Abstract
The aim of the present study was to investigate the effects of microRNA (miR-)146a on the cisplatin sensitivity of the non-small cell lung cancer (NSCLC) A549 cell line and study the underlying molecular mechanism. The differences in expression of miRNAs between A549 and A549/cisplatin (A549/DDP) cells were determined, and miR-146a was selected to study its effect on cisplatin sensitivity of A549/DDP cells. miR-146a mimic and inhibitor transient transfection systems were constructed using vectors, and A549/DDP cells were infected with miR-146a mimic and inhibitor to investigate growth, apoptosis and migration. The directed target of miR-146a was determined and the underlying molecular mechanism was validated in the present study. The results of the present study demonstrated that miR-146a was downregulated in NSCLC A549/DDP cells, compared with A549 cells. The overexpression of miR-146a induced apoptosis and inhibited the growth and invasion of A549/DDP cells, which resulted in increased cisplatin sensitivity in NSCLC cells. The JNK2 gene was determined as the direct target of miR-146a, and may be activated by the overexpression of miR-146a. Additionally, JNK2 activated the expression of p53 and inhibited B cell lymphoma 2. The upregulation of miR-146a increased cisplatin sensitivity of the A549 cell line by targeting JNK2, which may provide a novel method for treating NSCLC cisplatin resistance.
Keywords: microRNA-146a, JNK2, cisplatin resistance, non-small cell lung cancer, apoptosis
Introduction
Non-small cell lung cancer (NSCLC) accounts for ~80% of lung cancer cases and is the leading cause of cancer-associated mortalities worldwide in 2008 (1). In clinical practice, chemotherapy remains the principal treatment due to the poor prognosis of this type of malignancy. Cisplatin is the most commonly used in chemotherapy for patients with NSCLC (2). Although cisplatin treatment leads to initially successful responses, it typically results in the development of chemoresistance and results in therapeutic failure (3). Therefore, elucidating the molecular mechanisms underlying cisplatin resistance in NSCLC may contribute to identify novel therapeutic targets for attenuating cisplatin resistance, which has been a focus in research in recent years (4). Previous studies have demonstrated that inactivation of cell apoptosis signaling pathways, activation of cell survival signaling pathways, abnormal expression of tumor associated genes and non-coding RNAs contribute to the cisplatin resistance of NSCLC (5–7). MicroRNAs (miRNA/miR), small endogenous non-coding RNAs between 21 and 25 nucleotides, may regulate post-transcriptional gene expression by targeting the 3′-untranslated regions (3′-UTR) of mRNAs (8). Previous studies have revealed that miRNAs are involved in human cancer as diagnostic and prognostic cancer biomarkers, and exhibit therapeutic tools (9–11). Furthermore, it has been demonstrated that dysregulation of miRNAs may contribute to the chemoresistance in human tumor cells, including cisplatin resistance (12–14). c-Jun N-terminal kinases (JNKs) function as a signaling hub in mitogen-activated protein kinase (MAPK) signaling pathways and are involved in inflammation, tumorigenesis, and cell proliferation, differentiation, migration and apoptosis (15,16). JNK1, JNK2 and JNK3 are major isoforms of JNK proteins and are encoded by MAPK8, MAPK9 and MAPK10, respectively (17). JNK1 and JNK2 are widely expressed in numerous tissue types, whereas JNK3 is primarily located in the nervous system (17,18). The JNK-mediated signaling pathway is involved in a variety of processes in human cancers (19). A previous study has identified that chemotherapy resistance is not only the result of deregulation of miRNAs targeting drug efflux transporter, but is a multifactorial process (20). A number of miRNAs are associated with cell apoptotic and survival signaling pathways, and cells exhibiting altered expression profiles of pro- and anti-apoptotic miRNAs are typically involved in resistance to cytotoxic agents (21,22). miR-200c has been identified as a regulator of tumorigenesis and tumor metastasis, and demonstrated to attenuate P-glycoprotein 1-mediated multi-drug resistance and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer (23). Upregulation of miR-21 serves key roles in cisplatin-resistant ovarian cancer via JNK-1/c-Jun pathway (24). In recent years, miR-146a has been identified to exhibit functions in a number of types of disease; in particular, in the proliferation, metastasis and apoptosis of distinct types of cancer cells (25). For chemotherapy resistance, only Tomokuni et al (26) has revealed an increased expression of miR-146a in hepatocellular carcinoma cell lines resistant to interferon-α. The association between miR-146a and cisplatin resistance, and the underlying molecular mechanism, remain unknown.
To the best of our knowledge, the results of the present study identified that miR-146a was significantly downregulated in NSCLC cisplatin-resistant A549 cells and that miR-146a targeted the 3′UTR of the JNK2 gene directly, which affected the phosphorylated JNK-mediated signaling pathway (27). Furthermore, overexpression of miR-146a was demonstrated to downregulate the levels of cisplatin resistance via the JNK signaling pathway, resulting in increased sensitivity to cisplatin and induced apoptosis in vitro. Therefore, the present study may identify a novel mechanism for NSCLC cell cisplatin resistance and provide a new way of treating NSCLC cisplatin resistance, by targeting miR-146a.
Materials and methods
Cell culture
A549 cells and A549/cisplatin (A549/DDP) cells (Guangzhou Zixiutang Biotechnology Co., Ltd., Guangzhou, China) were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. All cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Target prediction
For miR-146a target prediction, PicTar (http://pictar.mdc-berlin.de/), miRBase (http://www.mirbase.org/), miRanda (http://www.microrna.org/microrna/home.do), Bibiserv (https://bibiserv.cebitec.uni-bielefeld.de/agenda) and TargetScan (http://www.targetscan.org/vert_71/) were used to identify and analyze potential targets of JNK2.
miRNA microarray analysis
Total RNA was extracted from culture cells with guanidine thiocyanate, 2-mercaptoethanol and chloroformin GenElute Mammalian Total RNA Purification kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 3 min on ice and the RNA concentration was read using the NanoDrop 2000 spectrophotometer. RNA quality was validated using a Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). Total RNA (100 ng) was used for cDNA synthesis. Subsequently, cDNA was labeled with fluorescence, fragmented and hybridized to the Affymetrix GeneChip Gene 1.0 ST Human Array (Thermo Fisher Scientific, Inc.). Following incubation at 45°C for 16 h, the arrays were washed with 1X saline sodium citrate (SSC), 0.1% SDS, 0.1X SSC, 0.1% SDS and 0.1X SSC and water for 2 min of each at room temperature, and then scanned (Affymetrix Model 3000 scanner; Thermo Fisher Scientific, Inc.). The data adjustments included data filtering, log2 transformation, gene centering. In addition, the signal was normalized using a Locally-weighted Regression filter.
RNA and miRNA extraction for reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated with buffer RLT and purified from A549 and A549/DDP cells using the RNeasy Mini kit (Qiagen Inc., Valencia, CA, USA) for 5 min on ice. miRNA was prepared using the miRcute miRNA isolation kit (Tiangen Biotech Co., Ltd., Beijing, China) and cDNA was synthesized from the RNA by reverse transcription employing the Qiagen OneStep RT-PCR kit (Qiagen GmbH, Hilden, Germany). Sequences of all the primers are presented in Table I. qPCR amplification was performed to enable the fluorescence-based quantitation of gene expression using the SYBR Green Quantitative RT-PCR kit (Sigma-Aldrich; Merck KGaA). U6 was regarded as the reference gene. The PCR volume and conditions were set up as previously described (28) The primers for miR-182-5p (catalog no., MIRAP00211), miR-106b (catalog no., MIRAP00129) and miR-146a (catalog no., MIRAP00183) were from the MystiCq® microRNA qPCR Assay Primer purchased for Sigma-Aldrich (Merck KGaA). In additon, 2−∆∆Cq (29) was applied for gene quantification. The primer sequences of U6 were: Forward, 5′-AAAGCAAATCATCGGACGACC and reverse, 3′-GTACAACACATTGTTTCCTCGGA.
Table I.
Primer sequences used in the present study.
| Primer | Sequence |
|---|---|
| miRNA-146a mimics | |
| Forward | 5′-UGAGAACUGAAUUCCAUGGGUU-3′ |
| Reverse | 5′-CCCAUGGAAUUCAGUUCUCAUU-3′ |
| miRNA-146a inhibitor | 5′-AACCCAUGGAAUUCAGUUCUCA-3′ |
| JNK2 primers | |
| Forward | 5′-CTGCGTCACCCATACATC-3′ |
| Reverse | 5′-TGGCGTTGCTACTTACTGC-3′ |
| p53 primers | |
| Forward | 5′-TGCGTGTGGAGTATTTGGATG-3′ |
| Reverse | 5′-TGGTACAGTCAGAGCCAACCAG-3′ |
| Bcl-2 primers | |
| Forward | 5′-CTGGTGGACAACATCGCTCTG-3′ |
| Reverse | 5′-GGTCTGCTGACCTCACTTGTG-3′ |
JNK2, Jun kinase 2; Bcl-2, B cell lymphoma 2; F, forward; R, reverse.
Transfection of plasmids, miRNA mimics and miRNA inhibitor
miR-146a mimic and inhibitor targeting JNK2 (1 ng) were synthesized by GenePharma, Inc. (Sunnyvale, CA, USA). While, the A549/DDP cells infected with empty plasmid were used as controls (MOCK group). The sequences are presented in Table I. The transfection in A549/DDP cells was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol (23).
Cell proliferation assay
A549/cisplatin (A549/DDP) cells (1×104 cells/well) were seeded in 96-well plates in RPMI-1640 medium. An MTT assay was performed using a Cell proliferation kit I (GE Healthcare Life Sciences, Little Chalfont, UK) according to the manufacturer's protocol. Subsequently, absorbance was determined at 570 nm using VersaMax (Molecular Devices; Thermo Fisher Scientific, Inc.) to estimate MTT-formazan production dissolved with dimethyl sulfoxide after 24, 48 and 72 h incubation. The OD490 was detected using a spectrodensitometer under 490 nm wavelength.
Apoptosis rate and cell cycle determination in vitro
The FITC Annexin V Apoptosis Detection kit with PI (BioLegend, Inc., San Diego, CA, USA) was used for cell apoptosis assay. After 72 h incubation, the transfected A549/cisplatin (A549/DDP) cells (1×105) were fixed with 2.5% glutaraldehyde for 30 min at room temperature. Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining flow cytometry was used to determine apoptotic rates, by identifying the relative amount of Annexin V-FITC positive and PI negative cells. Unstained cells and cells stained with Annexin V-FITC or PI alone were used as controls. PI staining flow cytometry was used to determine the cell cycle. CYTOSPEC™ version 7.0 from Purdue University Cytometry Laboratories was applied for data analysis.
Cell invasion assay
The cell invasion assay was performed using 24-well Transwell plates, as previously described (30). A total of 8×104 cells were seeded in serum-free RPMI-1640 medium in the upper and lower Transwell chambers and incubated for 24 h. Matrigel (BD Biosciences) was pre-coated on the upper side of the membrane and incubated at 37°C for 1 h for gel formation. Following the removal of the Transwell inserts, the cells that had not migrated through the filter and remained inside each Transwell were removed by wiping with a cotton swab. Cells that had migrated through the filter and adhered to the other side of the filter were stained with crystal violet at room temperature for 5 min for image capture, and counted using a light microscope with ×200 magnification (30).
JNK2-3′-UTR luciferase reporter assay
Human JNK2 cDNA, containing putative and mutant target sites for miR-146a, was chemically synthesized and inserted into a pMIR-REPORT™ vector (Ambion; Thermo Fisher Scientific, Inc.). For the luciferase reporter assay, miR-146a mimic, miRNA mimic control, miR-146a inhibitor and miRNA inhibitor control were transfected into wild-(pMIR-jnk2-wt) or mutant-type (pMIR-jnk2-mut) reporter vectors using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity was determined 48 h after transfection using a dual-luciferase assay kit (Promega Corporation, Madison, WI, USA) (31). Additionally, Renilla luciferase activity was applied for luciferase intensity normalization.
Protein extraction and western blot analysis
The total protein was extracted from A549/DDP cells and separated using SDS-PAGE (10% gel), as previously described (32). Subsequently, the gel was transferred to a polyvinylidene difluoride membrane (Solvay Chemicals, Brussels, Belgium) and blocked with 5% skim milk at room temperature for 1 h. The rabbit anti-JNK2 (catalog no., PA528664), -p53 (catalog no., PA527822) and -B cell lymphoma (Bcl-)2 primary antibodies (catalog no., PA520069) purchased from Wuhan Khayal Bio-Technology Co., Ltd. (Wuhan, China) were used at a dilution of 1:1,000. The incubation with primary antibodies was at room temperature for 1 h. Subsequently, the goat anti-rabbit immunoglobulin G secondary antibody conjugated with horseradish peroxidase (cat. no. Ab97051, Abcam, Cambridge, MA, USA) was used at a dilution of 1:10,000. In addition, the incubation with secondary antibodies was at room temperature for 45 min. The signal was visualized using the enhanced chemiluminescence kit (GE Healthcare Life Sciences). Image J 1.41 software (NIH, Bethesda, MD, USA) was used to compare the gray values between the proteins of interest and the internal control protein (β-actin), and between the phosphorylated protein and the total protein.
RT-qPCR analysis
The expression levels of JNK2, p53 and Bcl-2 mRNA in A549/DDP cells were assessed using RT-qPCR. Total RNA was extracted from the cells using the TRIzol method (Thermo Fisher Scientific, Inc.) at 4°C for 15 min. Subsequently, cDNA was synthesized from the RNA by reverse transcription using SYBR Green Quantitative RT-PCR kit (Sigma-Aldrich; Merck KGaA). PCR amplification was performed to enable fluorescence-based quantitation of gene expression. PCR reaction volumes were 10 µl and composed of cDNA (1 µl), primers (0.2 µl each), 2X Premix Ex Taq (5 µl) and H2O (3.6 µl). The primer sequences used are presented in Table I. For cDNA synthesis, samples were incubated at 40°C for 30 min, 98°C for 5 min and 5°C for 5 min. The PCR conditions were as follows: Pre-denaturation at 96°C for 5 min, followed by initiation at 94°C for 30 sec, annealing at 60°C for 30 sec and elongation at 78°C for 1.5 min for 35 cycles, after which samples were stored at 4°C. Additionally, 2−∆∆Cq (29) was applied for gene quantification. β-actin was selected as the internal reference.
Statistical analysis
All data are presented as the mean ± standard deviation. Statistical analysis was performed using GraphPad Prism version 5.01 (GraphPad Software, Inc., La Jolla, CA, USA) via Student's t-test. For all comparisons, P<0.05 was considered to indicate a statistically significant difference.
Results
miR-146a is downregulated in A549/CDDP cells compared with A549 cells
miRNA microarray analyzed 30 miRNAs and PicTar, miRBase, miRanda, Bibiserv and TargetScan were used to identify and analyze potential targets of JNK2. A total of 3 miRNAs, including miR-146a, miR-182-5p and miR-106b, were identified to be potential targets of JNK2 in >2 databases. In addition, the expression levels of these 3 miRNAs in A549 and A549/DDP cells were determined. As presented in Fig. 1, the expression levels of miR-182-5p, miR-106b and miR-146a were demonstrated to be downregulated in NSCLC A549/DDP cells, compared with A549 cells. Only the expression of miR-146a revealed statistical significance (P<0.05).
Figure 1.
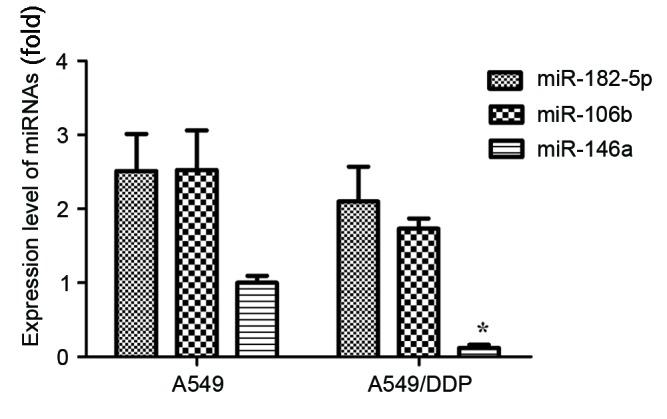
Relative expression of different miRNAs in A549 and A549/DDP cells. *P<0.05 vs. A549 group. miR, microRNA; DDP, cisplatin.
JNK2 is a direct target of miR-146a
The luciferase reporter assay was performed to determine whether miR-146a directly targets JNK2 by binding to the 3′-UTR of the JNK2 gene. Wild-type and mutant JNK2 3′UTR sequences, which contained the miR-146a binding site, were constructed and inserted into vectors (Fig. 2A). The reporter construct and miR-control, miR-146a mimics, miR-146a inhibitor and relative controls were transfected into the A549/DDP cell line. As presented in Fig. 2B, the luciferase reporter activity represented a significant decrease in JNK2 3′-UTR in the presence of miR-146a mimics in A549/DDP cells, compared with the miR-control. Furthermore, a significant increase was observed in the presence of miR-146a inhibitor (Fig. 2B). Therefore, JNK2 was identified as a direct target of miR-146a.
Figure 2.
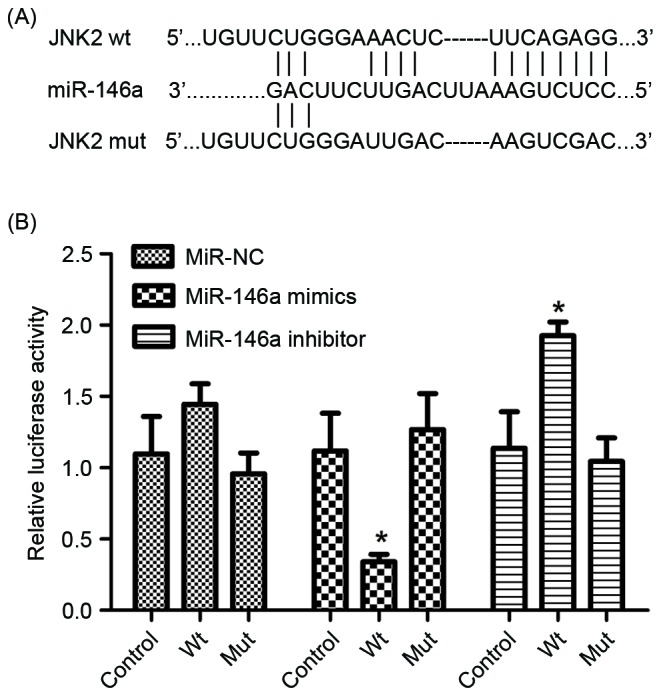
miR-146a targets JNK2 3′-UTR. (A) The box presents a miR-146a predicted binding site, 1184–1194, in JNK2 3′-UTR. (B) Luciferase reporter assay results demonstrated overexpression of miR-146a may reduce the expression of JNK2 since miR-146a could target and degrade the 3′-UTR of JNK2 mRNA. *P<0.05 vs. control group. miR, microRNA; JNK, Jun kinase; wt, wild type; mut, mutation type.
miR-146a inhibits proliferation and invasion, and induces apoptosis of A549/DDP cells
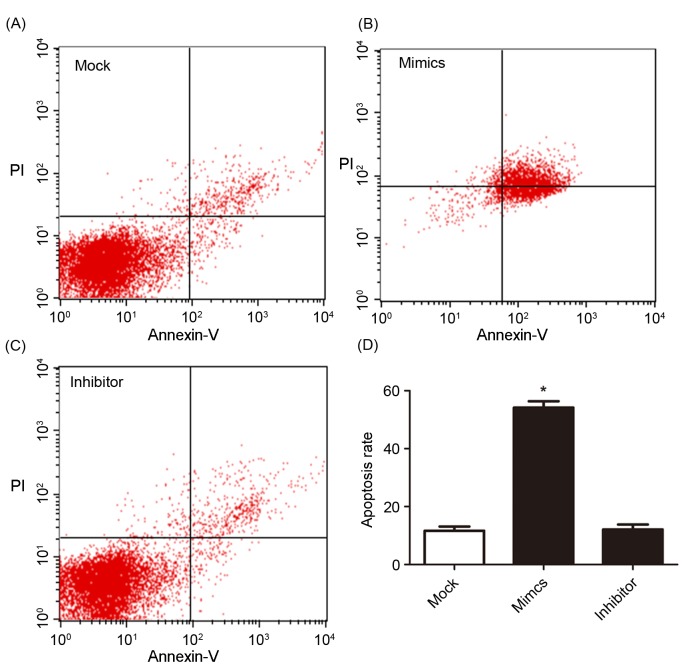
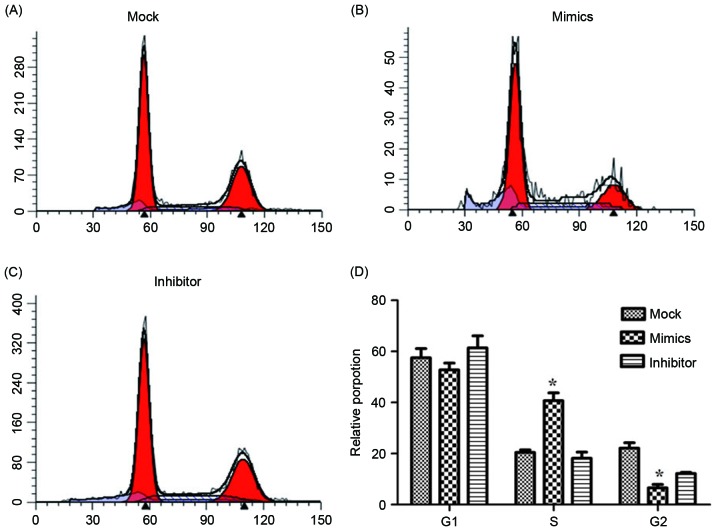
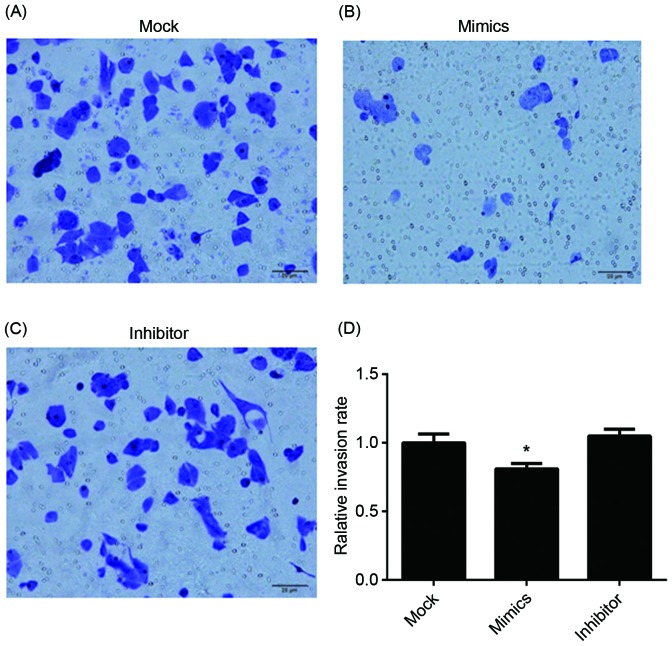
To investigate the effects of miR-146a on the cisplatin sensitivity, the A549/DDP cells were transfected with miR-146a mimics and inhibitors. The A549/DDP cells infected with empty plasmid were used as controls (MOCK group). The proliferation of cells was determined after 24, 48 and 72 h incubation, and the results are presented in Fig. 3. After 24 h incubation, upregulated miR-146a A549/DDP cells revealed a significantly decreased proliferation rate, compared with the other two groups. The cell apoptosis and cell cycle were determined using flow cytometry and are presented in Figs. 4 and 5. Fig. 4A, B and C present cell apoptosis in the mock, mimic and inhibitor groups, respectively, and Fig. 4D demonstrates the apoptosis rate of A549/DDP cells. The results of the present study revealed that, in the miR-146a overexpression group, the apoptosis rate was significantly increased compared with the other two groups. The relative proportion of A549/DDP cells in G1, S and G2 period were calculated and are presented in Fig. 5. Cells in the miR-146a mimic group were arrested in the S period, which resulted in cells failing to enter the G2 period and complete mitosis. For invasion, the results of the Transwell assay of infected A549/DDP cells are presented in Fig. 6. The relative invasion rate of A549/DDP cells in the miR-146a mimic group was significantly decreased, compared with the other two groups.
Figure 3.
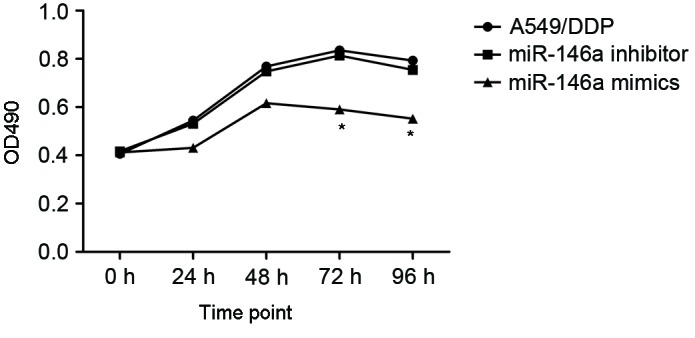
Proliferation rate of transfected A549/DDP cells after 24, 48, 72 and 96 h incubation. *P<0.05 vs. A549/DDP group. DDP, cisplatin; miR, microRNA; OD, optical density.
Figure 4.
Apoptosis of transfected A549/DDP cells. (A) Apoptosis results in the mock group, (B) the miR-146a mimic group and (C) the miR-146a inhibitor group. (D) Apoptotic rate of A549/DDP cells in different groups. *P<0.05 vs. mock group. DDP, cisplatin.
Figure 5.
Cell cycle analysis of transfected A549/DDP cells. (A) Cell cycle distribution in the mock group, (B) the miR-146a mimic group and (C) the miR-146a inhibitor group. (D) The proportion of A549/DDP cells in G1, S and G2 phases of mock, miR-146a mimic and miR-146a inhibitor group. *P<0.05 vs. mock group. DDP, cisplatin.
Figure 6.
Invasion of transfected A549/DDP cells. (A) Invasion photogram of the mock group, (B) the miR-146a mimic group and (C) the miR-146a inhibitor group. (D) The invasion rate of A549/DDP cells in mock, miR-146a mimic and miR-146a inhibitor group. *P<0.05 vs. mock group. DDP, cisplatin.
Expression level of tumor-associated factors
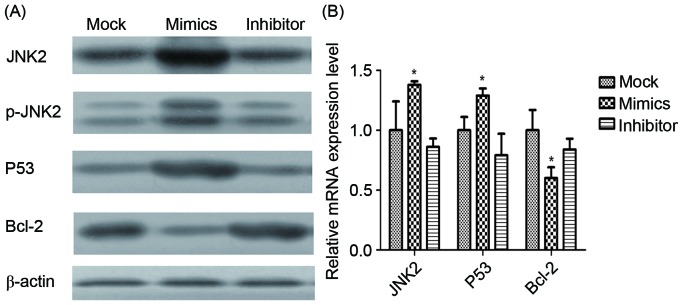
Expression levels of JNK2, P53 and Bcl-2 mRNA and proteins in infected A549/DDP cells were determined using RT-qPCR and western blot analysis (Fig. 7). JNK2, as the direct target of miR-146a, was identified to be significantly upregulated by miR-146a overexpression. Additionally, p53 was demonstrated to be significantly upregulated by miR-146a overexpression, whereas Bcl-2 was revealed to be significantly downregulated by miR-146a.
Figure 7.
mRNA and protein expression of different factors. (A) The protein expression of JNK2, p-JNK2, p53, Bcl-2 and β-actin in the mock, miR-146a mimic and miR-146a inhibitor groups, determined using western blot analysis. β-actin was used as the loading control. (B) The mRNA expression of JNK2, p53 and Bcl-2, determined using the reverse transcription-quantitative polymerase chain reaction. *P<0.05 vs. mock group. JNK2, Jun kinase 2; p-, phosphorylated; Bcl-2, B cell lymphoma 2.
Discussion
The results of the present study demonstrated that miR-146a was downregulated in NSCLC A549/DDP cells and may affect the phosphorylated JNK-mediated signaling pathway by targeting JNK2 gene 3′-UTR directly. Additionally, the overexpression of miR-146a was revealed to induce apoptosis, and inhibit the proliferative and invasive capabilities of A549/DDP cells, which resulted in an increased sensitivity of NSCLC cells to cisplatin. miR-146a, one of the most common downregulated miRNAs in tumor cells, may inhibit cell proliferation, invasion and migration, and induce apoptosis of cancer cells, thereby functioning as a tumor suppressor miRNA (33–35). To the best of our knowledge, only Tomokuni et al (26) has demonstrated that miR-146a suppresses the sensitivity to interferon-α in hepatocellular carcinoma cells. Furthermore, to the best of our knowledge, the present study was the first to investigate the association between miR-146a and cisplatin resistance, and the mechanism of its function.
Numerous miRNAs have been identified as differentially expressed in cisplatin-sensitive and cisplatin-resistant tumor cells; for example, miR-214 overexpression was associated with cisplatin resistance of ovarian cancer cells (36). The overexpression of miR-141 and downregulation of its target gene, the Kelch-Like ECH-Associated Protein 1, have been associated with cisplatin resistance in esophageal squamous cell carcinoma (37). In addition, overexpression of miR-21 in cisplatin resistant ovarian cancer cells has been identified to be a secondary event associated with the activation of the JNK-1/c-Jun pathway in these cells (24). miR-146a has been revealed to be upregulated in a number of types of cancer including papillary thyroid carcinoma, anaplastic thyroid cancer and cervical cancer (38,39), which suggests that miR-146a may function as an oncogenic miRNA. However, decreased expression of miR-146a was identified in prostate, pancreatic and gastric cancers (32,40–42). Therefore, miR-146a may have opposing functions in different types of cancer. In NSCLC, miR-146a was downregulated, inhibited cell growth and migration, and induced apoptosis (34). In the present study, miR-146a was downregulated in A549/DDP NSCLC cells compared with A549 NSCLC cells. Furthermore, the overexpression of miR-146a in the present study resulted in an increased apoptosis rate, and decreased proliferation and migration of A549/DDP cells, compared with control A549/DDP cells. Therefore, it may be concluded that miR-146a is associated with cisplatin resistance of A549 cells and the overexpression of miR-146a may attenuate cisplatin resistance of A549/DDP cells.
To study the underlying molecular mechanism of miR-146a in A549/DDP cells, the target of miR-146a was determined using a luciferase reporter assay. The results of the present study identified that JNK2 was a direct target of miR-146a. Cisplatin, a DNA-damaging drug, induces cell apoptosis by binding to DNA. JNK is involved in the regulation of apoptosis and responds to stress signals by phosphorylating transcription factors, including c-Jun and ATF-2 (43), or by activating other target proteins involved in the initiation or execution of apoptosis, including p53 (44), c-Myc (45) and proteins of the Bcl-2 family (46). The p53 gene, encoding a transcriptional factor, may induce apoptosis of tumor cells and serve as a tumor suppressor gene. The Bcl-2 protein, a member of the Bcl-2 family, may inhibit the cell apoptosis in a number of types of cancer (47). In the present study, expression of JNK2 and the phosphorylation protein p-JNK2 were upregulated by the overexpression of miR-146a, which indicated that JNK2 may be activated by miR-146a. Furthermore, JNK2 activated the expression of p53 and inhibited Bcl-2. The results of the present study demonstrated that miR-146a increased cisplatin sensitivity of NSCLC A549/DDP cells by targeting JNK2 and inducing cell apoptosis. The present study may provide a novel method for treating NSCLC cisplatin resistance, by targeting miR-146a.
Acknowledgements
The present study was supported by the Program for Social Development Research of Yinzhou District of Ningbo (grant no. 2013-107).
References
- 1.Eaton KD, Martins RG. Maintenance chemotherapy in non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8:815–821. doi: 10.6004/jnccn.2010.0058. [DOI] [PubMed] [Google Scholar]
- 2.Socinski MA, Crowell R, Hensing TE, Langer CJ, Lilenbaum R, Sandler AB, Morris D. American College of Chest Physicians: Treatment of Non-small Cell Lung Cancer, Stage IV: ACCP Evidence-based Clinical Practice Guidelines (2nd edition) Chest. 2007;132(3 Suppl):277S–289S. doi: 10.1378/chest.07-1381. [DOI] [PubMed] [Google Scholar]
- 3.Korita PV, Wakai T, Shirai Y, Matsuda Y, Sakata J, Takamura M, Yano M, Sanpei A, Aoyagi Y, Hatakeyama K, Ajioka Y. Multidrug resistance-associated protein 2 determines the efficacy of cisplatin in patients with hepatocellular carcinoma. Oncol Rep. 2010;23:965–972. doi: 10.3892/or_00000721. [DOI] [PubMed] [Google Scholar]
- 4.Rose MC, Kostyanovskaya E, Huang RS. Pharmacogenomics of cisplatin sensitivity in non-small cell lung cancer. Genomics Proteomics Bioinformatics. 2014;12:198–209. doi: 10.1016/j.gpb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Wang Z, Liu L, Chen L. Akt is the downstream target of GRP78 in mediating cisplatin resistance in ER stress-tolerant human lung cancer cells. Lung Cancer. 2011;71:291–297. doi: 10.1016/j.lungcan.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Edwards JK, Pasqualini R, Arap W, Calin GA. MicroRNAs and ultraconserved genes as diagnostic markers and therapeutic targets in cancer and cardiovascular diseases. J Cardiovasc Transl Res. 2010;3:271–279. doi: 10.1007/s12265-010-9179-5. [DOI] [PubMed] [Google Scholar]
- 10.Fabbri M. miRNAs as molecular biomarkers of cancer. Expert Rev Mol Diagn. 2010;10:435–444. doi: 10.1586/erm.10.27. [DOI] [PubMed] [Google Scholar]
- 11.Jackson A, Linsley PS. The therapeutic potential of microRNA modulation. Discov Med. 2010;9:311–318. [PubMed] [Google Scholar]
- 12.Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–531. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 13.Yu ZW, Zhong LP, Ji T, Zhang P, Chen WT, Zhang CP. MicroRNAs contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol. 2010;46:317–322. doi: 10.1016/j.oraloncology.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478–486. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Johnson GL, Nakamura K. The c-jun kinase/stress-activated pathway: Regulation, function and role in human disease. Biochim Biophys Acta. 2007;1773:1341–1348. doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhanasekaran DN, Johnson GL. MAPKs: Function, regulation, role in cancer and therapeutic targeting. Oncogene. 2007;26:3097–3099. doi: 10.1038/sj.onc.1210395. [DOI] [PubMed] [Google Scholar]
- 17.Cuevas BD, Abell AN, Johnson GL. Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene. 2007;26:3159–3171. doi: 10.1038/sj.onc.1210409. [DOI] [PubMed] [Google Scholar]
- 18.Bode AM, Dong Z. The functional contrariety of JNK. Mol Carcinog. 2007;46:591–598. doi: 10.1002/mc.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bubici C, Papa S. JNK signalling in cancer: In need of new, smarter therapeutic targets. Br J Pharmacol. 2014;171:24–37. doi: 10.1111/bph.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panczyk M. Pharmacogenetics research on chemotherapy resistance in colorectal cancer over the last 20 years. World J Gastroenterol. 2014;20:9775–9827. doi: 10.3748/wjg.v20.i29.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol. 2010;223:289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- 22.Haenisch S, Cascorbi I. miRNAs as mediators of drug resistance. Epigenomics. 2012;4:369–381. doi: 10.2217/epi.12.39. [DOI] [PubMed] [Google Scholar]
- 23.Sui H, Cai GX, Pan SF, Deng WL, Wang YW, Chen ZS, Cai SJ, Zhu HR, Li Q. miR200c attenuates P-gp-mediated MDR and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer. Mol Cancer Ther. 2014;13:3137–3151. doi: 10.1158/1535-7163.MCT-14-0167. [DOI] [PubMed] [Google Scholar]
- 24.Echevarría-Vargas IM, Valiyeva F, Vivas-Mejía PE. Upregulation of miR-21 in cisplatin resistant ovarian cancer via JNK-1/c-Jun pathway. PLoS One. 2014;9:e97094. doi: 10.1371/journal.pone.0097094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomokuni A, Eguchi H, Tomimaru Y, Wada H, Kawamoto K, Kobayashi S, Marubashi S, Tanemura M, Nagano H, Mori M, Doki Y. miR-146a suppresses the sensitivity to interferon-α in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2011;414:675–680. doi: 10.1016/j.bbrc.2011.09.124. [DOI] [PubMed] [Google Scholar]
- 27.Hansen PR, Holm AM, Svendsen UG, Olsen PS, Andersen CB. Apoptosis and formation of peroxynitrite in the lungs of patients with obliterative bronchiolitis. J Heart Lung Transplant. 2000;19:160–166. doi: 10.1016/S1053-2498(99)00115-1. [DOI] [PubMed] [Google Scholar]
- 28.Sui H, Zhou S, Wang Y, Liu X, Zhou L, Yin P, Fan Z, Li Q. COX-2 contributes to P-glycoprotein-mediated multidrug resistance via phosphorylation of c-Jun at Ser63/73 in colorectal cancer. Carcinogenesis. 2011;32:667–675. doi: 10.1093/carcin/bgr016. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci USA. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamiya T, Kobayashi Y, Kanaoka K, Nakashima T, Kato Y, Mizuno A, Sakai H. Fluorescence microscopic demonstration of cathepsin K activity as the major lysosomal cysteine proteinase in osteoclasts. J Biochem. 1998;123:752–759. doi: 10.1093/oxfordjournals.jbchem.a022001. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Vandenboom TG, II, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson M, Joyner J, Dildar K, et al. The role of miR-146a and novel Rhenium compounds on prostate cancer cell lines derived from African Americans and European American patients. Cancer Res. 2015;75:4840–4840. doi: 10.1158/1538-7445.AM2015-4840. [DOI] [Google Scholar]
- 36.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-6426. [DOI] [PubMed] [Google Scholar]
- 37.Imanaka Y, Tsuchiya S, Sato F, Shimada Y, Shimizu K, Tsujimoto G. MicroRNA-141 confers resistance to cisplatin-induced apoptosis by targeting YAP1 in human esophageal squamous cell carcinoma. J Hum Genet. 2011;56:270–276. doi: 10.1038/jhg.2011.1. [DOI] [PubMed] [Google Scholar]
- 38.Pacifico F, Crescenzi E, Mellone S, Iannetti A, Porrino N, Liguoro D, Moscato F, Grieco M, Formisano S, Leonardi A. Nuclear factor-{kappa}B contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J Clin Endocrinol Metab. 2010;95:1421–1430. doi: 10.1210/jc.2009-1128. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res. 2011;17:4277–4284. doi: 10.1158/1078-0432.CCR-10-2866. [DOI] [PubMed] [Google Scholar]
- 42.Hou Z, Xie L, Yu L, Qian X, Liu B. MicroRNA-146a is down-regulated in gastric cancer and regulates cell proliferation and apoptosis. Med Oncol. 2012;29:886–892. doi: 10.1007/s12032-011-9862-7. [DOI] [PubMed] [Google Scholar]
- 43.Hayakawa J, Depatie C, Ohmichi M, Mercola D. The activation of c-Jun NH2-terminal kinase (JNK) by DNA-damaging agents serves to promote drug resistance via activating transcription factor 2 (ATF2)-dependent enhanced DNA repair. J Biol Chem. 2003;278:20582–20592. doi: 10.1074/jbc.M210992200. [DOI] [PubMed] [Google Scholar]
- 44.Buschmann T, Potapova O, Bar-Shira A, Ivanov VN, Fuchs SY, Henderson S, Fried VA, Minamoto T, Alarcon-Vargas D, Pincus MR, et al. Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol Cell Biol. 2001;21:2743–2754. doi: 10.1128/MCB.21.8.2743-2754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noguchi K, Kitanaka C, Yamana H, Kokubu A, Mochizuki T, Kuchino Y. Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. J Biol Chem. 1999;274:32580–32587. doi: 10.1074/jbc.274.46.32580. [DOI] [PubMed] [Google Scholar]
- 46.Park J, Kim I, Oh YJ, Lee K, Han PL, Choi EJ. Activation of c-Jun N-terminal kinase antagonizes an anti-apoptotic action of Bcl-2. J Biol Chem. 1997;272:16725–16728. doi: 10.1074/jbc.272.27.16725. [DOI] [PubMed] [Google Scholar]
- 47.Lee DH, Ha JH, Kim Y, Jang M, Park SJ, Yoon HS, Kim EH, Bae KH, Park BC, Park SG, et al. A conserved mechanism for binding of p53 DNA-binding domain and anti-apoptotic Bcl-2 family proteins. Mol Cells. 2014;37:264–269. doi: 10.14348/molcells.2014.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]