Abstract
Previous studies have shown that the L-type amino acid transporter 1 (LAT1) is highly expressed in many types of cancer. Upregulated LAT1 expression is considered to be associated with cancer cell proliferation. In the present study, we investigated LAT1 expression in 210 patients with colorectal cancer (CRC) and 40 patients with colonic adenoma using an immunohistochemical method, and analyzed the associations between LAT1 expression and clinicopathological factors and prognosis. The biological significance of LAT1 was also examined under conditions with sub-normal amounts of essential amino acids using colon cancer cell lines. High expression of LAT1 was observed in 152 of 210 CRC patients (72.4%) and 12 of 40 patients with colonic adenoma (30%), and this difference in the frequency of LAT1 expression between CRC and adenoma was significant (P<0.001). High expression of LAT1 was associated with venous invasion (P=0.027). The restriction of amino acids suppressed cell proliferation in CRC cells with higher LAT1 expression, while cellular proliferation was not suppressed in the cells expressing lower levels of LAT1. Mammalian target of rapamycin (mTOR) expression was also downregulated under restricted availability of amino acids, suggesting that the restriction of amino acids induced an antitumor effect through inhibition of the LAT1/mTOR pathway. In summary, the present study demonstrated that LAT1 expression is frequently upregulated in CRC and is associated with cancer cell proliferation via the mTOR pathway.
Keywords: L-type amino acid transporter 1, colorectal cancer, mammalian target of rapamycin
Introduction
Amino acid transport systems play an important role in supplying amino acids to cells (1). To date, many amino acid transport systems in mammals have been identified, and are classified into several transporter families based on differences in substrate selectivity and ion dependence (1,2). Among these amino acid transporters, L-type amino acid transporter 1 (LAT1), which belongs to amino acid transport system L, is highly expressed in tumor cell lines (3,4). LAT1 is an Na+-independent amino acid transporter and transports large branched-chain and aromatic neutral amino acids, including the essential amino acids histidine, isoleucine, leucine, phenylalanine, tryptophan, tyrosine, methionine and valine (1,3,4). LAT1 forms heterodimeric complexes with the 4F2 cell-surface antigen heavy chain (4F2hc) via a disulfide bond (3). The expression of LAT1 mRNA is restricted to certain organs, such as the brain, spleen, placenta, and testis (3–5). Furthermore, it has been reported that LAT1 is highly expressed in cancer tissue of various types of cancer, including oral squamous cell carcinoma (6), esophageal carcinoma (7), gastric cancer (8), prostate cancer (9), non-small cell lung carcinoma (10), biliary tract cancer (11), pancreatic cancer (12), and breast cancer (13).
Although amino acids are essential nutrients due to their roles as substrates of protein synthesis and in cellular ATP generation, they also regulate translation, transcription, and cell growth via mammalian target of rapamycin (mTOR) (14). Previous studies suggest that the inhibition of mTOR by rapamycin can exert antitumor effects in various types of tumors in vitro and in vivo (15–17).
In the present study, we retrospectively examined LAT1 expression in CRC, and demonstrated that LAT1 expression in cancer tissues was increased when compared with that in nonmalignant tissues. In addition, we showed that the biological significance of upregulated LAT1 was associated with its contribution to cellular proliferation via the regulation of the mTOR pathway in vitro. We hypothesized that restriction of the amount of amino acids available for uptake into cancer cells through LAT1 could lead to similar antitumor effects. Taken together, the findings indicate that LAT1 may be a promising molecular target for the treatment of CRC in the future.
Materials and methods
Patients
This study was performed in accordance with ethical guidelines for clinical research with the approval of our institutional ethics committee. Informed consent was obtained from the individuals included in the study.
Primary CRC specimens were obtained from 210 patients (123 male and 87 female) who underwent curative resection at Fukushima Medical University between January 1990 and December 2007. The ages of the patients ranged from 24 to 89 years, with a mean age of 66 years. At the time of primary tumor resection, the carcinomas were staged according to the UICC classification: 7 cases were allocated to stage 0, 33 to stage I, 82 to stage II, 59 to stage III, and 29 to stage IV. Furthermore, colonic adenoma specimens were obtained from 40 patients (36 male and 4 female) who had undergone polypectomy at Fukushima Medical University between March 2007 and April 2009. The ages of the colonic adenoma patients ranged from 39 to 79 years, with a mean of 64.4 years.
Immunohistochemical staining
All specimens of CRC and colonic adenoma were fixed in formalin and embedded in paraffin. Two serial sections (thickness, 4 µm) were prepared: One was used for hematoxylin and eosin (H&E) staining, and the other was used for immunohistochemical staining of LAT1. Immunohistochemical staining was performed on the paraffin-embedded sections using a polymer peroxidase method. Briefly, deparaffinized and rehydrated sections were treated with 0.3% hydrogen peroxide in methanol for 30 min to block endogenous peroxidase activity, before sections were autoclaved in 0.01 M pH 6.0 citrate buffer for 5 min, and cooled for 30 min. After rinsing in PBS, the sections were incubated with affinity-purified anti-LAT1 antibodies (mouse monoclonal; dilution, 1:200; Cosmo Bio Co., Tokyo, Japan) overnight at 4°C. A further wash in PBS was followed by treatment with goat anti-mouse immunoglobulin antibodies conjugated to a peroxidase-labeled polymer (ENvision+kit; Dako Cytomation, Glostrup, Denmark) as the secondary antibody for 30 min at room temperature. Staining was visualized with diaminobenzidine (DAB), and was followed by counterstaining with hematoxylin.
Evaluation of LAT1 expression
The evaluation of staining was performed according to a previously described method with minor modifications (18). LAT1 expression of cells was considered positive only if distinct cell staining was present. The proportion of positively stained tumor cells was scored as follows: 1, ≤10% of tumor cells stained; 2, 11–25% stained; 3, 26–50% stained; and 4, ≥51% stained. Staining scores of 3 and 4 were classified as high expression of LAT1, whereas staining scores of 1 and 2 were classified as low expression of LAT1.
Cell culture
We used the cultured human colon cancer cell lines, including LoVo, SW48, SW620, and SW837, which were originally obtained from the American Type Culture Collection (Rockville, MD, USA) to examine LAT1 expression in primary and metastatic site of colorectal cancer. SW48 and SW837 cells were derived from primary colon and rectal cancer, respectively. LoVo and SW620 were derived from metastatic site of lymph node. The characteristics of theses cell lines are described on the following site: https://www.atcc.org/Products/Cells_and_Microorganisms/Cell_Lines/Human.aspx. The cells were grown at 37°C in the presence of 5% CO2 in the recommended media. In the experiments, the amino acid contents of the media were modified. These media were purchased from Wako (Osaka, Japan). Fetal bovine serum (Nichirei Bioscience, Tokyo, Japan) was added to all media at a concentration of 10%.
Protein extraction and western blot analysis
Cells (1×106) were seeded in 10 cm plates and allowed 1 day for attachment. Subsequently, the medium was removed and exchanged with RPMI-1640 (control) or with a medium containing 1/4 of the quantity of amino acids present in the control (amino acid-restricted medium). The cells were cultured for 72 h and harvested. Cultured cells were washed with cold PBS and lysed in RIPA buffer (1X TBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 0.004% sodium azide) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) containing 10 µl PMSF solution, 10 µl sodium orthovanadate solution, and 10 µl protease inhibitor cocktail solution per ml of 1X RIPA lysis buffer. Lysates were cleared by centrifugation at 15,000 rpm for 15 min at 4°C. Protein concentration was determined by the Bradford assay using a Quick Start™ Bradford Dye Reagent and Quick Start™ Bovine Serum Albumin Standard set (Bio-Rad, Hercules, CA, USA). The protein samples (15 µg) were then electrophoresed on a Novex 4–12% Tris-Glycine Gel (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the separated proteins were transferred to nitrocellulose membranes (iBlot™ Gel Transfer System; Invitrogen; Thermo Fisher Scientific, Inc.). The membranes were blocked with Super Block Blocking Buffers (Thermo Fisher Scientific, Inc.) and incubated with a 1:200 dilution of rabbit anti-LAT1 antibody (MBL, Nagoya, Japan) for 2 h or with a 1:200 dilution of rabbit anti-mTOR antibody (Cell Signaling Technology, Danvers, MA, USA) or mouse monoclonal anti-β-actin antibody (Santa Cruz Biotechnology, Inc.) for 1 h. The secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit antibodies (Santa Cruz Biotechnology, Inc.), applied at a 1:5,000 dilution for 30 min. The membranes were incubated with Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.) and scanned with a luminescent imaging analyzer LAS-4000 mini (Fujifilm, Tokyo, Japan).
Cell proliferation assay
The cell proliferation assay was performed using a Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). The cells were seeded at a concentration of 5×103 cells/well in 96-well plates. After allowing 1 day for attachment, the medium was removed and exchanged with RPMI-1640 (control) or the medium in which the amount of amino acids (histidine, isoleucine, leucine, phenylalanine, tryptophan, tyrosine, methionine, and valine) were adjusted to 1/4 of the amount of amino acids in the control medium. After culturing for each 24 h from the start, cells were incubated with 100 µl of 10% CCK-8 reagent for 4 h at 37°C. The color reaction was measured at 450 nm with a Bio Rad ELISA reader (Richmond, CA, USA).
Statistical analysis
Associations between categorical variables were evaluated with the χ2 test, Fisher's exact test or the Mann-Whitney U test. Differences at P<0.05 were considered significant. A survival analysis was conducted using the Kaplan-Meier method, and the log-rank test was used to determine the significance of differences between the survival curves. The period of overall survival (OS) was calculated from the time of surgery to the date of death from any cause, and was censored at the time of the last visit to our hospital or December 2014, whichever came first. All statistical analyses were performed using SPSS 11.0 software (SPSS, Inc., Chicago, IL, USA). The cell proliferation assays were performed in triplicate, and the results were presented as the mean ± standard deviation; statistical significance was analyzed with the Student's t-test for comparison of the two groups, and P<0.05 was considered to indicate a statistically significant difference.
Results
Immunohistochemical analysis of LAT1 in CRC and colonic adenoma
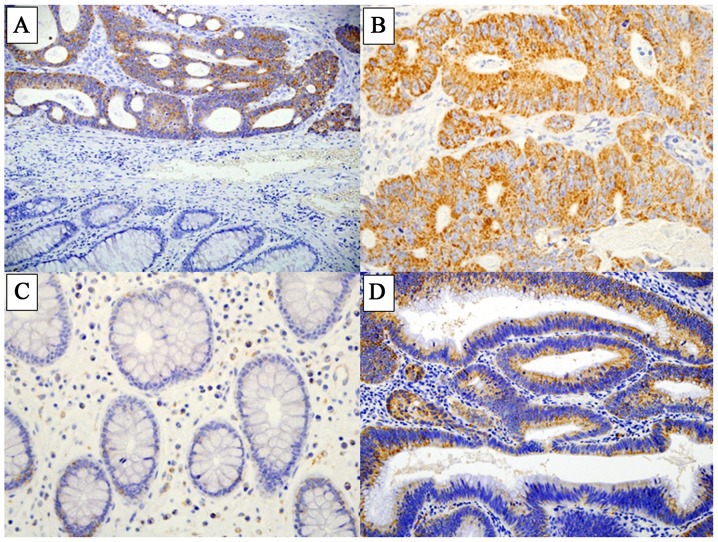
LAT1 expression was detected at high levels in CRC tissues, while few normal epithelial cells in the cancer-adjacent non-malignant tissue were positively stained for LAT1 (Fig. 1A-C). LAT1 was mostly expressed in the cellular membrane and cytoplasm. Among the 210 CRC cases, a staining score of 1 was recorded in 22 cases (10.5%), score 2 in 36 cases (17.1%), score 3 in 50 cases (23.8%), and score 4 in 102 cases (48.6%).
Figure 1.
Representative staining of L-type amino acid transporter 1 (LAT1) expression in colorectal cancer (CRC) tissues and colonic adenoma. (A) Conjugated images of cancer tissues and normal tissues (magnification, ×40). (B) High expression (score 4) of LAT1 in cancer tissues (magnification, ×200). (C) No LAT1 expression was observed in normal mucosa (magnification, ×200). (D) LAT1 expression (score 4) in colonic adenoma (magnification, ×200).
In the colonic adenoma specimens, LAT1 was expressed in the membrane and cytoplasm of adenomatous cells (Fig. 1D). The distribution of LAT1 scores among the 40 cases was as follows: Score 1, 19 cases (47.5%); score 2, 9 cases (22.5%); score 3, 7 cases (17.5%); and score 4, 5 cases (12.5%).
High expression of LAT1, indicated by a staining score of 3 or 4, was detected in 152 (72.4%) of the 210 CRC cases. In the 40 colonic adenoma cases, 12 cases (30.0%) showed high expression of LAT1. The incidence of high LAT1 expression was significantly different between CRC and adenoma (P<0.001).
Associations between LAT1 expression in CRC and clinicopathological factors and prognosis
High expression of LAT1 was significantly associated with the depth of invasion (Tis and T1 vs. T2-4; P=0.048) and venous invasion (P=0.027) in CRC tissues (Table 1). No significant associations could be determined between the level of LAT1 expression and other variables (patient sex or age, and tumor location, histology, lymphatic invasion, lymph node metastasis and stage) (Table I).
Table I.
Clinicopathologic correlation of LAT1 expression in CRC.
| LAT1 expression | |||
|---|---|---|---|
| Characteristics | High expression (n=152) (%) | Low expression (n=58) (%) | P-value |
| Gender | 0.53 | ||
| Male | 87 (57.2) | 36 (62.1) | |
| Female | 65 (42.8) | 22 (37.9) | |
| Age (years) | 0.89 | ||
| <60 | 46 (30.3) | 17 (29.3) | |
| ≥60 | 106 (69.7) | 41 (70.7) | |
| Tumor location | 0.54 | ||
| Right | 53 (34.9) | 16 (27.6) | |
| Left | 52 (34.2) | 24 (41.4) | |
| Rectum | 47 (30.9) | 18 (31.0) | |
| Histology | 0.57 | ||
| Well | 69 (45.4) | 26 (44.8) | |
| Moderate | 68 (44.7) | 24 (41.4) | |
| Poor | 5 (3.3) | 1 (1.7) | |
| Mucinous | 10 (6.6) | 7 (12.1) | |
| Depth | 0.048 | ||
| Tis | 2 (1.3) | 5 (8.6) | (Tis, T1 vs. T2-4) |
| T1 | 10 (6.6) | 5 (8.6) | |
| T2 | 17 (11.2) | 8 (13.8) | |
| T3 | 110 (72.4) | 38 (65.5) | |
| T4 | 13 (8.5) | 2 (3.5) | |
| Lymphatic invasion | 0.066 | ||
| Absent | 27 (17.8) | 17 (29.3) | |
| Present | 125 (82.2) | 41 (70.7) | |
| Venous invasion | 0.027 | ||
| Absent | 24 (15.8) | 17 (29.3) | |
| Present | 128 (84.2) | 41 (70.7) | |
| Lymph node metastasis | 0.42 | ||
| Negative | 89 (58.6) | 35 (60.3) | |
| Positive | 63 (41.4) | 19 (32.8) | |
| Unknown | 0 (0.0) | 4 (6.9) | |
| UICC stage | 0.21 | ||
| 0 | 2 (1.3) | 5 (8.6) | |
| I | 22 (14.5) | 11 (19.0) | |
| II | 63 (41.4) | 18 (31.0) | |
| III | 43 (28.3) | 16 (27.6) | |
| IV | 22 (14.5) | 7 (12.1) | |
LAT1, L-type amino acid transporter 1; CRC, colorectal cancer; UICC, Union for International Cancer Control.
Regarding OS in CRC patients, no significant difference was observed between patients with high expression of LAT1 (n=152) and those with low expression of LAT1 (n=58) (P=0.57) (Fig. 2).
Figure 2.
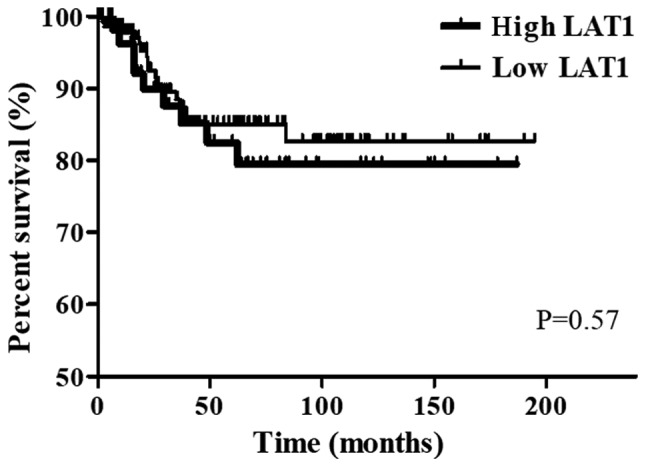
Kaplan-Meier curves showing overall survival in CRC patients with high LAT1 expression and low LAT1 expression. LAT1, L-type amino acid transporter 1; CRC, colorectal cancer.
Expression of LAT1 protein in CRC cell lines and cell proliferation under limited availability of essential amino acids
LAT1 expression was detected as a band of ~38 kDa in all four CRC cell lines (LoVo, SW48, SW620, and SW837) (Fig. 3A). The level of LAT1 expression varied depending on the cell line: LAT1 expression in SW620 cells was relatively weak in comparison with the other CRC cell lines (Fig. 3A).
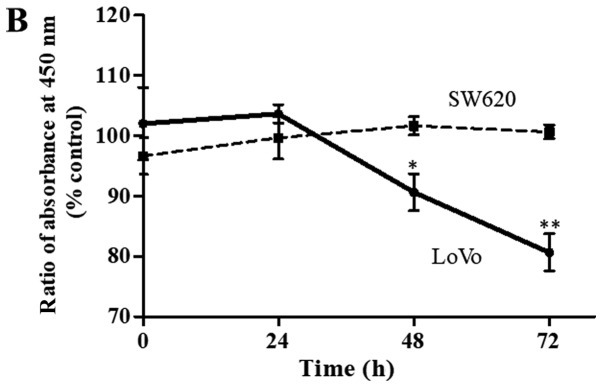
Figure 3.
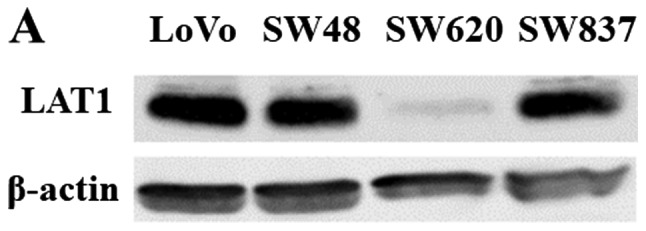
(A) LAT1 protein expression detected by western blotting in four CRC cell lines. β-actin was probed as an internal control. (B) Time-dependent effects of the restriction of essential amino acids on cell proliferation in LoVo cells (solid line) and SW620 cells (dotted line). LoVo and SW620 cells were incubated with the control medium or the amino acid-restricted medium (containing 1/4 of the quantity of amino acids present in the control medium) for 0–72 h. Each data point represents the mean ± standard deviation of the ratio of the absorbance at 450 nm in the amino acid-restricted medium relative to those in the control medium. The experiment was performed independently three times in triplicate. *P<0.01 vs. control; **P<0.001 vs. control. LAT1, L-type amino acid transporter 1; CRC, colorectal cancer.
Based on the aforementioned LAT1 expression data from the CRC cell lines, LoVo cells, which exhibited a relatively high level of LAT1 expression, and SW620 cells, which showed a relatively low level of LAT1 expression, were used for the cell proliferation assays with control medium or amino acid-restricted medium. The rate of LoVo cell proliferation in the amino acid-restricted medium was significantly lower than that in the control medium, while the SW620 cell proliferation rate did not change significantly (Fig. 3B).
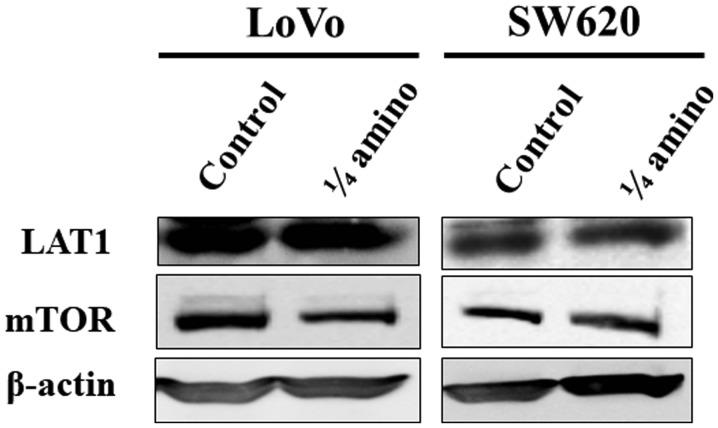
LAT1 and mTOR expression under limited availability of essential amino acids
To elucidate the mechanism underlying the growth suppression of CRC cell lines by restriction of amino acids, the expression levels of LAT1 and mTOR were examined in LoVo and SW620 cells cultured in the control medium or the amino acid-restricted medium for 72 h. LAT1 expression was not altered between the two groups in either of the cell lines. The limited availability of essential amino acids reduced the level of mTOR expression in LoVo cells, whereas no such alteration of mTOR expression was observed in the SW620 cells (Fig. 4).
Figure 4.
Effect of limited availability of essential amino acids on mTOR expression in LoVo and SW620 cells, which were incubated in the control medium or the amino acid-restricted medium (containing 1/4 of the quantity of amino acids present in the control medium) for 72 h. Whole cell lysates (30 µg of protein) were subjected to western blot analysis. LAT1, mTOR, and β-actin were probed. β-actin expression was used as an internal control. LAT1, L-type amino acid transporter 1; mTOR, mammalian target of rapamycin.
Discussion
Previous studies demonstrated that LAT1 was highly expressed in cancer tissue of various cancer types, including oral squamous cell cancer (6), esophageal cancer (7), gastric cancer (8), prostate cancer (9), non-small cell lung cancer (10), biliary tract cancer (11), pancreatic cancer (12), and breast cancer (13), using an immunohistochemical staining method, as in the present study. However, there have been few reports concerning LAT1 expression in CRC. In previous studies, high levels of LAT1 expression were detected in 35% of colon cancer cases (16/45 patients) (19) and 50% of rectal cancer cases (22/44 patients) (20). In the present study, we investigated the frequency of high LAT1 expression in 210 CRC patients, and found that LAT1 expression was highly expressed in >70% of cases. Increased LAT1 expression was frequently observed in the membrane and cytoplasm of cancer cells, while no LAT1 expression was detected in the normal mucosa. Regarding other types of cancer, the frequency of high LAT1 expression has been detected in 93 (59%) of 157 esophageal squamous cell carcinoma patients (7), 36 (41.4%) of 87 gastric cancer patients (8), 13 (24%) of 54 prostate cancer patients (9), 163 (51%) of 321 non small lung cancer patients (10), 89 (64%) of 139 biliary tract cancer patients (11), 51 (52.6%) in 97 pancreatic cancer patients (12), and 56 (43.4%) of 129 breast cancer patients (13). Thus, compared with other cancer types, the frequency of high LAT1 expression was relatively high in the cases of CRC examined in the present study. In addition to CRC, we found that LAT1 was also highly expressed in 30% of 40 cases of colonic adenoma examined. These findings suggested that LAT1 expression is elevated in the early/precursor stages of colon carcinogenesis, and may be associated with cellular proliferation. In addition, LAT1 expression was further enhanced in CRC tissues, indicating its possible involvement in the acquisition of malignant potential.
In the present study, high LAT1 expression was significantly associated with the depth of invasion and venous invasion, and showed no associations with any of the other clinicopathological factors investigated. No data regarding the association between the LAT1 expression in CRC and clinicopathological factors were reported in previous studies (19,20). Previous reports (7,8,10) indicated that LAT1 expression was associated with depth of invasion, lymph node metastasis, and venous invasion in esophageal squamous cell carcinoma, and with lymph node metastasis in gastric carcinoma and non-small cell lung cancer. Moreover, high LAT1 expression was reported to be associated with poor prognosis in several types of cancer (7,8,10–13). In the present study, no relationship between high LAT1 expression and lymph node metastasis, which might influence the prognosis, was identified in CRC. Additionally, no difference in prognosis between CRC patients with high LAT1 expression and those with low LAT1 expression was observed with regard to OS. Although the present study did not examine the Ki-67 labeling index, which has been widely used as a proliferation marker, several reports (8,10–12,19) have suggested that LAT1 expression is significantly associated with the Ki-67 labeling index in various cancers. Conflicting results (21–23) have been reported regarding the associations between the Ki-67 labeling index and clinical outcomes in CRC patients. Allegra et al (22), in a study of CRC patients who underwent curative resection between November 1977 and December 1990, reported that patients with high Ki-67 expression had significantly improved disease-free survival and OS rates than those with low levels of Ki-67. High LAT1 expression was consistently associated with cellular proliferation, which may result in the possibility of enhanced chemosensitivity. In the present study, we investigated LAT1 expression in patients with stage III or IV CRC who received 5FU-based monotherapy. Further investigation will be required to conclude the association between LAT1 expression and the prognosis of CRC patients treated with newer anticancer drugs, including oxaliplatin and irinotecan.
While amino acids that enter cancer cells through LAT1 (including leucine, isoleucine, phenylalanine, methionine, tyrosine, histidine, tryptophan, and valine) are essential nutrients since they are used as substrates for intracellular protein synthesis and ATP generation, they also regulate translation, transcription, and cell growth through mTOR (14). We have shown that the restriction of the availability of these eight different amino acids can suppress the viability of LoVo cells, which express a relatively high level of LAT1 protein; however, in SW620 cells, which show a relatively low protein expression level of LAT1, the cell viability did not differ between the control and amino acid-restricted media. Knockdown of LAT1 expression using an siRNA method may be valuable in further elucidating the function of LAT1 overexpression in cancer cells. In fact, a previous report revealed that the downregulation of LAT1 expression inhibited the growth, migration and invasion of gastric cancer cells (24). In the current study, we also demonstrated another LAT1-dependent method of cell growth suppression: A restriction of the availability of amino acids could suppress cell proliferation in the cancer cells with high-level LAT1 expression, whereas it did not affect cell growth in the cancer cells with low LAT1 expression, which may have a similar expression level to that in normal cells. In a previous study, the concentration of those eight amino acids in CRC tissue was approximately two times higher than that in colonic normal tissue (25). In normal tissues, LAT2 and other transporters facilitate the uptake of the amino acids (26). Therefore, the restriction of those amino acids may be selectively effective for the starvation of cancer cells with high LAT1 expression. Furthermore, we found that the restriction of the amino acids could suppress mTOR expression without alteration of LAT1 expression in LoVo cells. LAT1 is linked to mTOR via the intake of leucine, which stimulates the mTOR pathway and thereby affects cellular proliferation (27). Although previous studies (28,29) have demonstrated that essential amino acids activate the mTOR pathway, the exact mechanisms regulating mTOR expression remain unknown. However, the reason that the restriction of essential amino acids could inhibit cellular proliferation in cancer cells with a high level of LAT1 expression may be explained by the suppression of mTOR expression. Further experiments, including knock-down or overexpression of LAT1 and the use of rapamycin, will elucidate the role of the LAT1/mTOR pathway.
In the present study, we found that LAT1 expression was enhanced in approximately 70% of CRC and 30% of colonic adenoma tissues. Recently, LAT1 inhibitor compounds have been developed and have demonstrated growth inhibitory effects on HT-29 cells, which are derived from colon cancer, in in vivo and in vitro experiments (30). Furthermore, positron emission tomography (PET) has been attempted in lung cancer diagnosis using L-[3-18F]-α-methyltyrosine [(18F) FMT], an amino acid tracer for PET, since the uptake of [18F] FMT correlates with LAT1 expression (31). Accumulation of data, including the current results, will be useful for improving the diagnosis and treatment of CRC.
References
- 1.Christensen HN. Role of amino acid transport and countertransport in nutrition and metabolism. Physiol Rev. 1990;70:43–77. doi: 10.1152/physrev.1990.70.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Palacín M, Estévez R, Bertran J, Zorzano A. Molecular biology of mammalian plasma membrane amino acid transporters. Physiol Rev. 1998;78:969–1054. doi: 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- 3.Kanai Y, Segawa H, Miyamoto Ki, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98) J Biol Chem. 1998;273:23629–23632. doi: 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- 4.Yanagida O, Kanai Y, Chairoungdua A, Kim DK, Segawa H, Nii T, Cha SH, Matsuo H, Fukushima J, Fukasawa Y, et al. Human L-type amino acid transporter 1 (LAT1): Characterization of function and expression in tumor cell lines. Biochim Biophys Acta. 2001;1514:291–302. doi: 10.1016/S0005-2736(01)00384-4. [DOI] [PubMed] [Google Scholar]
- 5.Prasad PD, Wang H, Huang W, Kekuda R, Rajan DP, Leibach FH, Ganapathy V. Human LAT1, a subunit of system L amino acid transporter: Molecular cloning and transport function. Biochem Biophys Res Commun. 1999;255:283–288. doi: 10.1006/bbrc.1999.0206. [DOI] [PubMed] [Google Scholar]
- 6.Kim DK, Ahn SG, Park JC, Kanai Y, Endou H, Yoon JH. Expression of L-type amino acid transporter 1 (LAT1) and 4F2 heavy chain (4F2hc) in oral squamous cell carcinoma and its precursor lesions. Anticancer Res. 2004;24:1671–1675. [PubMed] [Google Scholar]
- 7.Honjo H, Kaira K, Miyazaki T, Yokobori T, Kanai Y, Nagamori S, Oyama T, Asao T, Kuwano H. Clinicopathological significance of LAT1 and ASCT2 in patients with surgically resected esophageal squamous cell carcinoma. J Surg Oncol. 2016;113:381–389. doi: 10.1002/jso.24160. [DOI] [PubMed] [Google Scholar]
- 8.Ichinoe M, Mikami T, Yoshida T, Igawa I, Tsuruta T, Nakada N, Anzai N, Suzuki Y, Endou H, Okayasu I. High expression of L-type amino-acid transporter 1 (LAT1) in gastric carcinomas: Comparison with non-cancerous lesions. Pathol Int. 2011;61:281–289. doi: 10.1111/j.1440-1827.2011.02650.x. [DOI] [PubMed] [Google Scholar]
- 9.Segawa A, Nagamori S, Kanai Y, Masawa N, Oyama T. L-type amino acid transporter 1 expression is highly correlated with gleason score in prostate cancer. Mol Clin Oncol. 2013;1:274–280. doi: 10.3892/mco.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, Hisada T, Tanaka S, Ishizuka T, Kanai Y, et al. Prognostic significance of L-type amino acid transporter 1 expression in resectable stage I–III nonsmall cell lung cancer. Br J Cancer. 2008;98:742–748. doi: 10.1038/sj.bjc.6604235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaira K, Sunose Y, Ohshima Y, Ishioka NS, Arakawa K, Ogawa T, Sunaga N, Shimizu K, Tominaga H, Oriuchi N, et al. Clinical significance of L-type amino acid transporter 1 expression as a prognostic marker and potential of new targeting therapy in biliary tract cancer. BMC Cancer. 2013;13:482. doi: 10.1186/1471-2407-13-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaira K, Sunose Y, Arakawa K, Ogawa T, Sunaga N, Shimizu K, Tominaga H, Oriuchi N, Itoh H, Nagamori S, et al. Prognostic significance of L-type amino-acid transporter 1 expression in surgically resected pancreatic cancer. Br J Cancer. 2012;107:632–638. doi: 10.1038/bjc.2012.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuya M, Horiguchi J, Nakajima H, Kanai Y, Oyama T. Correlation of L-type amino acid transporter 1 and CD98 expression with triple negative breast cancer prognosis. Cancer Sci. 2012;103:382–389. doi: 10.1111/j.1349-7006.2011.02151.x. [DOI] [PubMed] [Google Scholar]
- 14.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Le Bihan S, Marsaud V, Mercier-Bodard C, Baulieu EE, Mader S, White JH, Renoir JM. Calcium/calmodulin kinase inhibitors and immunosuppressant macrolides rapamycin and FK506 inhibit progestin-and glucocorticosteroid receptor-mediated transcription in human breast cancer T47D cells. Mol Endocrinol. 1998;12:986–1001. doi: 10.1210/mend.12.7.0128. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Adam RM, Santiestevan E, Freeman MR. The phosphatidylinositol 3′-kinase pathway is a dominant growth factor-activated cell survival pathway in LNCaP human prostate carcinoma cells. Cancer Res. 1999;59:2891–2897. [PubMed] [Google Scholar]
- 17.Luan FL, Hojo M, Maluccio M, Yamaji K, Suthanthiran M. Rapamycin blocks tumor progression: Unlinking immunosuppression from antitumor efficacy. Transplantation. 2002;73:1565–1572. doi: 10.1097/00007890-200205270-00008. [DOI] [PubMed] [Google Scholar]
- 18.Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, Hisada T, Ishizuka T, Kanai Y, Endou H, et al. L-type amino acid transporter 1 (LAT1) is frequently expressed in thymic carcinomas but is absent in thymomas. J Surg Oncol. 2009;99:433–438. doi: 10.1002/jso.21277. [DOI] [PubMed] [Google Scholar]
- 19.Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, Hisada T, Tanaka S, Ishizuka T, Kanai Y, et al. L-type amino acid transporter 1 and CD98 expression in primary and metastatic sites of human neoplasms. Cancer Sci. 2008;99:2380–2386. doi: 10.1111/j.1349-7006.2008.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebara T, Kaira K, Saito J, Shioya M, Asao T, Takahashi T, Sakurai H, Kanai Y, Kuwano H, Nakano T. L-type amino-acid transporter 1 expression predicts the response to preoperative hyperthermo-chemoradiotherapy for advanced rectal cancer. Anticancer Res. 2010;30:4223–4227. [PubMed] [Google Scholar]
- 21.Martins SF, Amorim R, Mota SC, Costa L, Pardal F, Rodrigues M, Longatto-Filho A. Ki-67 expression in crc lymph node metastasis does not predict survival. Biomed Res Int. 2015;2015:131685. doi: 10.1155/2015/131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allegra CJ, Paik S, Colangelo LH, Parr AL, Kirsch I, Kim G, Klein P, Johnston PG, Wolmark N, Wieand HS. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes' B and C colon cancer: A national cancer institute-national surgical adjuvant breast and bowel project collaborative study. J Clin Oncol. 2003;21:241–250. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 23.Jansson A, Sun XF. Ki-67 expression in relation to clinicopathological variables and prognosis in colorectal adenocarcinomas. APMIS. 1997;105:730–734. doi: 10.1111/j.1699-0463.1997.tb05078.x. [DOI] [PubMed] [Google Scholar]
- 24.Shi L, Luo W, Huang W, Huang S, Huang G. Downregulation of L-type amino acid transporter 1 expression inhibits the growth, migration and invasion of gastric cancer cells. Oncol Lett. 2013;6:106–112. doi: 10.3892/ol.2013.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 26.Segawa H, Fukasawa Y, Miyamoto K, Takeda E, Endou H, Kanai Y. Identification and functional characterization of a Na+-independent neutral amino acid transporter with broad substrate selectivity. J Biol Chem. 1999;274:19745–19751. doi: 10.1074/jbc.274.28.19745. [DOI] [PubMed] [Google Scholar]
- 27.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Campbell LE, Miller CM, Proud CG. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oda K, Hosoda N, Endo H, Saito K, Tsujihara K, Yamamura M, Sakata T, Anzai N, Wempe MF, Kanai Y, Endou H. L-type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 2010;101:173–179. doi: 10.1111/j.1349-7006.2009.01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaira K, Oriuchi N, Otani Y, Shimizu K, Tanaka S, Imai H, Yanagitani N, Sunaga N, Hisada T, Ishizuka T, et al. Fluorine-18-alpha-methyltyrosine positron emission tomography for diagnosis and staging of lung cancer: A clinicopathologic study. Clin Cancer Res. 2007;13:6369–6378. doi: 10.1158/1078-0432.CCR-07-1294. [DOI] [PubMed] [Google Scholar]