Abstract
MicroRNA-1470 (miR-1470) is overexpressed in esophageal squamous cell carcinoma (ESCC); however, its role and underlying molecular mechanism remain unknown. The aim of the present study was to explore the tumorigenic role and mechanism of miR-1470 overexpression in ESCC. The expression of miR-1470 in ESCC tissues and cell lines was detected using human miRNA microarrays and the reverse transcription-quantitative polymerase chain reaction, respectively. The effects of miR-1470 on cell proliferation, migration and senescence were determined using a Cell Counting Kit-8 assay, Transwell migration assay and β-galactosidase staining kit. Western blotting was used to analyze the expression levels of genes in the apoptosis signaling pathway. An increased expression level of miR-1470 was observed in ESCC tissues compared with that in paracancerous tissues. Knockdown of miR-1470 significantly suppressed proliferation, and down-regulated the cell cycle regulatory gene cyclin E1. It was also revealed that knockdown of miR-1470 significantly inhibited migration, and decreased the expression levels of matrix metalloproteinase 2 (MMP2), MMP13 and MMP14. Western blotting analysis revealed that knockdown of miR-1470 induced apoptosis by increasing B-cell lymphoma 2 (Bcl-2) expression. The results of the present study suggest that overexpression of miR-1470 in ESCC promotes cancer cell proliferation by accelerating the cell cycle and inhibiting apoptosis, and also enhances cancer cell migration by upregulating MMPs.
Keywords: microRNA-1470, proliferation, migration, senescence, esophageal squamous cell carcinoma
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most common malignant diseases globally, particularly in China, where it is the fourth most common cause of cancer-associated mortality (1–3). Currently, there are limited clinical approaches towards the early diagnosis and treatment of ESCC, resulting in a 10% 5-year survival rate for patients (4). Thus, clarification of the molecular mechanisms underlying the pathogenesis of ESCC and development of therapeutic strategies are urgently required.
MicroRNAs (miRNAs) are small non-coding RNAs (between 18 and 22 nucleotides in length), which serve a role in gene expression regulation primarily by repressing translation (5,6). Prior evidence supports the notion that miRNAs modulate proliferation, apoptosis, metastasis and metabolism of cancer (6,7). Numerous miRNAs are involved in esophageal tumorigenesis (8,9). However, the role of miRNA-1470 (miR-1470) in esophageal cancer progression remains undefined.
Bcl-2 is an apoptosis inhibitor (10). When Bcl-2 is suppressed, tumor cells undergo apoptosis and cease proliferation (11). Various moderators of apoptosis exert their effects through the caspase enzyme system. Among them, caspase-3 has been demonstrated to be the most markedly associated with apoptosis (12,13). Furthermore, decreased caspase-3 expression has been observed in several human malignancies including oral squamous cell carcinoma, non-small cell lung carcinoma, breast cancer, gastric cancer, and ESCC (14–17). Overexpressions of Bcl-2 and other apoptotic inhibitors serve the roles in therapeutic resistance of ESCC (18,19). Whereas decreased Bcl-2 expression promotes apoptotic responses to anticancer drugs, increased expression of Bcl-2 leads to resistance to chemotherapeutic drugs and radiation therapy. The inhibition of cell apoptosis is associated with the onset and development of cancer, as well as its sensitivity to chemotherapy (19–21).
In the present study, it was demonstrated that miR-1470 was upregulated in ESCC tissues, and knockdown of miR-1470 significantly inhibited proliferation and migration, and promoted senescence, of ESCC cells. Furthermore, knockdown of miR-1470 exhibited apoptosis-promoting activity, including downregulation of the anti-apoptotic protein Bcl-2 and upregulation of the pro-apoptotic Bcl-2-associated X protein (BAX). Taken together, the results of the present study suggest that miR-1470 affects cancer progression via the regulation of Bcl-2 expression and may be a promising therapeutic target for the treatment of ESCC.
Materials and methods
Cell culture and transfection
The ESCC cell lines including KYSE30, KYSE180, KYSE450, KYSE150 and KYSE510, provided by Dr Yutaka Shimada (Kyoto University, Kyoto, Japan), were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA). All cells were maintained at 37°C with 5% CO2. Homo sapiens (hsa)-miR-1470 inhibitor was designed and provided by Shanghai GenePharma, Co., Ltd. (Shanghai, China). Transfection was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc. Carlsbad, USA), according to the manufacturer's protocol. Transfected cells were incubated at 37°C with 5% CO2 for 48 h.
The sequence of the miR-1470 inhibitor was 5′-CGGGGUGCACGGGCGGAGGGC-3′; the sequence of the negative control was 5′-CAGUACUUUUGUGUAGUACAA-3′.
Cell proliferation assay
Cells were seeded in 96-well flat-bottomed plates, with each well containing 2×103 cells in 100 µl cell suspension. After incubation for 24, 48, 7 h and 9 6 h, cell viability was determined using the Cell Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol using a microplate read at 450 nm. Each experiment, with 6 replicates, was repeated in triplicate.
Cell migration assay
Cells (5×105 cells in serum-free RPMI-1640 medium) were seeded in the upper chamber. The wells in the lower chamber were filled with RPMI-1640 medium containing 20% FBS, and incubated at 37°C for 36 h. Cells on the upper membrane surface were wiped off using a cotton swab, and the lower membrane surface was fixed with methanol and stained with 0.1% crystal violet. Each experiment was repeated in triplicate.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
RT-qPCR was performed using the Premix Ex Taq kit (Takara Biotechnology Co., Ltd., Dalian, China) and a 7300 Real-Time PCR system (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The mRNA expression levels of genes of interest were normalized to the endogenous expression level of GAPDH. The following primer pairs were used for the PCR assay: Cyclin E1 (CCNE1) forward, 5′-TGACCTAAGGGACTCCCACAA-3′; CCNE1 reverse, 5′-TGATAATGTGGAGAGGGCAGC-3′; matrix metalloproteinase 2 (MMP2) forward, 5′-GTGATGGTTCCCCTGTTCACT-3′, MMP2 reverse, 5′-CATGGGGAATGGTTGAAGGGA-3′; MMP13 forward, 5′-TCCCTTGAGATATGGAAGGATGC-3′; MMP13 reverse, 5′-TAGTTCTTCCCTTGATGGCCG-3′; MMP14 forward, 5′-TGGATGCGAGTACCCCAAGA-3′, MMP14 reverse, 5′-GTAGCCCGGTTCTACCTTCAG-3′. A Hairpin-it™ miR-1470 RT-qPCR Primer Set (Shanghai GenePharma, Co., Ltd.) was used for the measurement of the relative quantity of hsa-miR-1470. The expression level of miR-1470 was normalized to the endogenous expression of U6.
Western blotting
Cells from each group were detached with trypsin, centrifuged at 1,000 × g for 5 min at 4°C and washed twice with pre-chilled PBS. Radioimmunoprecipitation Assay buffer (Beyotime Institute of Biotechnology, Haimen, China) was subsequently added to lyse cells and cells were incubated on ice for protein extraction. Protein concentration was determined using the Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). Equal amounts (10 µg per lane) of proteins were separated by SDS-PAGE (12% gel) and transferred onto a polyvinylidene difluoride membrane (Merck KGaA, Darmstadt, Germany). The membrane was soaked in 10% skimmed milk (in PBS, pH 7.2, containing 0.1% Tween-20) for 2 h at 4°C and incubated with an appropriate amount of primary antibody [working dilutions of antibodies: Bcl-2, 1:1,000, cat. no. 10783-1-AP; BAX, 1:1,000, cat. no. 23931-1-AP (both Proteintech Group, Inc., Chicago, IL, USA) pro-caspase-3, 1:1,000, cat. no. sc-7148; pro-caspase-12, 1:1,000, cat. no. sc-5627; β-actin, 1:400, cat. no. sc-47778), (Santa Cruz Biotechnology, Inc., Dallas, TX, USA; Proteintech Group, Inc., Chicago, IL, USA)] at 4°C overnight. Detection was performed using horseradish peroxidase-conjugated secondary antibodies (anti-mouse IgG, 1:10,000; cat. no. 474-1806 or anti-rabbit IgG, 1:10,000; cat. no. 5450-0010) for 2 h at room temperature (KPL, Inc., Gaithersburg, MD, USA) and Immobilon™ Western chemiluminescent HRP substrate kit (EMD Millipore, Billerica, MA, USA). Densitometry analysis was performed using ImageJ 1.46 software (National Institutes of Health, Bethesda, MD, USA).
Assessment of senescence
To assess senescence, β-galactosidase activity was measured using a Senescence β-Galactosidase Staining kit (Cell Signaling Technology, Inc., Danvers, MA, USA) and performed according to the manufacturer's protocol. Briefly, negative control (NC)-or miRNA-1470 inhibitor-transfected cells were washed twice with 1X PBS, fixed in 1X fixation buffer for 5 min, washed three times with 1X PBS and incubated with the staining mixture overnight at 37°C. β-galactosidase-positive cells were visualized as blue cells when examined using a light microscope (magnification, ×400). For NC- and miRNA-1470 inhibitor-transfected cells, a total of 100 cells were counted from different fields of the plate, and the percentage of blue cells was calculated.
Statistical analysis
All results were confirmed in at least three independent experiments, and data from one representative experiment are presented. Student's t-test was used to compare the means of experimental groups. All quantitative data are presented as the mean ± standard deviation. The analysis was performed using GraphPad Prism (version 5.01; GraphPad Software, La Jolla, CA, USA). For all comparisons, P<0.05 was considered to indicate a statistically significant difference.
Results
ESCC tissues overexpress miR-1470
In the present study, it was revealed that the expression level of miR-1470 was increased in ESCC tissues compared with their adjacent non-malignant tissues using Agilent Human miRNA Microarrays (Fig. 1A). The expression of miR-1470 was further evaluated in 5 ESCC cell lines (KYSE30, KYSE180, KYSE450, KYSE150 and KYSE510) using RT-qPCR. The results revealed that KYSE150 and KYSE510 exhibited increased expression levels compared with other cell lines (Fig. 1B).
Figure 1.
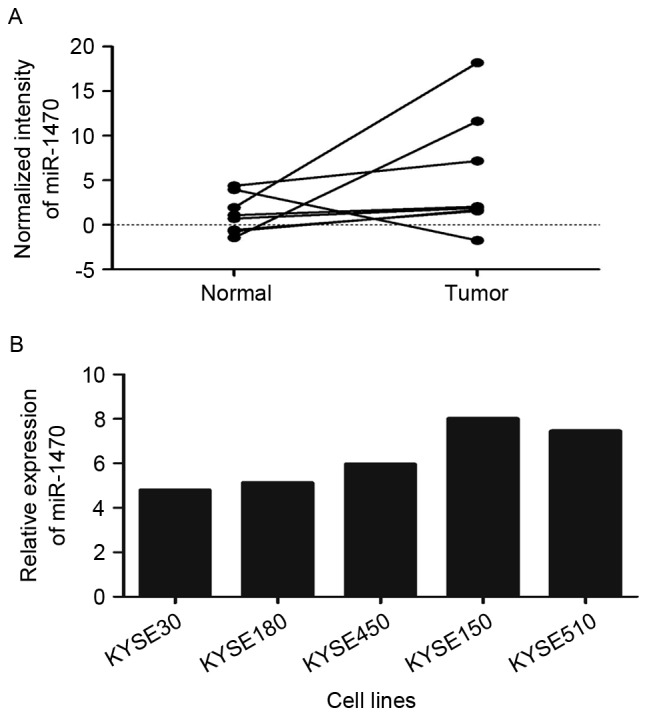
Upregulation of miR-1470 in ESCC tissues. (A) miR-1470 was upregulated in ESCC tissues when compared with normal tissues using microarray analysis. (B) The expression level of miR-1470 in KYSE30, KYSE180, KYSE450, KYSE150 and KYSE510 cells was detected using reverse transcription-quantitative polymerase chain reaction analysis. miR-1470 expression was normalized against an endogenous control U6. ESCC, esophageal squamous cell carcinoma; miR-1470, micro RNA-1470.
Knockdown of miR-1470 suppresses proliferation of ESCC cells by regulating cell cycle regulatory protein
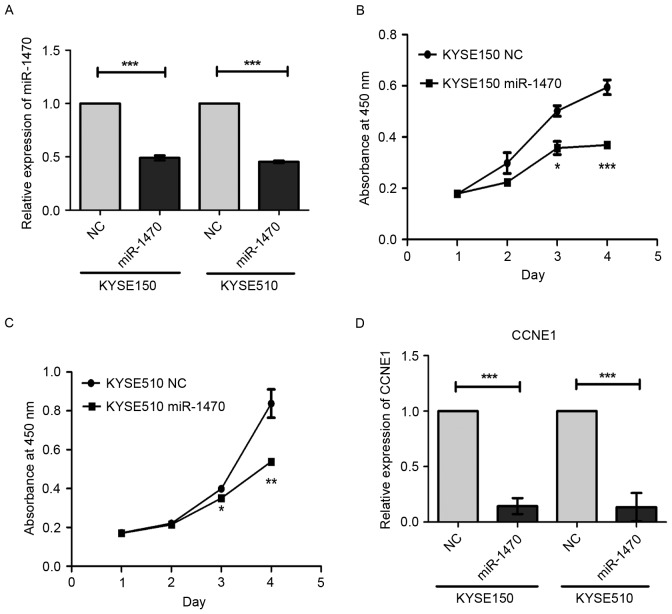
To explore the role of miR-1470 in esophageal carcinogenesis, the cell viability was evaluated using a CCK-8 assay. Knockdown of miR-1470 using the inhibitor significantly inhibited proliferation of KYSE150 and KYSE510 cells (Fig. 2A-C). The mRNA level of the cell cycle regulatory gene CCNE1 in miR-1470 inhibitor-transfected cells were significantly decreased (Fig. 2D). These observations indicate that miR-1470 may promote cell proliferation via up-regulating CCNE1 in ESCC.
Figure 2.
Downregulation of miR-1470 suppresses the proliferation and expression of cell cycle protein CCNE1 in KYSE150 and KYSE510 cells. (A) Level of miR-1470 in KYSE150 and KYSE510 cells with miR-1470 inhibitor and a negative control were detected using reverse transcription-quantitative polymerase chain reaction analysis. miR-1470 expression was normalized against an endogenous control U6. Cell proliferation was analyzed in miR-1470-knockdown and negative control (B) KYSE150 and (C) KYSE510 cells. (D) CCNE1 mRNA level in KYSE150 and KYSE510 cells following treatment with an miR-1470 inhibitor. *P<0.05, **P<0.01, ***P<0.001.
Knockdown of miR-1470 inhibits the migration of ESCC cells by decreasing the expression of MMPs
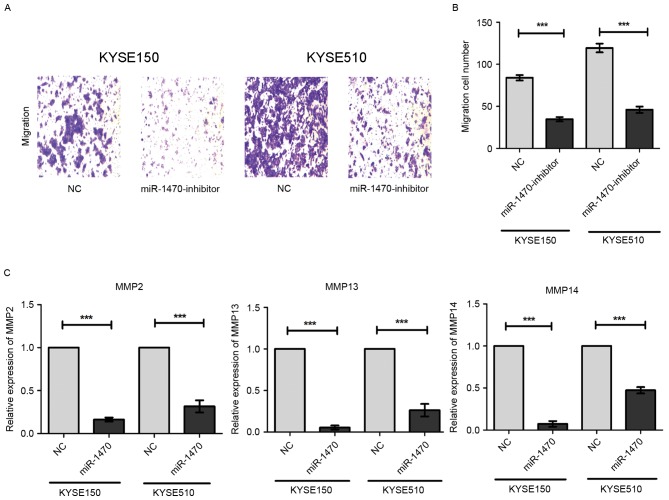
Using a Transwell assay, it was identified that knockdown of miR-1470 led to a decrease in the number of migratory KYSE150 and KYSE510 cells (Fig. 3A). Quantification and statistical analysis of the decrease in migratory cell numbers revealed that the difference was statistically significant (Fig. 3B). MMPs, a family of zinc-binding proteins, have been demonstrated to serve a role in tumor cell metastasis owing to their ability to degrade the extracellular matrix. Using RT-qPCR, we found that knockdown of miR-1470 significantly decreased the expression levels of MMP2, MMP13 and MMP14. Taken together, these data suggest that down-regulated miR-1470 may suppress the migration of ESCC cells via inhibition of MMPs.
Figure 3.
miR-1470 inhibits the migration of ESCC cells. (A) Migration of cells was analyzed. Representative images of the cells which had travelled through the micropore membrane. (B) Quantification of the number of migratory cells. ***P<0.001. (C) The mRNA expression levels of MMP2, MMP13 and MMP14 were detected using the reverse transcription-quantitative polymerase chain reaction. All three MMPs examined revealed significantly different expression levels in cells transfected with miR-1470 inhibitor when compared with those transfected with NC. ***P<0.001.
Suppression of miR-1470 induces senescence and activates apoptosis pathway in ESCC cells
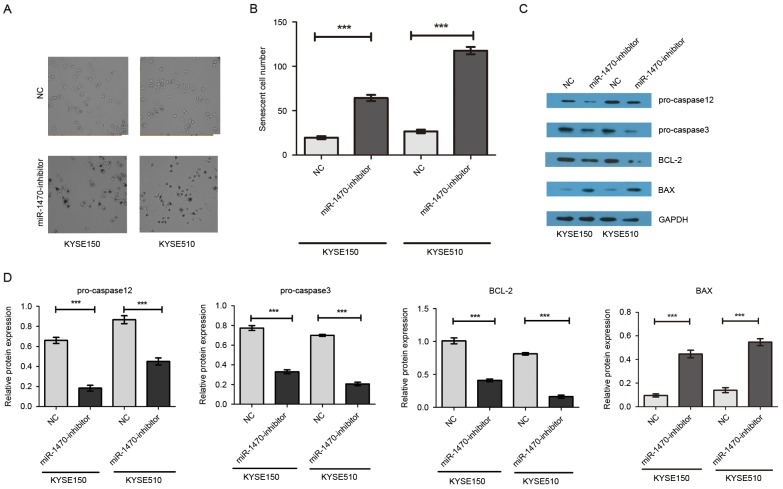
To investigate the effect of miR-1470 on cell senescence, morphological evaluation of KYSE150 and KYSE510 cells was performed following knockdown of miR-1470 expression. The results revealed an elongated phenotype resembling cellular senescence (Fig. 4A and B). As presented in Fig. 4C and D, compared with the control group, knockdown of miR-1470 markedly decreased anti-apoptotic protein Bcl-2, pro-caspase-12, pro-caspase-3 and increased pro-apoptotic protein BAX. These results suggest that suppression of miR-1470 may induce cell apoptosis.
Figure 4.
miR-1470 induces cell senescence via the apoptotic Bcl-2 signaling pathway. (A) Cell senescence was analyzed by β-galactosidase staining at 48 h post-transfection. Representative images of senescent cells. (B) Number of senescent cells. ***P<0.001. (C) The expression levels of pro-caspase-12, pro-caspase-3, Bcl-2 and BAX were determined by western blotting at 48 h post-transfection and normalized to that of β-actin. (D) The levels of pro-caspase-12, pro-caspase-3 and Bcl-2 were significantly decreased and that of BAX was significantly increased in cells transfected with miR-1470 inhibitor when compared with the cells transfected with NC. Results represent the mean ± standard deviation of at least three independent experiments. ***P<0.001.
Discussion
Previous studies have demonstrated that miRNAs are frequently dysregulated in numerous types of human cancer. As the roles of miRNAs in cancer are gradually investigated, their potential as a therapeutic target has generated interest with regard to the development of novel strategies for treating cancer (22–24). Previous studies have characterized the miRNA expression profiles associated with distinct stages of ESCC, and dysregulation of miRNAs has been demonstrated to serve a role in esophageal carcinogenesis (25–28). miR-375 inhibits tumor growth and metastasis in ESCC through the suppression of insulin-like growth factor 1 receptor (27). Cui et al (29) demonstrated that targeting oncogenic phospholipase Cε1 using miR-145 impaired tumor proliferation and metastasis of ESCC. Although Nie et al (30) demonstrated that miR-1470 mediated lapatinib-induced p27 up-regulation by targeting c-jun, the expression status, role and underlying molecular oncogenic mechanism of miR-1470 in esophageal cancer progression remain to be established.
miR-1470 is located at 19p13.12. In the present study, upregulation of miR-1470 in ESCC tissues was revealed using microarray. Functional experiments further revealed that downregulation of miR-1470 significantly inhibited the proliferation of ESCC cells, and also decreased the level of the cell cycle regulatory gene CCNE1. To the best of our knowledge, prior to the present study there has been no reported association between miR-1470 and CCNE1. The results of the present study indicate that miR-1470 positively regulated CCNE1. The results suggest that upregulation of miR-1470 expression in ESCC promoted cell proliferation by accelerating cell cycle transition.
Metastasis is the primary cause of mortality in patients with ESCC. It was identified that downregulated miR-1470 expression inhibited the migration of ESCC cells in vitro. MMPs are proteolytic enzymes that serve a role in the transformation and progression of tumors at all stages, particularly during invasion and metastasis (31,32). In the present study, downregulation of miR-1470 significantly suppressed the expressions of MMP2, MMP13 and MMP14. The results suggested that overexpression of miR-1470 in ESCC promoted the migration of tumor cells via increasing the expression levels of MMPs.
Senescence generally contributes to the elimination of damaged cells by phagocytic cells, and to the subsequent promotion of tissue remodeling, in a similar manner to apoptosis (33). In the present study, knockdown of miR-1470 significantly induced cell senescence and activated the apoptotic signaling pathway.
In conclusion, it was demonstrated that downregulation of miR-1470 significantly inhibited ESCC cell proliferation and migration. The identification of miR-1470 provides insight into the pathogenesis of ESCC, and may represent a potential therapeutic target for the treatment of ESCC.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81460425 and 81760526), the Yunnan Provincial Research Foundation for Basic Research, China (grant no. 2013FD012), the Foundation for the Talents of Kunming University of Science and Technology (grant no. KKSY201226099) and the Open Project Program of State Key Laboratory of Molecular Oncology (SKL-KF-2017-11).
References
- 1.Zhao P, Dai M, Chen W, Li N. Cancer trends in China. Jpn J Clin Oncol. 2010;40:281–285. doi: 10.1093/jjco/hyp187. [DOI] [PubMed] [Google Scholar]
- 2.Mariette C, Piessen G, Triboulet JP. Therapeutic strategies in oesophageal carcinoma: Role of surgery and other modalities. Lancet Oncol. 2007;8:545–553. doi: 10.1016/S1470-2045(07)70172-9. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Ell C, Lorenz D. Diagnosis and treatment of oesophageal carcinoma: Changes in every respect. Viszeralmedizin. 2015;31:314. doi: 10.1159/000441536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acunzo M, Croce CM. MicroRNA in Cancer and Cachexia-A Mini-Review. J Infect Dis. 2015;212(Suppl 1):S74–S77. doi: 10.1093/infdis/jiv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Qin X, Li Y, Zhang X, Niu R, Zhang H, Cui A, An W, Wang X. MiR-133a suppresses the migration and invasion of esophageal cancer cells by targeting the EMT regulator SOX4. Am J Transl Res. 2015;7:1390–1403. [PMC free article] [PubMed] [Google Scholar]
- 9.Mao Y, Li L, Liu J, Wang L, Zhou Y. MiR-495 inhibits esophageal squamous cell carcinoma progression by targeting Akt1. Oncotarget. 2016;7:51223–51236. doi: 10.18632/oncotarget.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llambi F, Green DR. Apoptosis and oncogenesis: Give and take in the BCL-2 family. Curr Opin Genet Dev. 2011;21:12–20. doi: 10.1016/j.gde.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulda S, Meyer E, Debatin KM. Inhibition of TRAIL-induced apoptosis by Bcl-2 overexpression. Oncogene. 2002;21:2283–2294. doi: 10.1038/sj.onc.1205258. [DOI] [PubMed] [Google Scholar]
- 12.Thoms HC, Dunlop MG, Stark LA. p38-mediated inactivation of cyclin D1/cyclin-dependent kinase 4 stimulates nucleolar translocation of RelA and apoptosis in colorectal cancer cells. Cancer Res. 2007;67:1660–1669. doi: 10.1158/0008-5472.CAN-06-1038. [DOI] [PubMed] [Google Scholar]
- 13.Törmänen-Näpänkangas U, Soini Y, Kahlos K, Kinnula V, Pääkkö P. Expression of caspases-3, −6 and −8 and their relation to apoptosis in non-small cell lung carcinoma. Int J Cancer. 2001;93:192–198. doi: 10.1002/ijc.1315. [DOI] [PubMed] [Google Scholar]
- 14.Li YH, Wang C, Meng K, Chen LB, Zhou XJ. Influence of survivin and caspase-3 on cell apoptosis and prognosis in gastric carcinoma. World J Gastroenterol. 2004;10:1984–1988. doi: 10.3748/wjg.v10.i13.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SX, Chai L, Cai ZG, Jin LJ, Chen Y, Wu HR, Sun Z. Expression of survivin and caspase 3 in oral squamous cell carcinoma and peritumoral tissue. Asian Pac J Cancer Prev. 2012;13:5027–5031. doi: 10.7314/APJCP.2012.13.10.5027. [DOI] [PubMed] [Google Scholar]
- 16.Nassar A, Lawson D, Cotsonis G, Cohen C. Survivin and caspase-3 expression in breast cancer: Correlation with prognostic parameters, proliferation, angiogenesis, and outcome. Appl Immunohistochem Mol Morphol. 2008;16:113–120. doi: 10.1097/PAI.0b013e318032ea73. [DOI] [PubMed] [Google Scholar]
- 17.Hsia JY, Chen CY, Chen JT, Hsu CP, Shai SE, Yang SS, Chuang CY, Wang PY, Miaw J. Prognostic significance of caspase-3 expression in primary resected esophageal squamous cell carcinoma. Eur J Surg Oncol. 2003;29:44–48. doi: 10.1053/ejso.2002.1338. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Yang H, Wen J, Luo K, Liu Q, Huang Y, Zheng Y, Tan Z, Huang Q, Fu J. NHE9 induces chemoradiotherapy resistance in esophageal squamous cell carcinoma by upregulating the Src/Akt/β-catenin pathway and Bcl-2 expression. Oncotarget. 2015;6:12405–12420. doi: 10.18632/oncotarget.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickson AW, Meng XW, Kaufmann SH. Anticancer therapy: Boosting the bang of Bim. J Clin Invest. 2008;118:3582–3584. doi: 10.1172/JCI37553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 21.Stauber RH, Mann W, Knauer SK. Nuclear and cytoplasmic survivin: Molecular mechanism, prognostic, and therapeutic potential. Cancer Res. 2007;67:5999–6002. doi: 10.1158/0008-5472.CAN-07-0494. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Wang C, Shan S, Liu X, Jiang Z, Ren T. TLR4/ROS/miRNA-21 pathway underlies lipopolysaccharide instructed primary tumor outgrowth in lung cancer patients. Oncotarget. 2016;7:42172–42182. doi: 10.18632/oncotarget.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xicola RM, Bontu S, Doyle BJ, Rawson J, Garre P, Lee E, de la Hoya M, Bessa X, Clofent J, Bujanda L, et al. Association of a let-7 miRNA binding region of TGFBR1 with hereditary mismatch repair proficient colorectal cancer (MSS HNPCC) Carcinogenesis. 2016;37:751–758. doi: 10.1093/carcin/bgw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bimonte S, Leongito M, Barbieri A, Del Vecchio V, Falco M, Giudice A, Palaia R, Albino V, Di Giacomo R, Petrillo A, et al. The therapeutic targets of miRNA in hepatic cancer stem cells. Stem Cells Int. 2016;2016:1065230. doi: 10.1155/2016/1065230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu BL, Xu LY, Du ZP, Liao LD, Zhang HF, Huang Q, Fang GQ, Li EM. MiRNA profile in esophageal squamous cell carcinoma: Downregulation of miR-143 and miR-145. World J Gastroenterol. 2011;17:79–88. doi: 10.3748/wjg.v17.i1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z, Li J, Tian L, Zhou C, Gao Y, Zhou F, Shi S, Feng X, Sun N, Yao R, et al. MiRNA expression profile reveals a prognostic signature for esophageal squamous cell carcinoma. Cancer Lett. 2014;350:34–42. doi: 10.1016/j.canlet.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 28.Winther M, Alsner J, Tramm T, Baeksgaard L, Holtved E, Nordsmark M. Evaluation of miR-21 and miR-375 as prognostic biomarkers in esophageal cancer. Acta Oncol. 2015;54:1582–1591. doi: 10.3109/0284186X.2015.1064161. [DOI] [PubMed] [Google Scholar]
- 29.Cui XB, Li S, Li TT, Peng H, Jin TT, Zhang SM, Liu CX, Yang L, Shen YY, Li SG, et al. Targeting oncogenic PLCE1 by miR-145 impairs tumor proliferation and metastasis of esophageal squamous cell carcinoma. Oncotarget. 2016;7:1777–1795. doi: 10.18632/oncotarget.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie W, Song W, Zhang W, Wang Y, Zhu A, Shao J, Guan X. miR-1470 mediates lapatinib induced p27 upregulation by targeting c-jun. J Cell Physiol. 2015;230:1630–1639. doi: 10.1002/jcp.24910. [DOI] [PubMed] [Google Scholar]
- 31.Banday MZ, Sameer AS, Mir AH, Mokhdomi TA, Chowdri NA, Haq E. Matrix metalloproteinase (MMP)-2, −7 and −9 promoter polymorphisms in colorectal cancer in ethnic Kashmiri population-A case-control study and a mini review. Gene. 2016;589:81–89. doi: 10.1016/j.gene.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 32.Serra R, Grande R, Gallelli L, Rende P, Scarcello E, Buffone G, Caliò FG, Gasbarro V, Amato B, de Franciscis S. Carotid body paragangliomas and matrix metalloproteinases. Ann Vasc Surg. 2014;28:1665–1670. doi: 10.1016/j.avsg.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 33.Muñoz-Espín D, Serrano M. Cellular senescence: From physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]