Abstract
MicroRNAs (miRs) are a class of short non-coding RNAs that serve an essential role in the tumorigenesis of gastric cancer (GC). MiR-203a has been reported as a tumor repressor in various types of human cancer. In the present study, the function of miR-203a on the proliferation of GC cells was investigated. Bioinformatics analyses revealed that miR-203a targets the 3′-untranslated region of E2F transcription factor 3 (E2F3) messenger RNA. A luciferase reporter assay and western blot analysis were performed to confirm whether E2F3 was a target of miR-203a. The relative luciferase activity was decreased when overexpressing miR-203a with E2F3-wild type pmirGLO-3′-untranslated region vector, compared with the control group in HEK293 cells. Overexpression of miR-203a suppressed cell proliferation and colony formation of SGC-7901 and AGS GC cells. Inhibition of miR-203a promoted the proliferation of GC cells. Collectively, the results indicated that miR-203a may function as a tumor suppressor in GC by targeting E2F3.
Keywords: microRNA-203a, gastric cancer, proliferation, E2F transcription factor 3
Introduction
Gastric cancer (GC) is one of the most frequently occurring malignant tumors worldwide (1). A high proportion of patients with GC are diagnosed at a late stage, when extensive invasion and lymphatic metastasis may have already occurred (2,3). Even following surgical resection, mortality due to recurrent disease as a result of metastasis and drug resistance is frequent (4,5). To investigate useful diagnostic biomarkers for early GC diagnosis, understanding the underlying mechanisms of GC is critical.
MicroRNAs (miRNAs or miRs) are a class of small, non-coding RNAs (~22 nucleotides in length) that serve key roles in various biological processes, including cell proliferation, metabolism, differentiation and apoptosis, at the post-transcriptional level (6,7). MiRNAs interact with a messenger RNA (mRNA) target to interfere with its translation into protein or to promote mRNA degradation, typically at the 3′-untranslated region (UTR) (8,9). MiRNAs may be used as molecular biomarkers for the diagnosis of cancer; it has been reported that numerous miRNAs are associated with the carcinogenesis of various types of cancer (10).
MiR-203a has been reported to be associated with various types of tumor, including esophageal squamous cell carcinoma (11), hepatocellular carcinoma (12), non-small cell lung cancer (13) and colorectal carcinoma (14). However, the function of miR-203a in GC remains unclear. The aim of the present study was to explore the function of miR-203a in GC carcinogenesis. The present study demonstrated that overexpression of miR-203a could suppress the proliferation of GC cells. In addition, E2F transcription factor 3 (E2F3) was identified as a direct target of miR-203a by luciferase activity assay.
Materials and methods
Cell culture
SGC-7901, AGS and HEK293 cells were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were grown in Dulbecco's modified Eagle's medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences). Cells were incubated at 37°C in a humidified atmosphere of 5% CO2 in air.
Plasmid vector constructs
Synthetic oligonucleotides containing the 3′-UTR sequences of wild-type (Wt) and mutant (Mut) E2F3 were cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI, USA). The miR-203a inhibitor (5′-CTAGTGGTCCTAAACATTTCAC-3′); and inhibitor-control (5′-TGACTGTACTGACTCGACTG-3′) were synthesized by Sangon Biotech Co., Ltd., Shanghai, China; the miR-203a mimic (sense, 5′-GUGAAAUGUUUAGGACCACUAG-3′; and antisense, 5′-AGUGGUCCUAAACAUUUCACUU-3′), miR-control (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′). Small interfering (si) RNA targeting E2F3 (si-E2F3) and a si-control (15) were purchased from Shanghai GenePharma Co., Ltd., Shanghai, China).
Luciferase activity assay
RegRNA (regrna.mbc.nctu.edu.tw/html/prediction.html) was used to predict the potential target genes for miR-203a. To determine whether E2F3 was a direct target of miR-203a, E2F3 Wt sense, 5′-CCTGTGGCACCCATCACCATTTCAA-3′ and antisense, 5′-TTGAAATGGTGATGGGTGCCACAGG-3′; and E2F3 Mut sense, 5′-CCTGTGGCACCCATCACCATAAGAA-3′ and antisense, 5′-TTCTTATGGTGATGGGTGCCACAGG-3′. Wt or Mut E2F3 3′-UTR were cloned into pmirGLO plasmids (Promega Corporation) and Wt and Mut E2F3 3′-UTR pmirGLO plasmids were co-transfected with miR-203a into HEK293 cells using Lipofectamine 2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Another group of HEK293 cells were co-transfected with miR-203a and an empty pmirGLO plasmid as a control. After 48 h, a luciferase activity assay was performed using a Dual-Luciferase Reporter (DLR) Assay System (Promega Corporation), according to the manufacturer's protocol.
Cell proliferation assay
Cells were plated at a density of 5,000 cells/well in 96-well plates. miR-203a mimic, miR-control, miR-203a inhibitor, inhibitor-control, si-E2F3 or si-control plasmids were transfected with Lipofectamine 2000 into SGC-7901 or AGS cells. An MTT assay with FLUOstar OPTIMA (BMG Labtech GmbH, Ortenberg, Germany) was used to measure the extent of cell proliferation. The optical density was measured at 492 nm at 24, 48 and 72 h post-transfection.
Colony formation assays
SGC-7901 cells were seeded into 12-well plates at a density of 1000 cells/ml, 2ml/well. AGS cells were seeded into 6-well plates at adensity of 1000 cells/ml, 2ml/well. The cells were transfected with miR-203a, miR-control, miR-203a inhibitor, inhibitor-control, si-E2F3 or si-control. The cells were incubated in the aforementioned conditions for 14 days and washed with PBS, then stained with 0.1% crystal violet for 30 min at 37°C. Images of the colonies were captured by Quantity One (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Western blot analysis
SGC-7901 cells were transfected with miR-203a or miR-control. After 48 h, the cells were lysed in radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 1 h at 4°C. Cell lysates were then suspended by centrifugation at 12,000 × g at 4°C for 20 min. The protein concentration was determined with a BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Proteins were separated with 10% SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked with 5% skimmed milk in TBS containing 0.05% Tween-20 for 2 h at room temperature. The membrane was then incubated at 4°C overnight with primary antibodies against E2F3 (1:300; bs-1722R; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China) and β-actin (١:١,500; sc-47778; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), followed by incubation with goat anti-mouse IgG (cat. no. 115-035-003; 1:1,000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) or goat anti-rabbit antibody IgG (cat. no. 111-035-144; 1:1,000; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. Immobilon Western Chemiluminescent horseradish-peroxidase substrate (EMD Millipore, Billerica, MA, USA) were used to visualize the protein bands.
Statistical analysis
Data are expressed as the mean ± standard error from three independent experiments. Student's t-tests with SPSS 13.0 software (SPSS Inc., Chicago, IL, USA) were utilized for statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-203a suppresses the proliferation of GC cells
SGC-7901 and AGS cells were transfected with a miR-203a mimic or miR-control plasmid and an MTT assay was used to examine the proliferation of these cells. The MTT assay revealed that the proliferation of SGC-7901 cells was significantly inhibited by miR-203a overexpression (Fig. 1A; P<0.01), whereas the proliferation of AGS cells was not significantly affected (Fig. 1B). Similarly, colony formation ability was assessed for SGC-7901 and AGS cells transfected as described above. Fewer colonies were observed in miR-203a mimic-transfected SGC-7901 (Fig. 1C; P<0.01) and AGS (Fig. 1D; P<0.01) cells, compared with miR-control-transfected cells. This result demonstrates that miR-203a inhibits the proliferation of GC cells.
Figure 1.
miR-203a induces inhibition of proliferation. To determine the rate of proliferation of (A) SGC-7901 and (B) AGS gastric cancer cells transfected with miR-203a mimic or miR-control, an MTT assay was performed. In addition, a colony formation assay was performed to assess the growth of (C) SGC-7901 and (D) AGS cells subsequent to transfection with miR-203a mimic or miR-control (Student's t-test; **P<0.01 vs. miR control). miR, microRNA; OD, optical density.
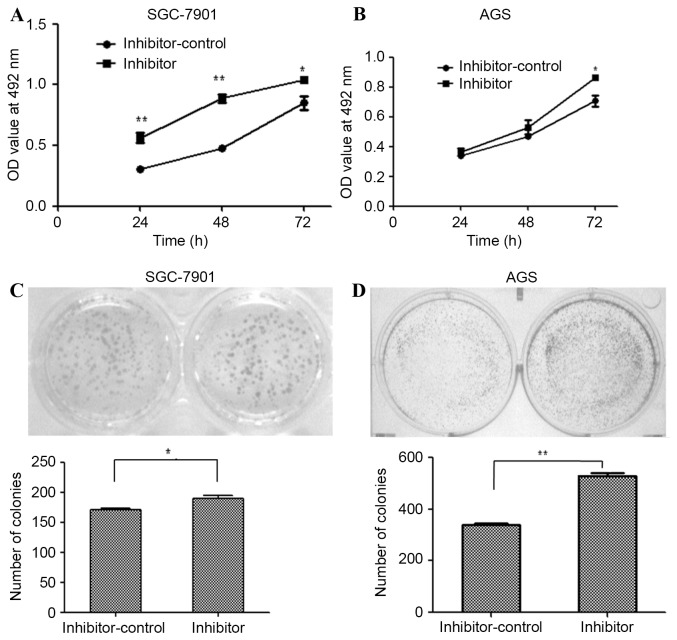
Inhibition of miR-203a increases the proliferation of GC cells
miR-203a inhibitor oligonucleotides were used to silence endogenous miR-203a expression in SGC-7901 and AGS cells. The effect of this inhibitor on SGC-7901 and AGS proliferation was determined by an MTT assay at 24, 48 and 72 h post-transfection. The miR-203a inhibitor increased the proliferation of SGC-7901 (Fig. 2A; P<0.01) and AGS cells (Fig. 2B; P>0.05), as determined by the MTT assays. The formation of colonies was upregulated by transfection with the inhibitor in SGC-7901 (Fig. 2C; P<0.01) and AGS (Fig. 2D; P<0.01) cells. Taken together, these results demonstrate that inhibition of miR-203a contributes to the proliferation of GC cells.
Figure 2.
Inhibition of miR-203a enhances gastric cancer cell proliferation. An MTT assay was used to determine the rate of proliferation of (A) SGC-7901 and (B) AGS gastric cancer cells transfected with a miR-203a-inhibitor or a control small interfering RNA. A colony formation assay was performed to detect the proliferation of (C) SGC-7901 and (D) AGS cells subsequent to transfection with the inhibitor or the control Student's t-test; (*P<0.05, **P<0.01 vs. inhibitor-control). miR, microRNA; OD, optical density.
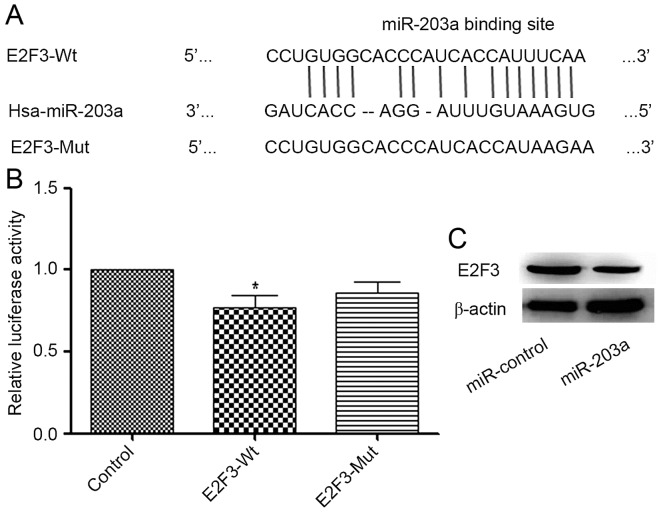
E2F3 is a direct target of miR-203a
RegRNA (regrna.mbc.nctu.edu.tw/html/prediction.html) was used to identify potential target genes of miR-203a. A binding site for miR-203a in the E2F3 3′-UTR was identified (Fig. 3A). A luciferase reporter assay demonstrated that the addition of miR-203a decreased the luciferase activity of E2F3-Wt significantly when compared with that of the control group, whereas the change in the luciferase activity of E2F3-Mut was not significant (Fig. 3B). To further investigate the association between miR-203a and E2F3, a western blot analysis was performed. The result demonstrated that the protein expression of E2F3 was decreased in the miR-203a-overexpressing group compared with that in the control group (Fig. 3C). Collectively, these data indicate that E2F3 is a direct target of miR-203a.
Figure 3.
E2F3 is a direct target of miR-203a. (A) Sequences of E2F3-Wt and E2F3-Mut 3′-UTRs, including miR-203a-binding sites. (B) A miR-203a mimic was co-transfected with pmirGLO-E2F3-Wt-3′-UTR, pmirGLO-E2F3-Mut-3′-UTR or empty pmirGLO plasmid vectors into HEK293 cells. Relative luciferase activity was quantified 48 h post-transfection. (C) Cells were transfected with miR-203a or miR-control. After 48 h, the level of expression of E2F3 was determined by a western blot analysis. β-actin was used as a control. Student's t-test; *P<0.05 vs. control). E2F3, E2F transcription factor 3; miR, micro; Wt, wild-type; Mut, mutant; UTR, untranslated region; Hsa, Homo sapiens.
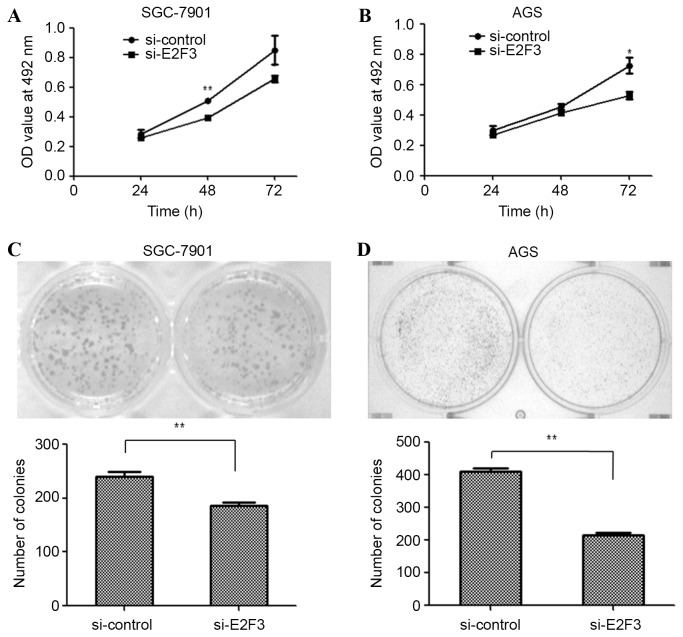
Silencing the expression of E2F3 inhibits the proliferation of GC cells
To examine the effects of E2F3 on the proliferation of SGC-7901 and AGS cells, the cells were transfected with si-E2F3 or a si-control. An MTT assay was performed to examine the proliferation of SGC-7901 and AGS cells. The MTT assay revealed that cell proliferation was inhibited in si-E2F3-transfected SGC-7901 (Fig. 4A; P<0.01 at 48 h) and AGS (Fig. 4B; P<0.05 at 72 h) cells. Similarly, the colony formation assay was employed in si-E2F3- and si-control-transfected SGC-7901 and AGS cells. The results revealed that fewer colonies were formed by si-E2F3-transfected SGC-7901 (Fig. 4C; P<0.01) and AGS cells (Fig. 4D; P<0.01), compared with si-control-transfected cells. The data suggest that miR-203a suppresses GC cell proliferation by downregulating E2F3.
Figure 4.
Silencing the expression of E2F3 inhibited proliferation in gastric cancer cells. To determine the rate of proliferation of gastric cancer (A) SGC-7901 and (B) AGS cells transfected with si-E2F3 or si-control, an MTT assay was performed. A colony formation assay was performed to detect the growth capabilities of (C) SGC-7901 and (D) AGS cells subsequent to transfection with si-E2F3 or si-control (Student's t-test; *P<0.05, **P<0.01). E2F3, E2F transcription factor 3; si, small interfering; OD, optical density.
Discussion
E2F3 is a member of the E2F family, which serves a key function in the control of cell cycle progression (16). It has been reported that E2F3 regulates cell cycle-dependent gene expression during the G1/S transition (17). E2F3 may function as an oncogene in GC (18). Previous studies have reported that overexpression of E2F3 is a frequent oncogenic event in human tumorigenesis and that this may be regulated by miRNA (19,20). In the present study, E2F3 was confirmed to be a direct target of miR-203a with luciferase activity and western blot assays. In previous studies, a number of miRNAs have been categorized as oncogenes or tumor-suppressor genes; and may contribute to cancer progression by interfering with mRNA stability or protein translation to regulate the gene expression of oncogenic or tumor-suppressive proteins (21).
miR-203 has been reported to be involved in several types of cancer (22,23) and may function as a tumor suppressor involved in proliferation, apoptosis, invasion and migration (24). For example, miR-203a suppresses the development of hepatocellular carcinoma cells by targeting homeobox D3 (25). Additionally, miR-203 inhibits proliferation and migration of lung cancer cells by targeting PKCα (26). Therefore, miR-203a may serve a critical function in the development of cancer. The present study suggested that miR-203a may serve an important role in suppressing GC proliferation and may regulate the expression of E2F3. The results of the present study provide an improved understanding of the tumor suppressive role of miR-203a during GC progression.
In summary, the present study demonstrated that the overexpression of miR-203a suppresses GC cell proliferation by targeting and reducing the relative expression level of the cell cycle-regulatory protein E2F3. The effect on GC cell proliferation following transfection with si-E2F3 was comparable to the effect of miR-203a overexpression. Furthermore, a miR-203a inhibitor upregulated the proliferation of GC cells. The tumor-suppressive function of miR-203a may provide new insight for potential therapies for GC. In the future, investigation of the E2F3-associated signaling pathway is to be performed by our group in order to explore more deeply the mechanism by which miR-203a suppresses the proliferation of GC cells.
Acknowledgements
The present study was supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region (grant no. 2013211A114).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Dassen A, Lemmens VE, van de Poll-Franse LV, Creemers GJ, Brenninkmeijer SJ, Lips DJ, Wurff Vd AA, Bosscha K, Coebergh JW. Trends in incidence, treatment and survival of gastric adenocarcinoma between 1990 and 2007: A population-based study in the Netherlands. Eur J Cancer. 2010;46:1101–1110. doi: 10.1016/j.ejca.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. doi: 10.1017/erm.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SJ, Wang YG, Lee HW, Kang HG, La SH, Choi IJ, Irimura T, Ro JY, Bresalier RS, Chun KH. Up-regulation of neogenin-1 increases cell proliferation and motility in gastric cancer. Oncotarget. 2014;5:3386–3398. doi: 10.18632/oncotarget.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohtsu A. Chemotherapy for metastatic gastric cancer: Past, present, and future. J Gastroenterol. 2008;43:256–264. doi: 10.1007/s00535-008-2177-6. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 9.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 10.Shah PP, Hutchinson LE, Kakar SS. Emerging role of microRNAs in diagnosis and treatment of various diseases including ovarian cancer. J Ovarian Res. 2009;2:11. doi: 10.1186/1757-2215-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Dong Z, Liang J, Guo Y, Guo X, Shen S, Kuang G, Guo W. Methylation-mediated repression of potential tumor suppressor miR-203a and miR-203b contributes to esophageal squamous cell carcinoma development. Tumour Biol. 2016;37:5621–5632. doi: 10.1007/s13277-015-4432-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Wu J, Liu M, Yin H, He J, Zhang B. Downregulation of miRNA-30c and miR-203a is associated with hepatitis C virus core protein-induced epithelial-mesenchymal transition in normal hepatocytes and hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2015;464:1215–1221. doi: 10.1016/j.bbrc.2015.07.107. [DOI] [PubMed] [Google Scholar]
- 13.Lin QH, Zhang KD, Duan HX, Liu MX, Wei WL, Cao Y. ERGIC3, which is regulated by miR-203a, is a potential biomarker for non-small cell lung cancer. Cancer Sci. 2015;106:1463–1473. doi: 10.1111/cas.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kara M, Yumrutas O, Ozcan O, Celik OI, Bozgeyik E, Bozgeyik I, Tasdemir S. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–86. doi: 10.1016/j.gene.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Chang S, Zhao Z, et al. MicroRNA-214 suppresses the proliferation of human hepatocellular carcinoma cells by targeting E2F3. Oncol Lett. 2015;10:3779–3784. doi: 10.3892/ol.2015.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julian LM, Vandenbosch R, Pakenham CA, Andrusiak MG, Nguyen AP, McClellan KA, Svoboda DS, Lagace DC, Park DS, Leone G, et al. Opposing regulation of Sox2 by cell-cycle effectors E2f3a and E2f3b in neural stem cells. Cell Stem Cell. 2013;12:440–452. doi: 10.1016/j.stem.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Sharma N, Timmers C, Trikha P, Saavedra HI, Obery A, Leone G. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281:36124–36131. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Li H, Zhang R, Liu J, Liu J. MicroRNA-449a inhibits proliferation and induces apoptosis by directly repressing E2F3 in gastric cancer. Cell Physiol Biochem. 2015;35:2033–2042. doi: 10.1159/000374010. [DOI] [PubMed] [Google Scholar]
- 19.Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ, Sauter G. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23:5616–5623. doi: 10.1038/sj.onc.1207749. [DOI] [PubMed] [Google Scholar]
- 20.Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26:356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiemer EA. The role of microRNAs in cancer: No small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Lee SA, Kim JS, Park SY, Kim HJ, Yu SK, Kim CS, Chun HS, Kim J, Park JT, Go D, et al. miR-203 downregulates Yes-1 and suppresses oncogenic activity in human oral cancer cells. J Biosci Bioeng. 2015;120:351–358. doi: 10.1016/j.jbiosc.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Shen B, Chen T, Dong P. miR-203 is involved in the laryngeal carcinoma pathogenesis via targeting VEGFA and Cox-2. Onco Targets Ther. 2016;9:4629–4637. doi: 10.2147/OTT.S96053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu G, Lai P, Liu M, Xu L, Guo Z, Liu H, Li W, Wang G, Yao X, Zheng J, et al. miR-203a regulates proliferation, migration, and apoptosis by targeting glycogen synthase kinase-3β in human renal cell carcinoma. Tumour Biol. 2014;35:11443–11453. doi: 10.1007/s13277-014-2476-x. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Sun H, Wang X, Hou N, Zhao L, Tong D, He K, Yang Y, Song T, Yang J, et al. EGR1 mediates miR-203a suppress the hepatocellular carcinoma cells progression by targeting HOXD3 through EGFR signaling pathway. Oncotarget. 2016;7:45302–45316. doi: 10.18632/oncotarget.9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, Wang N, Zhang S, Zen K, Zhang C, et al. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PloS One. 2013;8:e73985. doi: 10.1371/journal.pone.0073985. [DOI] [PMC free article] [PubMed] [Google Scholar]