Abstract
Long non-coding RNA (Lnc)TCF7 is a novel lncRNA that is involved in tumorigenesis. Previous studies have revealed that lncTCF7 serves an essential role in maintaining cancer stem cell self-renewal; however, the functions of lncTCF7 in colorectal cancer (CRC) remain unknown. Therefore, the present study aimed to investigate the role of lncTCF7 in CRC. LncTCF7 was upregulated in 52/58 CRC tissues, and its expression correlated with tumor size, lymph metastasis and tumor-node-metastasis stage in CRC. Knocking down lncTCF7 in colon cancer cell lines decreased cell proliferation, migration and invasion, while lncTCF7 overexpression showed opposite changes. In addition, lncTCF7 promoted cell proliferation in vivo. LncTCF7 activated the Wnt/β-catenin signaling pathway, which is essential for cancer development. Survival analysis revealed that patients with higher expression of lncTCF7 had significantly worse prognosis compared with patients with low expression. These findings indicate that lncTCF7 regulates CRC progression and support the notion of lncTCF7 as a CRC prognostic marker.
Keywords: CRC, TCF7, LncTCF7, EMT, Wnt/β-catenin
Introduction
Colorectal cancer (CRC) is the third most common cancer and the third leading cause of cancer-related death in the world (1). There has been remarkable progress in molecular subtyping of CRC in the past decade and much of this information has been used for clinical purposes. For example, K-RAS mutation status can predict treatment response to cetuximab. The latest National Comprehensive Cancer Network clinical guideline recommends BRAF and mismatch repair genes to be examined in routine tumor testing (2). However, cancer is known to be a heterogeneous disease and thus identification and understanding of additional prognostic factors would be helpful in guiding treatments that would ultimately affect survival.
Long non-coding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides (nt) that are 5′ capped and 3′ polyadenylated. lncRNAs function in a wide range of biological processes and can regulate gene expression in cis or trans by diverse mechanisms. LncRNA-mediated biology has been implicated in many cellular processes such as cell proliferation, invasion and so on. One particularly important lncRNA is lncTCF7 which has been reported to maintain cancer stem cell self-renewal throughing Wnt/β-catenin signaling pathway (3).
In the present study we have examined lncTCF7 expression in tissue samples from CRC patients, assessed molecular mechanisms responsible for its involvement in CRC tumorigenesis, and assessed its potential value as a biomarker in predicting CRC prognosis.
Materials and methods
Tissue samples
The CRC surgical samples were obtained from part of surgical patients in the Department of Surgery at Peking University First Hospital between August 2011 and December 2012. These were confirmed to be colorectal adenocarcinoma histologically by experienced pathologists. Clinicopathologic staging was determined according to the CRC Guideline of the National Comprehensive Cancer Network (2). Written informed consents were obtained from these patients.
Cell culture
The cell lines DLD1 and LoVo were purchased from the American Type Culture Collection. DLD1 and LoVo were cultured in DMEM and F-12K (both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) culture media, respectively, at 37°C with 5% CO2. The media were supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml of penicillin, and 100 µg/ml of streptomycin (both Keygen Biotech, Nanjing, China).
Cell transfection
The plasmid expressing short hairpin RNA (shRNA) specifically targeting the lncTCF7 (shRNA-lncTCF7) and its scrambled RNA (shRNA-nc) control were constructed by GenePharma Co. (Shanghai, China). The shRNA sequence is 5′-CTTTGGATAAGATCTACGTTT-3′. The plasmid over-expressing lncTCF7 (oe-lncTCF7) and its empty vector (oe-Vec) were obtained from GenePharma Co. Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used for the transfection. Cell clones stably expressing the shRNA and oe-lncTCF7 were selected with G418.
RNA extraction and qRT-PCR
Total RNA was extracted from the surgical samples and cultured cells using TRIzol reagent (Takara Bio, Dalian, China). RQ1 RNase-Free DNase (Promega, Madison, WI, USA) was used to remove DNA. RNA reverse transcription to cDNA was performed with RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) and quantitative real-time PCR (qRT-PCR) analysis was performed with SYBR-Green PCR Master Mix (Life) according to the manufacturer's instructions. The results were normalized to the expression of 18S and β-actin. The primers used for qRT-PCR are listed below: lncTCF7 Forward, AGGAGTCCTTGGACCTGAGC and Reverse, AGTGGCTGGCATATAACC-AACA; 18S Forward, GTAACCCGTTGAACCCCATT and Reverse, CCATCCAATCG-GTAGTAGCG; β-actin Forward, CATGTACGTTGCTATCCAGGC and Reverse, CTCC-TTAATGTCACGCACGAT. The ΔCq value was used to represent the expression level of lncRNA in quantitative qRT-PCR. For each amplicon designed, the ΔCq value was normalized using the equation: ΔCq=Cq (target)-Cq (18S/β-actin).
Western blot analysis
Lysate was extracted from surgical samples or cultured cells by a lysis buffer, and the protein content was assayed by a protein assay (BCA) kit (Thermo Fisher Scientific, Inc.). Protein was fractionated in 10% SDS-PAGE and transferred onto a PVDF membrane (Roche, Mannheim, Germany). Rabbit monoclonal antibodies against E-cadherin, N-cadherin, Vimentin, Slug, c-Myc, Cyclin D1 (Cell Signaling Technology, Inc., Danvers, MA, USA), MMP7 (Abcam, Cambridge, UK) and TCF7 (Abgent Inc., San Diego, CA, USA) were used as the primary antibodies. The antibody against GAPDH (Cell Signaling Technology, Inc.) was used for loading control. Secondary antibody was then added and proteins of interest were visualized by using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.).
Cell proliferation
The effect of lncTCF7 on proliferation of CRC cell lines was measured with Cell Counting kit-8 (CCK-8) (Dojindo). After transfection of shRNA-lncTCF7, shRNA-nc, oe-lncTCF7, or oe-Vec for 48 h, DLD1 and LoVo cells were re-suspended, seeded at 2×103/100 µl/well in a 96-well plate, and incubated for 24 or 48 h. CCK-8 reagent was added to each well. The plate was incubated for another 2 h at 37°C, and the absorbance at 450 nm was measured.
Transwell assays
The migration ability of CRC cells was assayed by a Transwell plate with a filter of 8 µm pores (BD Biosciences, San Diego, CA, USA). A total of 5×105 cells in serum-free medium were added into the upper insert. A total of 600 µl of medium containing 20% FBS was added into the lower compartment. After incubation for 48 h, the cells on the upper surface of the filter were wiped, and the cells adherent on the undersurface of the filter were fixed in methanol, stained with crystal violet and counted with a microscope. Three visual fields were selected randomly for cell counting. An invasion assay was performed in a similar manner except that the top chamber was pre-coated with 50 µl Matrigel (BD Biosciences).
RNA-FISH
A fragment of 550 bp lncTCF7 cDNA was amplified from the lncTCF7 plasmid by using a high fidelity DNA polymerase (Takara) as the DNA template. From this template, a fluorescein labeled DNA as the lncTCF7 FISH probe was made with fluorescein-12-dUTP (Roche) and Klenow DNA polymerase following the manufacturer's protocol. CRC tissue with its adjacent normal tissue embedded in OCT (Sakura Finetek, Inc., Torrance, CA, USA) was cut to sections of 4 µm thickness. The slide was soaked in proteinase K for 5 min and washed in 2× SSC twice. A FISH hybridization solution (Beijing Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China) containing 30 ng/µl lncTCF7 FISH probe DNA was applied to the slide which was subsequently incubated at 37°C for 16 h. The slide was then washed in 0.4× SSC/0.001% NP-40 at 560C for 5 min followed by a second wash in 2× SSC/0.0001% NP-40 for another 2 min. The slide was covered with a drop of mounting medium containing DAPI, and observed under a fluorescence microscope.
Tumor xenograft model
Male BALB/c nude mice of 4–5 weeks age were purchased from Beijing Vital River Co. (Beijing, China) and maintained in the Experimental Animal Center of Peking University First Hospital. DLD1 cells (1×107 cells) stably expressing shRNA-lncTCF7, or oe-lncTCF7, or respective control vectors were mixed with 50% Matrigel (BD Biosciences), and were injected subcutaneously into the right flank of a nude mouse (n=6/group). Tumor volume was measured every 2 days and calculated as width2 x length/2. After two weeks, the mice were sacrificed, the tumors were removed and weight was measured.
Statistical analysis
Two-tailed Student's t-test was used for data comparison. χ2 test was performed to evaluate the association between lncTCF7 level and clinicopathological parameters. Kaplan-Meier method was used to estimate overall survival of the CRC patients. P<0.05was considered to indicate a statistically significant difference. Each experiment was repeated at least 3 times independently.
Results
lncTCF7 is upregulated in CRC tissues
lncRNAs have been reported to play a role in tumorigenesis and progression and aberrant expression of lncRNAs has been associated with worse cancer prognosis. We found that lncTCF7 was significantly increased in cancer tissue in comparison with peri-tumor control.
In 58 CRC cases, 52 cases showed a higher lncTCF7 expression levels in tumor than in peri-tumor mucosa and the median ratio increased 3-fold (Fig. 1A). Using RNA-FISH, we examined the location of lncTCF7 in two pairs of specimens as shown in Fig. 1B, more abundant lncTCF7 RNA was seen in cancer tissue compared with peri-tumor mucosa and it was specifically localized in the nuclei of CRC specimens. In Fig. 1C, increased TCF7 protein expression was also noted by western blot in ten pairs of specimens.
Figure 1.
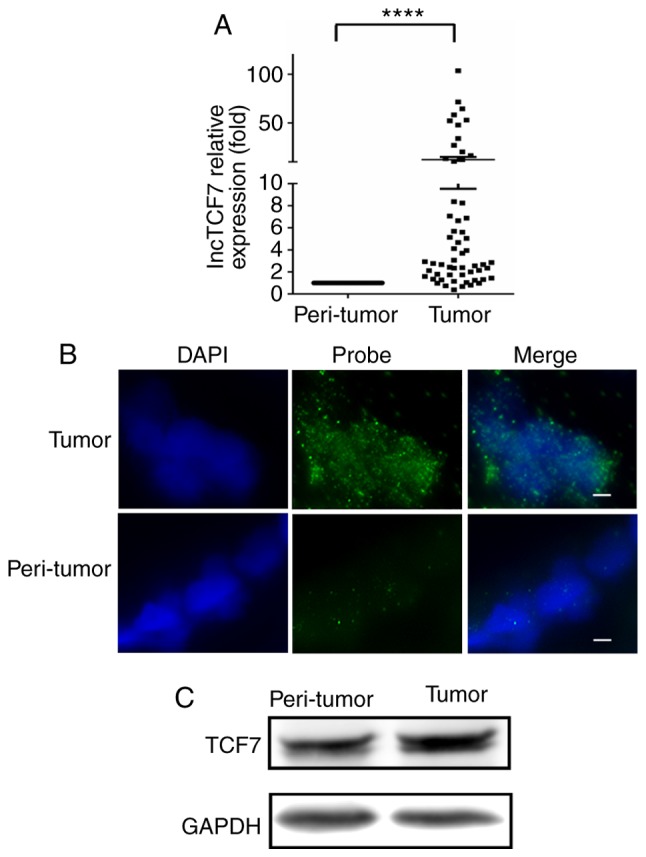
LncTCF7 level is highly expressed in human CRC tissues. 58 CRC patient surgery samples were studied. Paired cancer tissues and adjacent normal mucosal tissues from each patient were subjected to the following experiments: (A) Total RNA from each tissue was harvested to detect lncTCF7 level by qRT-PCR. 18S RNA served as internal control. Error bar, mean ± SEM (****P<0.0001). compared to peri-tumor tissue. (B) lncTCF7 level and localization was studied by RNA FISH assay on tumor and peri-tumor tissue. Frozen sectioned tissues on slides were permeabilized and hybridized with FITC-labeled LncTCF7 probe. Nuclei were stained with DAPI. Green, lncTCF7; Blue nuclei. Scale bar, 25 µm. (C) TCF7 protein levels in tumor and peri-tumor samples were represented by western blot analysis on representative case. LncTCF7, Long non-coding RNA TCF7, CRC, colorectal cancer.
lncTCF7 stimulates CRC cell migration and invasion in vitro
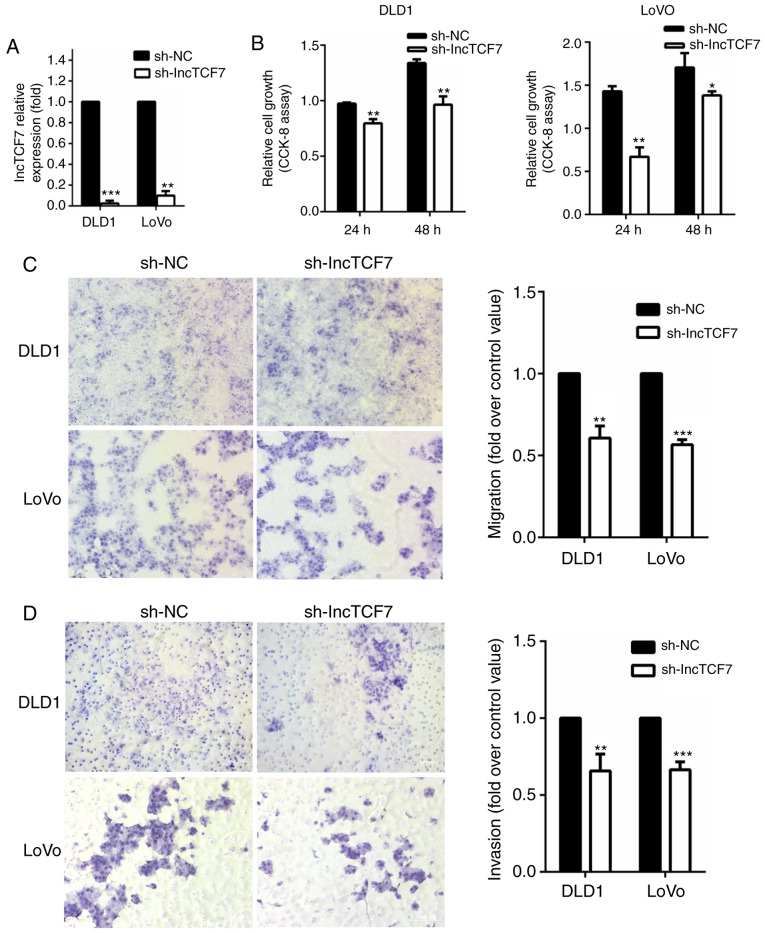
The DLD1 and LoVo cells transfected with shRNA-lncTCF7 had reduced expression of lncTCF7 compared to cells transfected with shRNA-nc as measured by qRT-PCR (Fig. 2A). As shown in Fig. 2B, suppression of lncTCF7 led to a mild decrease in cell growth. Cell migration was assessed by Transwell assay (Fig. 2C). In both DLD-1 and Lovo cell lines, there was a 30–40% decrease in migrated cell numbers in shRNA-lncTCF7 group compared to that in control. In addition, reduced expression of lncTCF7 also inhibited invasion by 25~30% (Fig. 2D).
Figure 2.
LncTCF7 knockdown inhibits CRC cell migration and invasion. Two CRC cell lines, DLD1 and LoVo, were used for the following experiments. sh-RNA targeting LncTCF7, and non-specific control, shRNA-nc were transfected stably into CRC cells (A) LncTCF7 expression was knocked down by specific sh-RNA. (B) Proliferation of CRC cells assessed by CCK8 assays. (C) Cancer cell migration was measured by Transwell assay. Cells that had moved through upper chamber were counted. (D) Cell invasion was detected by Matrigel assay. Cells penetrating through Matrigel were counted. The values represent the means ± SD. *P<0.05, **P<0.01, ***P<0.001 compared with shRNA-nc. shRNA, short hairpin RNA.
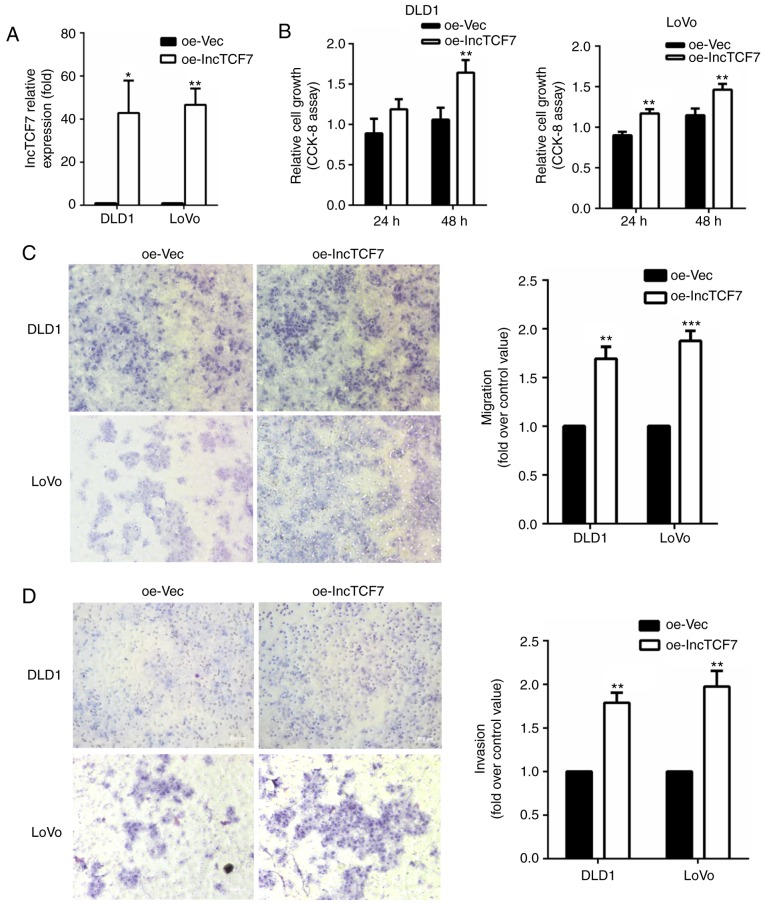
We also measured cell proliferation and migration after overexpressing lncTCF7 in DLD-1 and Lovo lines (Fig. 3A). As shown in Fig. 3B and C, increased lncTCF7 stimulated CRC cell proliferation and migration significantly. Fig. 3D showed that overexpression of lncTCF7 increased cell invasion rate by 80~100%.
Figure 3.
LncTCF7 overexpression stimulates CRC cell migration and invasion. DLD1 and LoVo cell lines were transfected with overexpression vector oe-lncTCF7, or empty vector, oe-Vec. (A) The lncTCF7 expression was increased by oe-lncTCF7 transfection. (B) Proliferation of CRC cells were determined by CCK8 assays. (C) Cancer cell migration was measured by Transwell assay. Cells that had moved through the upper chamber were counted. (D) Cell invasion was detected by Matrigel assay. Cells penetrating through Matrigel were counted. The values represented are the means ± SD. *P<0.05, **P<0.01, ***P<0.001 compared with oe-Vec. lncTCF7, long non-coding RNA TCF7; CRC, colorectal cancer.
Taken together, these in vitro experiments support the hypothesis that lncTCF7 promotes tumor cell migration and invasion.
LncTCF7 promotes tumorigenesis in vivo
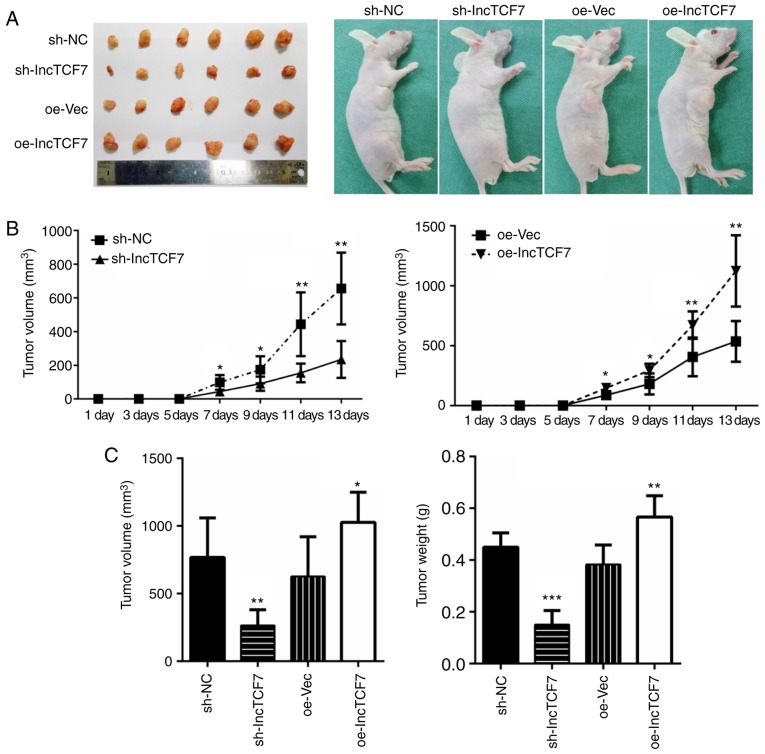
Next we detected how lncTCF7 influences tumorigenesis in vivo. 1×107 cells were inoculated into nude mice and tumor growth was monitored for 14 days (Fig. 4A). At the sixth day, a palpable tumor could be recognized. As shown in Fig. 4, reduced expression of lncTCF7 in the shRNA-lncTCF7 group led to significantly smaller tumor volume and weight (Fig. 4A, C, left), and a slower tumor growth (Fig. 4B, left). On the other hand, overexpression of lncTCF7 with oe-lncTCF7 resulted in faster growth (Fig. 4B, right), and bigger tumor size (Fig 4C). These data indicate that lncTCF7 promotes CRC tumorigenesis in vivo.
Figure 4.
lncTCF7 promotes tumorigenesis in vivo (A) lncTCF7 knockdown or overexpressing stable DLD1 line cells were injected subcutaneously into nude mice, (n=6 for each groups). The tumors were dissected for mass and volume measurement at the end of second week. (B) Tumor growth curves. Tumor size was calculated every other day 5 days after injection. (C) Tumor weight and volume were measured at the end point. The values represent the means ± SD. *P<0.05, **P<0.01, ***P<0.001. lncTCF7, long non-coding RNA TCF7.
LncTCF7 regulates epithelial-mesenchymal transition by activating the Wnt pathway
Given the finding that lncTCF7 promotes CRC cell migration and invasion, we attempted to investigate the underlying molecular mechanisms. Epithelial-mesenchymal transition, EMT, is pivotal in cancer metastasis and invasion (4) and the Wnt signaling pathway is one of the key pathways in regulating EMT (5). Thus, we set out to examine if lncTCF7 affects the Wnt signal pathway.
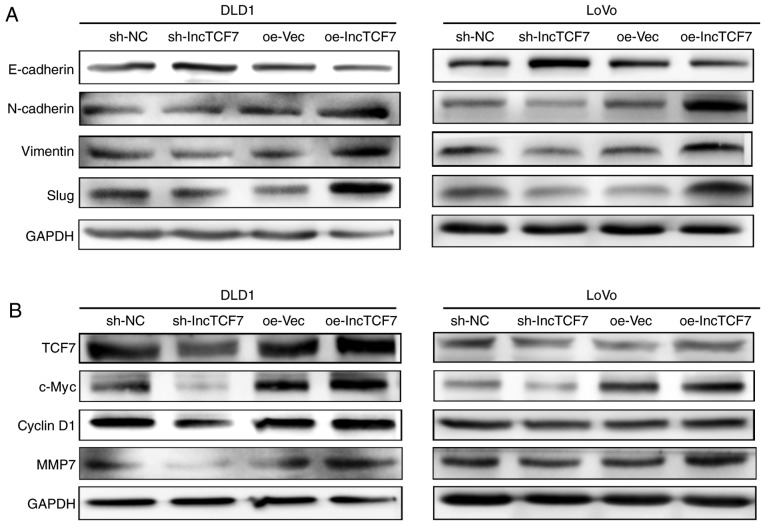
First, the epithelial and mesenchymal markers were assayed by western blot analysis. Fig. 5A shows that lncTCF7 inhibited the expression of E-cadherin, an epithelial marker, but enhanced the expression of N-cadherin, Vimentin and slug, three classic mesenchymal markers, in DLD1 and LoVo cells.
Figure 5.
LncTCF7 initiate EMT by activating Wnt/β-catenin signaling pathway. DLD1 and LoVo cell lines were transfected with lncTCF7, shRNA-nc, oe-LncTCF7, or oe-Vec (A) The expression of E-cadherin, N-cadherin, vimentin and slug in DLD1 and LoVo cells were detected by western blot. GAPDH was used as an internal control. (B) The expression of TCF7, c-Myc, CyclinD1, and MMP7 in DLD1 and LoVo cells were detected by western blot. GAPDH was used as an internal control. shRNA, short hairpin RNA; lncTCF7, long non-coding RNA TCF7; oe-lncTCF7, over-expressing lncTCF7.
Next, we assessed the state of the Wnt pathway upon lncTCF7 manipulation by measuring several downstream effects. As shown in Fig. 5B, reduced expression of lncTCF7 resulted in reduced expression of c-Myc, Cyclin D1 and MMP7 while overexpressing lncTCF7 demonstrated opposite effects.
LncTCF7 correlates with poor prognosis of CRC patients
To test the value of lncTCF7 as a prognostic biomarker, we studied the association between lncTCF7 expression and clinical profiles of CRC patients. Demographic and clinical data were reviewed (Table I). A total of 58 cases were categorized according to LncRNA expression level. Group of lncTCF7 was divided according to median expression fold compared to its peri-tumor. As shown in Table I, there were significant differences in tumor size, lymph node metastasis, and TNM stage between two groups.
Table I.
Clinicopathologic characteristics of lncTCF7 expression in CRC patients.
| Clinicopathological variables | N | High expressiona | Low expressiona | χ2 | P-value |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 28 | 14 | 14 | 0 | 1 |
| Female | 30 | 15 | 15 | ||
| Age (years)a | |||||
| <64 | 29 | 12 | 17 | 1.724 | 0.189 |
| ≥64 | 29 | 17 | 12 | ||
| Tumor sizea (cm) | |||||
| <4.5 | 25 | 8 | 17 | 5.695 | 0.017 |
| ≥4.5 | 33 | 21 | 12 | ||
| Lymph metastasis | |||||
| Yes | 27 | 18 | 9 | 5.613 | 0.018 |
| No | 31 | 11 | 20 | ||
| TNM classification | |||||
| I–II | 27 | 8 | 19 | 8.385 | 0.004 |
| III–IV | 31 | 21 | 10 |
Group of lncTCF7 expression was performed according to median. Lnc, long non-coding RNA; CRC, colorectal cancer; TNM, tumor node metastasis.
Furthermore, Kaplan-Meier survival analysis revealed worse prognosis in the high lncTCF7 group. The overall 3-year survival rate was about 68% in the high group, and 89% in the low group (Fig 6). The median follow-up time is 47 months. Log rank test confirmed that the difference is statistically significant (P=0.0441 with a hazard ratio of 3.513, 95% confidence interval 1.032 to 9.996).
Figure 6.
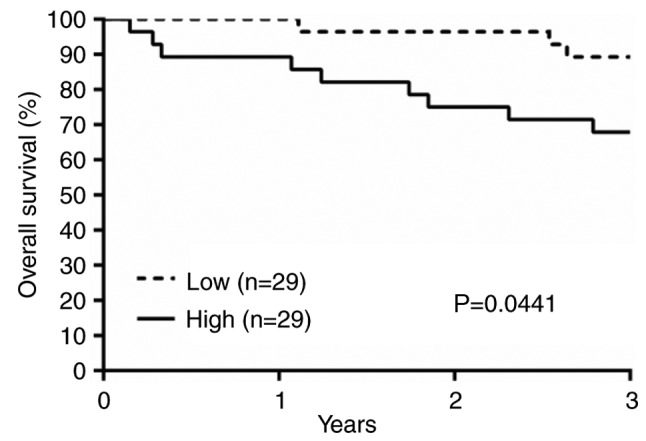
LncTCF7 expression is related to CRC prognosis Kaplan-Meier overall survival curves of patients with low vs. high lncTCF7 expression. Differences between the two groups were evaluated by the log-rank test (P=0.0441). CRC, colorectal cancer; lncTCF7, long non-coding RNA TCF7.
Disscusion
LncRNAs contribute not only to homeostasis, but also to pathologic processes via regulating expression of many genes. LncRNAs regulate gene expression by influencing chromatin remodeling, transcriptional control, post-transcriptional processing, and protein metabolism (6). It has been thought that lncRNA dysregulation is associated with tumorigenesis and their aberrant expression has important pathogenic consequences such as initiating metastasis. It is known that high expression of HOTAIR, H19, SAMMSON, AFAP-AS1, UPAT (7–11) correlate with a worse cancer prognosis. High expression of lncRNAs including MALAT1, HOTAIR, CCAL, 91H (12–15) were reported to correlate with a poor prognosis in CRC patients while lower lncRNA Loc285194 expression was associated with a better survival (16). These findings suggested that lncRNAs may serve as diagnostic or prognostic markers and could potentially be useful as treatment targets.
LncTCF7 expression is associated with cancer stem cell self-renewal (3). However, the function of lncTCF7 in CRC remains largely unknown. Our study investigated lncTCF7 expression in surgical samples from CRC patients and found that lncTCF7 level increased on average about 10 times in CRC tissue. In addition, lncTCF7 promoted cell proliferation, migration and invasion in vitro and tumorigenesis in nude mice inoculation experiments.
EMT is a process which closely relates with cancer invasion and metastasis (17,18). There are several signal pathways regulating EMT (19,20). One of them is Wnt/β-catenin signaling cascade (17). Interestingly, lncTCF7 promoted expression of the mesenchymal markers N-cadherin, vimentin and slug, and suppressed expression of the epithelial marker E-cadherin. Given the close link between lncTCF7 and Wnt pathway, we therefore hypothesized that lncTCF7 activated Wnt/β-catenin signaling and subsequently regulated EMT and resulted in tumorigenesis.
TCF7 is one of TCF/LEF members which plays an essential role in Wnt/β-catenin pathway (21–23). It has been known that lncTCF7 recruits the SWI/SNF complex to the promoter of TCF7 to facilite its expression (3). We found that lncTCF7 is enriched in the CRC tissues nucleus. Our research demonstrated that lncTCF7 promotes the expression of TCF7. Therefore, lncTCF7 increased expression of TCF7 to promote the activation Wnt signal pathway. The activation of Wnt/β-catenin cascade enhanced expression of downstream members. In this study, lncTCF7 upregulated c-Myc, Cyclin D1 and MMP7 levels in CRC cells.
Lastly, lncTCF7 expression is significantly elevated in CRC samples compared to normal colon epithelium. lncTCF7 level affects TNM staging and survival and CRC patients with high lncTCF7 levels have worse prognosis. This study thus supports the value of lncTCF7 as a CRC prognostic marker.
Acknowledgments
The present study is supported by the grant on potential therapeutic approaches for surgical infection and the underlying mechanisms from China Health and Medical Development Foundation. The authors would like to thank Professor Ding-fang Bu and Qiang Zhu for their technical assistance for RNA-FISH assay, and Mrs. Yuanyuan Ma for the animal study assistance.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.National Comprehensive Cancer Network: NCCN clinical practice guidelines in oncology. https://www.nccn.org/professionals/physician_gls/f_guidelines.asp https://www.nccn.org/professionals/physician_gls/f_guidelines.asp
- 3.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Liu CC, Cai DL, Sun F, Wu ZH, Yue B, Zhao SL, Wu XS, Zhang M, Zhu XW, Peng ZH, Yan DW. FERMT1 mediates epithelial-mesenchymal transition to promote colon cancer metastasis via modulation of β-catenin transcriptional activity. Oncogene. 2017;36:1779–1792. doi: 10.1038/onc.2016.339. [DOI] [PubMed] [Google Scholar]
- 5.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mondal T, Kanduri C. Noncoding RNA scaffolds in pluripotency. Circ Res. 2012;110:1162–1165. doi: 10.1161/RES.0b013e318257c489. [DOI] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J, Dong N, He J, Sun Q, Lv G, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother. 2016;81:152–159. doi: 10.1016/j.biopha.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Taniue K, Kurimoto A, Sugimasa H, Nasu E, Takeda Y, Iwasaki K, Nagashima T, Okada-Hatakeyama M, Oyama M, Kozuka-Hata H, et al. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc Natl Acad Sci USA. 2016;113:1273–1278. doi: 10.1073/pnas.1500992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z, Protivankova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510–1515. doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 15.Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, Li R, Ying H, Wang F, Liu X, et al. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9:e103022. doi: 10.1371/journal.pone.0103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZJ, Liu HL, Zhou HC, Wang GC. TIPE2 inhibits hypoxia-induced Wnt/β-catenin pathway activation and EMT in glioma cells. Oncol Res. 2016;24:255–261. doi: 10.3727/096504016X14666990347356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin B, Wang W, Meng XX, Du G, Li J, Zhang SZ, Zhou BH, Fu ZH. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer. 2016;16:863. doi: 10.1186/s12885-016-2904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Deng Q, Liang Z, Liu Z, Geng H, Zhao L, Zhou Q, Liu J, Ma J, Wang D, et al. Cigarette smoke extract induces epithelial-mesenchymal transition of human bladder cancer T24 cells through activation of ERK1/2 pathway. Biomed Pharmacother. 2016;86:457–465. doi: 10.1016/j.biopha.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Cho YH, Cha PH, Kaduwal S, Park JC, Lee SK, Yoon JS, Shin W, Kim H, Ro EJ, Koo KH, et al. KY1022, a small molecule destabilizing Ras via targeting the Wnt/β-catenin pathway, inhibits development of metastatic colorectal cancer. Oncotarget. 2016;7:81727–81740. doi: 10.18632/oncotarget.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui L, Guan Y, Qu Z, Zhang J, Liao B, Ma B, Qian J, Li D, Li W, Xu GT, Jin Y. WNT signaling determines tumorigenicity and function of ESC-derived retinal progenitors. J Clin Invest. 2013;123:1647–1661. doi: 10.1172/JCI65048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Wang W, Wang X. Roles of transcriptional factor 7 in production of inflammatory factors for lung diseases. J Transl Med. 2015;13:273. doi: 10.1186/s12967-015-0617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]