blot detection of protein expression of target genes
Tissue samples or cells were placed in ice with 100 µl lysis solution for 30 min, followed by 30 min spinning in a 4°C centrifuge at 12,000 × g. After that, bicinchoninic acid (BCA) assay was used for protein quantitation. Total 50 µg protein was loaded in 10% dodecyl sulfate, sodium salt-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to the poly vinylidene fluoride (PVDF) membrane. After 2 h blocking, LMO1, Bcl-2 and Bax antibodies were added followed by overnight incubation at 4°C. The blot was rinsed with TBST before and after the addition of HRP-Goat anti-mouse IgG with 1 h incubation at room temperature. Chemiluminescence was applied for color causing and imaging. The band integrated optical density value (IOD) of loaded proteins was then read. The ratios of IOD of each target proteins to that of the internal reference (GAPDH) were represented as the density of target proteins.
Statistical analysis
All assay results were analyzed using SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA). The individual relations of each protein in various tissues between their related clinicopathological characteristics were measured using χ2 test. The association of the proteins with each other was measured by the Spearman's rank-order correlation. Kaplan-Meier (K-M) survival analysis and COX multivariate model were employed for analysis of the relation between LMO1 expression and patient prognosis. Comparison of the cell viability, level of protein was evaluated with analysis of variance (ANOVA) or t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of LMO1, Bcl-2 and Bax in gastric cancerous and adjacent tissues
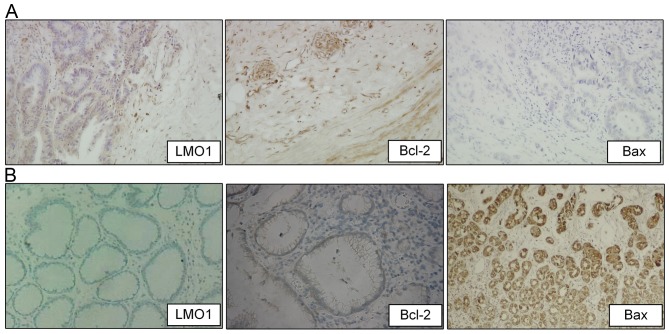
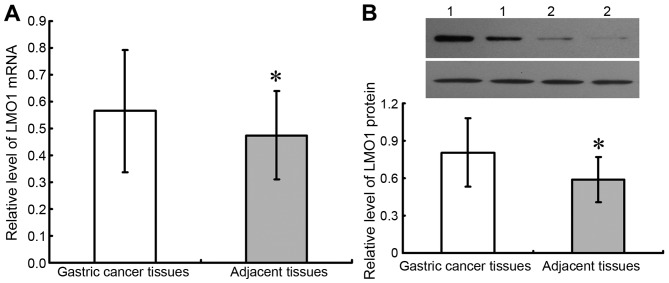
In gastric cancer tissues, the positive expression rates of LMO1, Bcl-2 and Bax protein were 75.00% (72/96), 71.88% (69/96) and 38.54% (37/96), respectively, compared to the rates of that in adjacent tissue samples at 28.00% (14/50), 30.00% (15/50), 76.00% (38/50), respectively. The result showed that LMO1 and Bcl-2 proteins had higher positive rates in gastric cancer tissue than that in adjacent tissue (P<0.05), whereas Bax saw a lower rate in gastric cancer tissue than it was in adjacent benign tissue (Table I; Fig. 1). Results of fresh tissues from qRT-PCR and western blot were demonstrated as Fig. 2, which showed that expressions of LMO1 mRNA and protein were higher in cancer tissues than in adjacent normal tissues (P<0.05).
Table I.
Expression of LMO1, Bcl-2, Bax proteins in gastric cancer and tumor-adjacent tissues.
| LMO1 | Bcl-2 | Bax | |||||
|---|---|---|---|---|---|---|---|
| Tissue type | n | Negative | Positive | Negative | Positive | Negative | Positive |
| Gastric cancer | 96 | 24 | 72 | 27 | 69 | 59 | 37 |
| Tumor-adjacent tissues | 50 | 36 | 14 | 35 | 15 | 12 | 38 |
| χ2 | 30.001 | 23.596 | 18.466 | ||||
| P-value | <0.001 | <0.001 | <0.001 | ||||
LMO1, LIM domain only 1; Bcl-2, apoptosis regulator Bcl-2; Bax, apoptosis regulator Bax.
Figure 1.
Expression of LMO1, Bcl-2, and Bax proteins in gastric cancer and adjacent tissues (Immunohistochemistry). Immunohistochemistry (magnification ×200) was utilized to detect expression of Bcl-2, and Bax protein in gastric cancer and adjacent tissues, and positive staining of LMO1, Bcl-2, and Bax proteins were located in cytoplasm. Expressions of LMO1, Bcl-2, and Bax proteins in cancer tissues were illustrated as (A), and in adjacent tissues were illustrated as (B). LMO1, LIM domain only 1; Bcl-2, apoptosis regulator Bcl-2; Bax, apoptosis regulator Bax.
Figure 2.
Expression of LMO1 mRNA and protein in gastric cancer and adjacent tissues (qRT-PCR and western blot). qRT-PCR and western blot were utilized to test expression of LMO1 mRNA and protein in fresh gastric cancer tissues and adjacent normal tissues, and it was demonstrated that levels of LMO1 mRNA (A) and protein (B) in cancer tissues were higher than in adjacent normal tissues. 1, Gastric cancer tissues; 2, Adjacent normal tissues. *P<0.05, compared with gastric cancer tissues. LMO1, LIM domain only 1; mRNA, messenger RNA.
Correlation of LMO1, Bcl-2 and Bax protein expression in gastric cancer tissues
The Spearman's rank-order correlation was applied to analyze protein expression of LMO1, Bcl-2 and BAX in gastric cancer cells. The results indicated a positive correlation between LMO1 and Bcl-2 protein (r=0.3024; P=0.003), whereas a negative one was shown between LMO1 and Bax protein (r=−0.2334; P=0.022). Bax protein expression was also negatively correlated to that of Bcl2 (r=−0.3445; P<0.001).
Association between LMO1, Bcl-2 and Bax protein and the clinicopathological characteristics in gastric cancer
The χ2 test result showed that LMO1 protein was associated with TNM cancer staging and lymphatic metastasis. Bcl-2 protein was shown in relation to the depth of tumor infiltration, while Bax protein was associated with lymphatic metastasis (P<0.05) (Table II).
Table II.
Relationship between LMO1, Bcl-2, Bax proteins and biological characteristics of gastric cancer patients.
| LMO1 | Bcl-2 | Bax | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Biological characteristics (n) | Negative (24) | Positive (72) | χ2 (P-value) | Negative (27) | Positive (69) | χ2 (P-value) | Negative (59) | Positive (37) | χ2 (P-value) |
| Sex | |||||||||
| Male (66) | 17 | 49 | 0.065 (0.799) | 17 | 49 | 0.586 (0.444) | 39 | 20 | 1.393 (0.238) |
| Female (30) | 7 | 23 | 10 | 20 | 20 | 17 | |||
| Age, years | |||||||||
| ≥60 (55) | 15 | 40 | 0.355 (0.551) | 17 | 38 | 0.494 (0.482) | 36 | 19 | 0.868 (0.351) |
| <60 (41) | 9 | 32 | 10 | 31 | 23 | 18 | |||
| Diameter, cm | |||||||||
| ≥5 (58) | 14 | 44 | 0.058 (0.810) | 14 | 44 | 1.152 (0.283) | 39 | 19 | 2.069 (0.150) |
| <5 (38) | 10 | 28 | 13 | 25 | 20 | 18 | |||
| Serosal invasion | |||||||||
| Negative (31) | 9 | 22 | 0.397 (0.529) | 13 | 18 | 4.320 (0.038) | 20 | 11 | 0.181 (0.671) |
| Positive (65) | 15 | 50 | 14 | 51 | 39 | 26 | |||
| TNM staging | |||||||||
| I–II (28) | 11 | 17 | 4.303 (0.038) | 10 | 18 | 1.126 (0.289) | 15 | 13 | 1.038 (0.308) |
| III (68) | 13 | 55 | 17 | 51 | 44 | 24 | |||
| Differentiation | |||||||||
| Well (67) | 16 | 51 | 0.148 (0.700) | 15 | 52 | 3.611 (0.057) | 40 | 27 | 0.289 (0.591) |
| Poor (29) | 8 | 21 | 12 | 17 | 19 | 10 | |||
| Lymphatic metastasis | |||||||||
| Positive (71) | 13 | 58 | 6.508 (0.011) | 18 | 53 | 1.037 (0.309) | 48 | 23 | 4.350 (0.037) |
| Negative (25) | 11 | 14 | 9 | 16 | 11 | 14 | |||
LMO1, LIM domain only 1; Bcl-2, apoptosis regulator Bcl-2; Bax, apoptosis regulator Bax; TNM, tumor node metastasis.
The relation between LMO1 protein and patient prognosis in gastric cancer
In the K-M survival analysis, patients with positive LMO1 expression had a lower survival rate than those with negative expression (χ2=9.025; P=0.003) (Fig. 3). Cox multivariate analysis suggests that LMO1 expression and TNM staging were independent risk factors of gastric cancer prognosis (P<0.05) (Table III).
Figure 3.
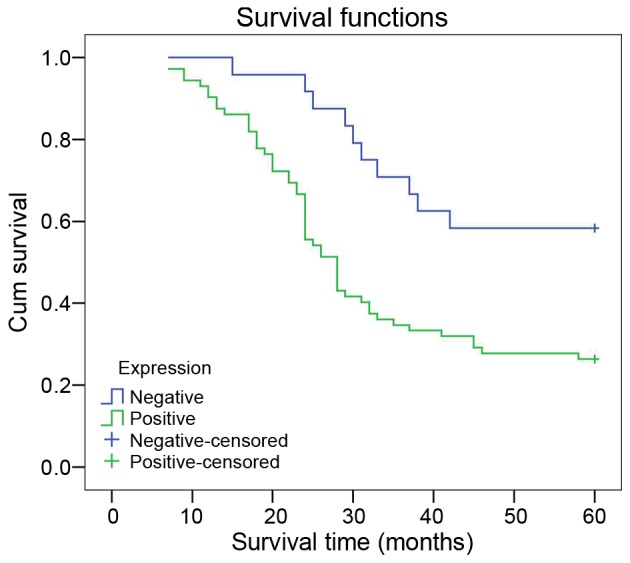
Kaplan-Meier survival curve of gastric cancer patients with different expression of LMO1 protein. It was illustrated that survival rate of gastric cancer patients with positive LMO1 protein was lower than those with negative LMO1 protein, and the Kaplan-Meier survival curve was displayed as Fig. 2. LMO1, LIM domain only 1.
Table III.
Results of Cox risk model for survival of gastric cancer patients.
| 95.0% CI for Exp(B) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | B | SE | Wald | df | Sig | Exp(B) | Lower | Upper |
| LMO1 expression | 1.082 | 0.425 | 6.477 | 1 | 0.011 | 2.952 | 1.283 | 6.794 |
| Lymphatic metastasis | −0.112 | 0.317 | 0.125 | 1 | 0.723 | 0.894 | 0.480 | 1.664 |
| TNM stages | 0.698 | 0.345 | 4.105 | 1 | 0.043 | 2.010 | 1.023 | 3.949 |
| Invasion | 0.143 | 0.306 | 0.218 | 1 | 0.640 | 1.153 | 0.634 | 2.099 |
| Sex | −0.264 | 0.314 | 0.707 | 1 | 0.400 | 0.768 | 0.415 | 1.420 |
| Age | −0.250 | 0.336 | 0.551 | 1 | 0.458 | 0.779 | 0.403 | 1.506 |
| Differentiation | 0.274 | 0.277 | 0.981 | 1 | 0.322 | 1.316 | 0.765 | 2.264 |
| Diameter | 0.090 | 0.313 | 0.082 | 1 | 0.775 | 1.094 | 0.592 | 2.019 |
LMO1, LIM domain only 1; TNM, tumor node metastasis; df, degrees of freedom; SE, standard error; CI, confidence interval; Sig, significance; Exp(B), exponentiation of the B coefficient.
Expression of LMO1 mRNA and protein in gastric cell lines
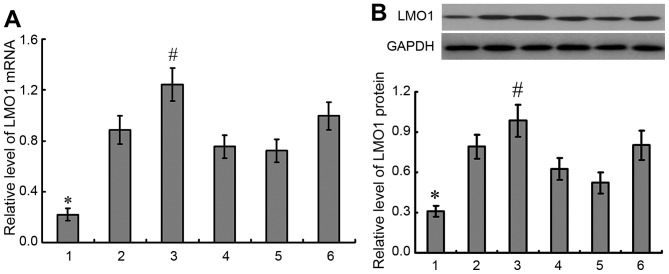
Expressions of LMO1 mRNA and protein were detected in 5 gastric cancer cell lines and gastric epithelial cell line GES-1, in which higher expressions of LMO1 mRNA and protein were demonstrated in gastric cancer cell lines than in GES-1 cells, and highest expressions of LMO1 mRNA and protein were detected in MKN45 cells (P<0.05) (Fig. 4). Then MKN45 cell line was selected for further studies.
Figure 4.
Expression of LMO1 mRNA and protein in gastric cell lines. A total of 5 gastric cancer cell lines and gastric epithelial cell line GES-1 were selected to detected expression of LMO1 gene with qRT-PCR (A) and western blotting (B). It was demonstrated that there was higher expression of LMO1 in gastric cancer cell lines than in GES-1 cells, and highest expression of LMO1 gene was detected in MKN45 cells. 1, GES-1; 2, mixed gastric adenocarcinoma cells MKN28 (which are a derivative of MKN74 cells); 3, MKN45; 4, SGC-7901; 5, BGC823; 6, MGC803. *P<0.01, compared with gastric cancer cell lines; #P<0.01, compared with other gastric cell lines. LMO1, LIM domain only 1.
Inhibitory effect of LMO1-siRNA on LMO1 in MKN45 cells
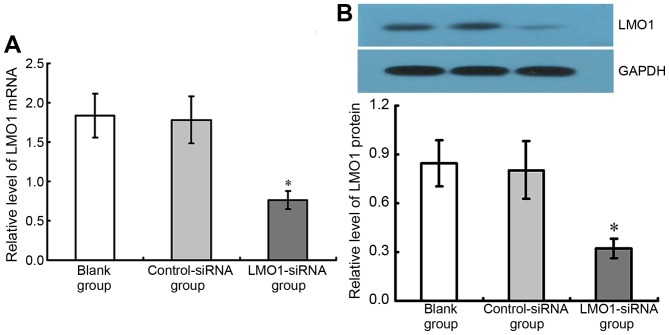
48 h after transfection of LMO1-siRNA (40 µmol/l), mRNA and protein expression levels of LMO1 in MKN45 cells, with high expression of LMO1 protein, were detected using RT-qPCR (Fig. 4A) and western blotting (Fig. 4B). The results show that the group with LMO1-siRNA transfection had significant lower LMO1 mRNA and protein expression rates than Control-siRNA group and Blank group (P<0.05). The Control-siRNA group and Blank group presented no significant difference in their LMO1 expression (P<0.05) (Fig. 5).
Figure 5.
Effect of LMO1-siRNA on LMO1 gene of MKN45 cells. LMO1-siRNA was transfected into MKN45 cells, and the variation of LMO1 (A) mRNA and (B) protein expression was demonstrated. It was illustrated that level of LMO1 was significantly downregulated after LMO1-siRNA transfection (P<0.05). *P<0.05 vs. Blank group or Control-siRNA group. LMO1, LIM domain only 1; mRNA, messenger RNA.
Change in cell viability of MKN45 cells after LMO1-siRNA transfection
The results of CCK8 assay indicate that the cell viability in the group treated with LMO1-siRNA transfection (40 µmol/l) was significantly lower than that in Control-siRNA group and Blank group (Fig. 6). 48 h after interference, LMO1-siRNA group saw a relative OD value at 0.598±0.097, while the value of Control-siRNA group and Blank group was 0.977±0.162 and 1.162±0.140, respectively.
Figure 6.
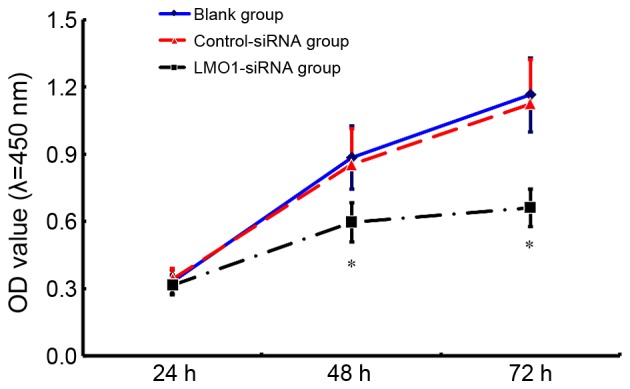
Variation in viability of MKN45 cells after of LMO1-siRNA transfection. The viability of MKN45 cells varied with different time after ANXA9-siRNA transfection (Fig. 5). Compared with Control-siRNA group and Blank group, viability of MKN45 cells treated with LMO1-siRNA group decreased more significantly. *P<0.05 vs. Blank group or Control-siRNA group. LMO1, LIM domain only 1.
Variation in cell apoptosis of MKN45 cells after LMO1-siRNA transfection
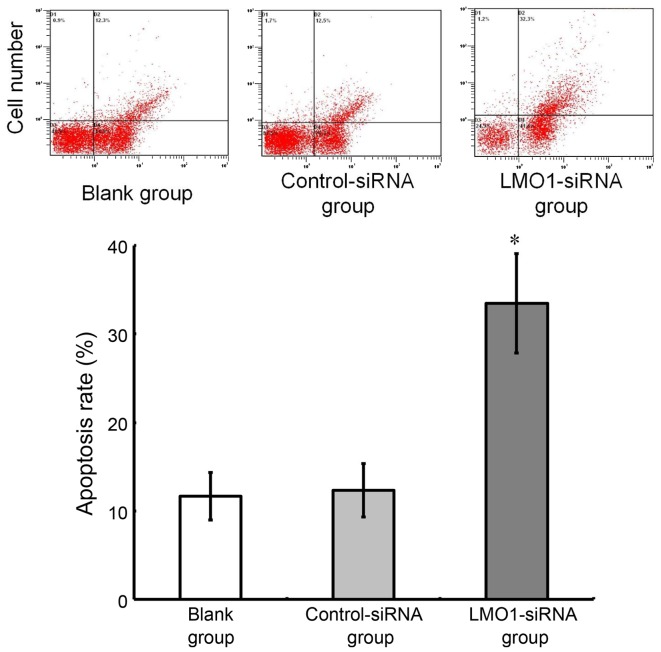
As analyzed using flow cytometry, the cell apoptotic rate was 31.46±5.22% in LMO1-siRNA group, while the rate in Control-siRNA group and Blank group was 13.45±1.51% and 12.68±1.32%, respectively. The cell apoptotic rate was higher in the LMO1-siRNA group than that in Control-siRNA group and Blank group (P>0.05). The Control-siRNA group and Blank group showed no significant difference in the cell apoptotic rate (P>0.05) (Fig. 7).
Figure 7.
Effect of LMO1-siRNA on apoptosis of MKN45 cells. MKN45 cells were transfected with LMO1-siRNA, and then were subjected to flow cytometry (FCM). Images of cell apoptosis were illustrated as Fig. 6A, and result of comparison was demonstrated Fig. 6B, which showed that apoptosis rate of MKN45 cells increased significantly after LMO1-siRNA transfection. *P<0.05 vs. Blank group or Control-siRNA group. LMO1, LIM domain only 1.
Expression of Bcl-2 and Bax mRNA and protein in MKN45 cells transfected with LMO1-siRNA
The results of qRT-PCR and western blot assay demonstrate that Bcl-2 protein expression considerably decreased in LMO1-siRNA treated group comparted with Control-siRNA group and Blank group, whereas Bax was found the contrary (P<0.05). There was no significant difference between Control-siRNA group and Blank group (P<0.05) (Fig. 8).
Figure 8.
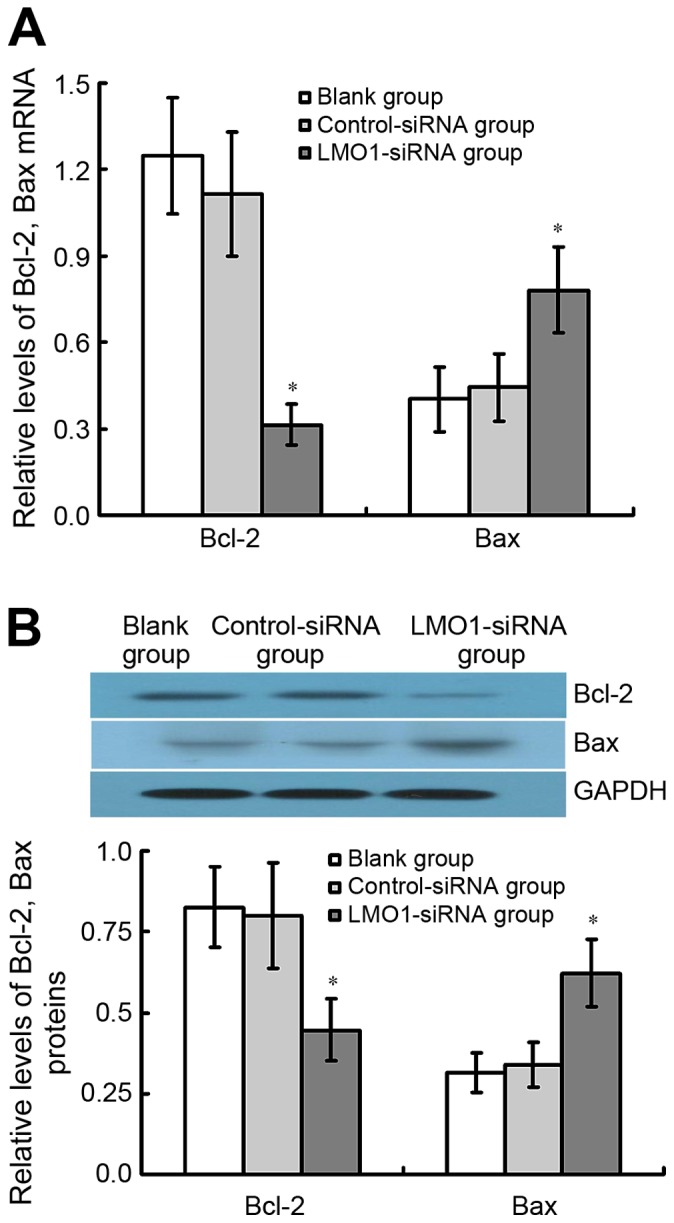
Variation of Bcl-2, Bax genes in MKN45 cells after of LMO1-siRNA transfection. After transfected with LMO1-siRNA, cells were subjected to qRT-PCR and western blottong assays, to detect the (A) mRNA or (B) protein expression levels of Bcl-2, Bax genes. Results showed that expression of Bcl-2 decreased, while Bax increased in MKN45 cells. *P<0.05 vs. Blank group or Control-siRNA group. LMO1, LIM domain only 1; Bcl-2, apoptosis regulator Bcl-2; Bax, apoptosis regulator Bax; mRNA, messenger RNA.
Discussion
One major cause of rapid gastric cancer aggressiveness is the strong anti-apoptotic ability of the tumor cells which increase survival of gastric cells and therefore promote tumor progression and metastasis (10,11). Thus, in order to improve the outcome of treatment, the anti-apoptotic ability may need to be inhibited and this may promote apoptosis in gastric cancer cells resulting in decreased tumor progression. As a variety of genes and channels are involved in the anti-apoptotic process (12,13), identifying the major functional genes is crucial. Although to search and identify novel genetic markers has become a hot topic for gastric carcinoma research, no major breakthrough has been achieved.
LMO1 is one of the novel genes which has been reported in recent studies that is associated with a variety of malignancies such as leukemia, colorectal cancer and lung cancer in terms of tumor progression, metastasis and apoptosis (8,14,15). However, only a few studies focused on the relation between LMO1 and gastric cancer. The present study found higher LMO1 expression in gastric cancer tissue comparing with that in adjacent mucosa. Similarly, in MKN45 gastric cancer cell line, the expression level was higher compared with that in GES-1 gastric epithelial cell line and other gastric cancer cell lines. These results suggested that LMO1 may have effect on progression of gastric carcinoma. Furthermore, the results indicated that LMO1 protein expression is associated with TNM staging and lymph node metastasis. In addition to that, LMO1 positive patients have a lower survival rate and the positive expression is an independent risk factor in prognosis. Thus, LMO1 can be suggested as one of the prognostic marker in gastric carcinoma, although additional experiments of a larger number of samples are needed.
In order to understand the potential mechanism of LMO1 in gastric cancer apoptosis, Bcl-2 and Bax protein were included in the present study. Bcl-2 which plays a vital role in the mitochondrial pathways of apoptosis can suppress cell apoptosis (16,17), whereas Bax functions as a pro-apoptotic gene which can induce cell apoptosis if overexpressed (18,19). A heterodimer of Bcl-2 and Bax can determine the cell apoptotic status by altering its binding ratio (20). Some studies report that inhibiting Bcl-2 expression but promoting Bax expression may induce tumor cell apoptosis in gastric carcinoma (21,22). The present study which compares the expression level of Bcl-2 and Bax in gastric cancer tissues to those in adjacent normal tissues, finds that Bcl-2 had higher expression level in the former tissues while Bax was seen an opposite result. Because of the association between Bcl-2 protein expression and gastric cancer filtration in addition to the relation of Bax protein expression with gastric cancer lymph node metastasis, Bcl-2 and Bax are thus suggested in relation to the progression of gastric cancer. In this study, LMO1 was found in positive correlation to Bcl-2, but negative correlation to Bax. LMO1 probably promote gastric cancer invasion and metastasis through regulation of Bcl-2 and Bax protein. With regard to better understand the mechanism of LMO1 in gastric cancer cell death, the present study adopted RNA interference technique (23) to suppress endogenous expression of LMO1 in MKN45 gastric cancer cell line. The results showed that the effective inhibition of LMO1 in gastric cancer cells leads to a decrease of tumor cell activity and an increase of cell apoptosis. Furthermore, the expression of Bcl-2 in MKN45 decreases while the expression of Bax increases significantly. All of these results indicated that LMO1 in gastric cancer may promote Bcl-2 expression and inhibit Bax expression although more experiments in vivo at molecular level are needed.
In conclusion, LMO1 was found increased in gastric cancer tissues and cell lines, probably affecting gastric cancer apoptosis through regulation of Bcl-2 and Bax. However, due to a limited number sample, only a partial experiment in vitro was accomplished. We consider enlarging the scale of experiment and completing the follow-up data in the urther study. Meanwhile, more work should be employed for further investigation of LMO1 in relation to other genetic molecule regulation. Animal experiments are also favored to ensure the accuracy of the study results.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Somi MH, Ghojazadeh M, Bagheri M, Tahamtani T. Clinicopathological factors and gastric cancer prognosis in the iranian population: A meta-analysis. Asian Pac J Cancer Prev. 2015;16:853–857. doi: 10.7314/APJCP.2015.16.3.853. [DOI] [PubMed] [Google Scholar]
- 3.Son T, Hyung WJ, Kim JW, Kim HI, An JY, Cheong JH, Choi SH, Noh SH. Anatomic extent of metastatic lymph nodes: Still important for gastric cancer prognosis. Ann Surg Oncol. 2014;21:899–907. doi: 10.1245/s10434-013-3403-x. [DOI] [PubMed] [Google Scholar]
- 4.Marin JJ, Al-Abdulla R, Lozano E, Briz O, Bujanda L, Banales JM, Macias RI. Mechanisms of resistance to chemotherapy in gastric cancer. Anticancer Agents Med Chem. 2016;16:318–334. doi: 10.2174/1871520615666150803125121. [DOI] [PubMed] [Google Scholar]
- 5.Kang HC, Kim IJ, Park JH, Shin Y, Ku JL, Jung MS, Yoo BC, Kim HK, Park JG. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin Cancer Res. 2004;10:272–284. doi: 10.1158/1078-0432.CCR-1025-3. [DOI] [PubMed] [Google Scholar]
- 6.Kumar R, Kaur M, Silakari O. Physiological modulation approaches to improve cancer chemotherapya review. Anticancer Agents Med Chem. 2014;14:713–749. doi: 10.2174/18715206113136660364. [DOI] [PubMed] [Google Scholar]
- 7.Frenzel LP, Patz M, Pallasch CP, Brinker R, Claasen J, Schulz A, Hallek M, Kashkar H, Wendtner CM. Novel X-linked inhibitor of apoptosis inhibiting compound as sensitizer for TRAIL-mediated apoptosis in chronic lymphocytic leukaemia with poor prognosis. Br J Haematol. 2011;152:191–200. doi: 10.1111/j.1365-2141.2010.08426.x. [DOI] [PubMed] [Google Scholar]
- 8.Beuten J, Gelfond JA, Piwkham D, Pollock BH, Winick NJ, Collier AB, III, Tomlinson GE. Candidate gene association analysis of acute lymphoblastic leukemia identifies new susceptibility locus at 11p15 (LMO1) Carcinogenesis. 2011;32:1349–1353. doi: 10.1093/carcin/bgr091. [DOI] [PubMed] [Google Scholar]
- 9.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 10.Sugita H, Iida S, Inokuchi M, Kato K, Ishiguro M, Ishikawa T, Takagi Y, Enjoji M, Yamada H, Uetake H, et al. Methylation of BNIP3 and DAPK indicates lower response to chemotherapy and poor prognosis in gastric cancer. Oncol Rep. 2011;25:513–518. doi: 10.3892/or.2010.1085. [DOI] [PubMed] [Google Scholar]
- 11.Hu X, Zhang F, Luo D, Li N, Wang Q, Xu Z, Bian H, Liang Y, Lu Y, Zheng Q, Gu J. URI promotes gastric cancer cell motility, survival, and resistance to adriamycin in vitro. Am J Cancer Res. 2016;6:1420–1430. [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Soung YH, Lee JW, Park WS, Kim SY, Cho YG, Kim CJ, Seo SH, Kim HS, Nam SW, et al. Inactivating mutation of the pro-apoptotic gene BID in gastric cancer. J Pathol. 2004;202:439–445. doi: 10.1002/path.1532. [DOI] [PubMed] [Google Scholar]
- 13.Mashima T, Oh-hara T, Sato S, Mochizuki M, Sugimoto Y, Yamazaki K, Hamada J, Tada M, Moriuchi T, Ishikawa Y, et al. p53-defective tumors with a functional apoptosome-mediated pathway: A new therapeutic target. J Natl Cancer Inst. 2005;97:765–777. doi: 10.1093/jnci/dji133. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Yan P, Jing N, Yang J. LMO1 is a novel oncogene in colorectal cancer and its overexpression is a new predictive marker for anti-EGFR therapy. Tumour Biol. 2014;35:8161–8167. doi: 10.1007/s13277-014-2066-y. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Yang J, Wang J, Guo H, Jing N. LMO1 is a novel oncogene in lung cancer, and its overexpression is a new predictive marker for anti-EGFR therapy. Med Oncol. 2014;31:99. doi: 10.1007/s12032-014-0099-0. [DOI] [PubMed] [Google Scholar]
- 16.Cheng Y, Zhao G, Zhang S, Nigim F, Zhou G, Yu Z, Song Y, Chen Y, Li Y. AS1411-induced growth inhibition of glioma cells by up-regulation of p53 and downregulation of Bcl-2 and Akt1 via nucleolin. PLoS One. 2016;11:e0167094. doi: 10.1371/journal.pone.0167094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy TL, Garikapati KR, Reddy SG, Reddy BV, Yadav JS, Bhadra U, Bhadra MP. Simultaneous delivery of Paclitaxel and Bcl-2 siRNA via pH-Sensitive liposomal nanocarrier for the synergistic treatment of melanoma. Sci Rep. 2016;6:35223. doi: 10.1038/srep35223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil J, Ramsey D, Szmida E, Leszczynski P, Pawlowski P, Bebenek M, Sasiadek MM. The BAX gene as a candidate for negative autophagy-related genes regulator on mRNA levels in colorectal cancer. Med Oncol. 2017;34:16. doi: 10.1007/s12032-016-0869-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu L, Wang Z, He SY, Zhang SF, Luo HJ, Zhou K, Li XF, Qiu SP, Cao KY. Bax-interacting factor-1 inhibits cell proliferation and promotes apoptosis in prostate cancer cells. Oncol Rep. 2016;36:3513–3521. doi: 10.3892/or.2016.5172. [DOI] [PubMed] [Google Scholar]
- 20.Adefolaju GA, Theron KE, Hosie MJ. BAX/BCL-2 mRNA and protein expression in human breast MCF-7 cells exposed to drug vehicles-methanol and dimethyl sulfoxide (DMSO) for 24 hrs. Niger Med J. 2015;56:169–174. doi: 10.4103/0300-1652.160349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim EM, Kim J, Park JK, Hwang SG, Kim WJ, Lee WJ, Kang SW, Um HD. Bcl-w promotes cell invasion by blocking the invasion-suppressing action of Bax. Cell Signal. 2012;24:1163–1172. doi: 10.1016/j.cellsig.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Zhu P, Zhang J, Zhu J, Shi J, Zhu Q, Gao Y. MiR-429 induces gastric carcinoma cell apoptosis through Bcl-2. Cell Physiol Biochem. 2015;37:1572–1580. doi: 10.1159/000438524. [DOI] [PubMed] [Google Scholar]
- 23.Baker M. RNA interference: From tools to therapies. Nature. 2010;464:1225. doi: 10.1038/4641225a. [DOI] [PubMed] [Google Scholar]