Abstract
The human ether-a-go-go-related potassium channel 1 (hERG1) is a component of the voltage-gated Kv11.1 potassium channel, which has been recently indicated to have a crucial role in the tumorigenesis of multiple tumors, including pancreatic carcinoma. Pancreatic carcinoma is one of the most malignant human cancer types, which has an extremely poor prognosis. The present study demonstrated that the expression levels of hERG1 were markedly elevated in pancreatic cancer tissues and pancreatic cancer cell lines, and that the abnormal hERG1 expression was significantly associated with the proliferation and invasion ability of pancreatic cancer. Furthermore, hERG1 was identified to be a direct target of miR-493, which is generally reduced in pancreatic cancer tissues and cell lines. These findings provide a novel insight into the regulatory mechanism of miR-493/hERG1 in pancreatic cancer cell proliferation and invasion, which may aid the development of novel diagnostic and therapeutic strategies for pancreatic cancer in the future.
Keywords: human ether-a-go-go-related potassium channel 1, microRNA-493, pancreatic cancer, proliferation, invasion
Introduction
Pancreatic cancer (PC) is one of the most aggressive malignancies in the world with an extremely low 5-year survival rate (1,2). To date, detection and diagnosis of PC is difficult on account of the lack of diagnostic markers. When the patients were diagnosed with PC, most of them developed an aggressive form of disease which limits the potential for therapeutic intervention (3). Although surgery is the primary way of treatment, pharmacological treatments are also an important adjuvant to surgery. During the past decades, studies have tried to understand the undelying molecular and signaling mechanisms that regulate the development of PC so that better therapy strategies may be developed (4–6).
At this stage, plenty of researches have demonstrated that, during the the occurrence and development of PC, several genetic and epigenetic changes have taken place, such as DNA methylation, microRNA (miRNAs) expression profile and so on (7–9). However, minimal improvements have been made in the prevention and treatment of PC due to the lack of a fundamental target. Therefore, there is an urgent need to develope a new associated factors or novel therapeutic targets in PC.
The human ether-a-go-go-related potassium channel (hERG), also known as KCNH2 or Kv11.1, plays an important role on the terminal repolarization in human ventricular myocytes (10). Several clinically successful drugs have a tendency to inhibit hERG, leading to the risk of adverse drug reactions with QT interval prolongation syndrome or sudden death during use (11,12). In recent years, increasing evidence has demonstrated that plasma membrane ether-a-go-go-related potassium channel 1 (hERG1) potassium (K+) channels are required for cell proliferation and have essential roles in many crucial cellular events such as apoptosis, migration and invasion (13–15). However, research examining the function of hERG1 in human PC are rare and the underlying mechanisms regulating hERG1 expression in PC progression remian largely unknown.
miRNAs are highly conserved small noncoding RNAs that recognize and bind to the 3′UTR of targeted mRNAs resulting in translational repression. More than half of the known miRNAs have been shown to participate in human tumorigenesis and/or metastasis by directly targeting oncogenes or tumor suppressor genes (16,17). Numerous profiling studies over the past decade have shown that the miRNAs play pivotal roles in multiply human cancers, including PC (18–20), which thereby being potential cancer biomarkers in PC. Based on the previous research about the miRNA expression profile in PC (21) and the result that we profiled Targetscan and miRBase databases to explore miRNAs which has putative binding sites with hERG1 gene (KCNH2), we selected miR-493 as a potential regulator in PC progression.
In the present study, we provide evidences that miR-493 was essential in the regulation of proliferation and invasion of PC cells, and the underlying molecular mechanism potentailly lied in the regulation of hERG channel expression, which shed a new light in understanding of how miRNAs act in tumorigenesis and provide novel therapeutic strategies in PC treatment.
Materials and methods
Tissue samples
Tissue samples were collected from patients diagnosed with PC and underwent surgery at The Third Affiliated Hospital of Harbin Medical University (Harbin, China). The samples were frozened and stored at −80°C until total RNA or protein was extracted. All patients have signed the consent to the research. The Research Ethics Committee of Harbin Medical University (Harbin, China) approved the study.
Cell culture and transfection
Human PC cell lines PANC-1 and CFPAC-1 and the human pancreatic cell line (HPDE) were purchased from the Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in DMEM (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified atmosphere of 5% CO2. miR-493 mimic and antisense oligodeoxyribonucleotide (AMO) were obtained from GenePharma (Shanghai, China). All the cell transfections were performed according to the kit instruction (X-tremeGENE; Roche Diagnostics, Indianapolis, IN, USA). Cells were studied within 8 h of harvest.
Luciferase assay
As miR-493 was identified as the potent regulator of HERG1 levels by Targetscan database, next it was examined whether HERG1 is a direct target of miR-493. Briefly, the miR-493-binding site in the 3′-UTR region of HERG1 (wild or mutant type) was cloned into the downstream of the firefly luciferase gene in a pGL3-promoter vector (Promega, Madison, WI, USA). Next, the plasmid DNA (wild-type or mutant pGL3-hERG1-3′ UTR; 500 ng/ml) and miR-297 mimics (100 nM) were transfected into cells using Lipofectamine 2000 for 48 h. Firefly luciferase activity was analyzed by Dual-Luciferase Reporter Assay System according to the manufacturer's protocol (Promega) and was normalized to the Renilla luciferase activity.
Cell proliferation assay
The cell proliferation was determined by MTT assay. Briefly, cells were seeded in 96-well plates at 1×105 cells/well and maintained for 24 h to allow cell adhesion. Subsequently, the cells were incubated with 30 µl of MTT (5 mg/ml) for 4 h. Afterwards, the foramazan crystals were dissolved in 100 µl DMSO, and the absorbance was measured at 570 nm by using a microplate reader (Olympus, USA). The viability of treated samples was assessed by comparison with negative control. Each experiment was tested in triplicate.
In vitro cell invasion assays
The invasion assay was performed with 24-well plates coated with 100 µl of Corning Matrigel Basement Membrane Matrix (BD Biosciences, San Diego, CA, USA). Cells were seeded on to the Matrigel coated wells (3×104 cells/cm2). After 24 h, the cells migrated to the bottom surface of the membrane were fixed in 4% paraformaldehyde in PBS. Once fixed, the cells were stained with crystal violet for 10 min at room temperature and then the pictures were captured by using a fluorescence microscope (Nikon Corp., Tokyo, Japan). Data were expressed as the average number of cells per insert.
RNA extraction and real-time quantitative PCR
Total RNA from mouse tissues was extracted using TRIzol reagent (Invitrogen, the Netherlands). The concentration of RNA was detected by a NanoDrop Spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Synthesis of hERG1 cDNA was carried out using a universal reverse transcription kit (Takara, Dalian, China), while a poly(A) tail was added to the miRNAs for miR-493 cDNA synthesis. Real-time PCR was performed using the SYBR-Green PCR Master Mix (Roche) and the ABI 7500 Real-Time PCR System (Life Technologies, Grand Island, NY, USA). PCR amplification was performed in a total volume of 20 µl with 2 µl cDNA, 6 µl DEPC, 10 µl SYBR-Green Master Mix, 1 µl forward primer and 1 µl reverse primer. PCR conditions were as follows: 40 cycles of 95°C for 15 sec, 60°C for 15 sec, and 72°C for 45 sec. After circle reaction, the threshold cycle (Ct) was determined, and relative miR-493 and hERG1 were calculated based on the Ct values and normalized by U6 or GAPDH level respectively in each sample.
Western blot analysis
For the western blot analysis, the total protein of the tissues and cells were harvested with radioimmunoprecipitation assay buffer (RIPA) containing 1% protease inhibitor (Sigma, St. Louis, MO, USA) and the protein concentration was determined by BCA Protein Assay kit (Thermo Scientific, USA) Samples were loaded to 8% SDS-PAGE gels and transferred onto PVDF (Pall Life Sciences, Port Washington, NY, USA) membrane by electrophoresis. After 2 h blocking with 5% nonfatmilk in PBS, membranes were incubated with hERG1 antibody (1:1,000 dilution) or monoclonal GAPDH antibody (1:3,000) overnight at 4°C with gentle shaking. Afterwards, secondary antibody labelled with fluorochrome dyes (Alexa Fluor 800; LI-COR Biosciences, Lincoln, NE, USA) was used. Finally, immunoreactivity was detected with the Odyssey fluorescent scanning system (LI-COR Biosciences) and the band densities were quantified by densitometry, using Scion Image software (Scion, Frederick, MD, USA). Data were normalized to GAPDH expression levels.
Statistical analysis
Data are expressed as mean ± standard error of mean (mean ± SEM) in triplicate experiments and analyzed with SPSS 13.0 software. A one-way analysis of variance or Student's t test was used to determine the significance of differences between control and test groups. P≤0.05 was considered to indicate a statistically significant difference (two-tailed).
Results
hERG1 is upregulated in human PC
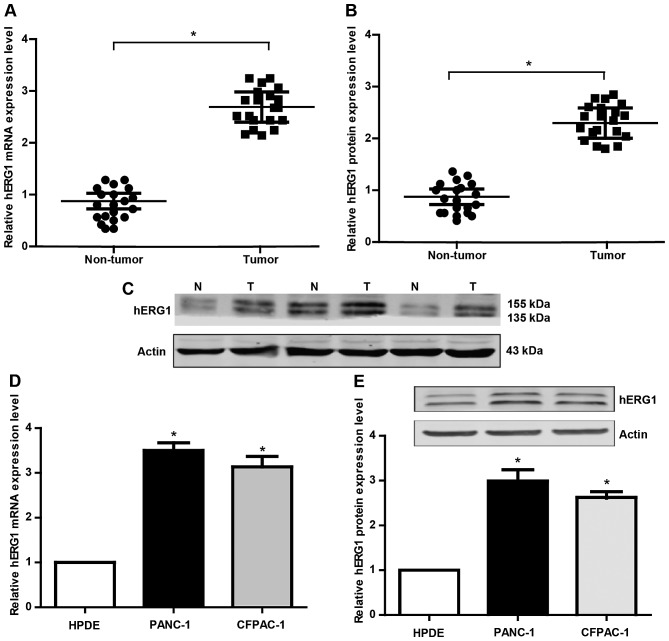
In order to explore the role of hERG1 in pancreatic carcinogenesis, the mRNA and protein expression of hERG1 was detected by qRT-PCR and western blot analyses. Twenty pairs of PC patient tissues and matched noncancerous tissues were collected in this research. There were 14 males and 6 females with a medium age at 63 years old. As shown in Fig. 1A-B, the hERG1 was found to be significantly upregulated in PC tissues compared to the noncancerous tissues. Meanwhile, we also evaluated hERG1 levels in HPDE, PANC-1, CFPAC-1 cells. The results (Fig. 1D and E) showed that the expression level of hERG1 in the PC cell lines (PANC-1, CFPAC-1) is obviously higher than that in the normal HPDE. These findings suggested that hERG1level may has a potential correlation with the pathogenesis of PC.
Figure 1.
The expression patterns of hERG1 in PC tissues and cell lines. (A and B) qRT-PCR and western blot analysis of hERG1 expression in PC tissues and the paired adjacent noncancerous tissues (n=20). (C) Part of the representative western blot results of hERG1 in PC tissues and paired adjacent noncancerous tissues. (D and E) The mRNA and protein expression of hERG1 in normal HPDE and PC cell lines (PANC-1, CFPAC-1). n=3; *P<0.05. hERG1, human ether-a-go-go-related potassium channel 1; PC, pancreatic cancer; HPDE, human pancreatic cell line.
MiR-493 is downregulated in PC and directly targets hERG1
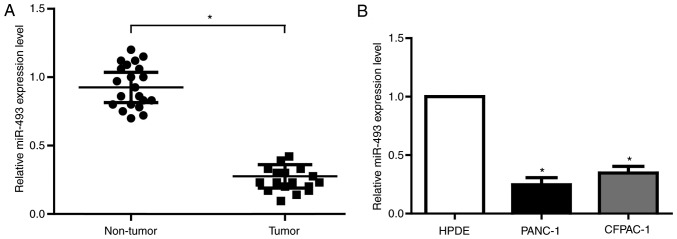
miRNA is one of the most important epigenetic regulators in human cancers. Based on the predictive results of bioinformatics tools and the expression profile of miRNA from previous study in PC, we found that miR-493 has potential biding sites with the 3′UTR of KCNH2 mRNA, which is the coding gene for hERG1. To validate the actual relationship of miR-493 and hERG1 in PC, we firstly explored the level of miR-493 in 20 pairs of PC tissues and matched noncancerous tissues. As is expected, miR-493 showed obvious decrease in all the tumor tissues (Fig. 2A). The results from PC cell lines were in accordance with that in PC tissues (Fig. 2B). Therefore, miR-493 is most likely to be the important regulator of hERG1 in PC cells.
Figure 2.
(A) The relative expression of miR-493 in PC tissues and paired adjacent noncancerous tissues (n=20). (B) The relative expression of miR-493 in PC cell lines. n=3. *P<0.05. HPDE, human pancreatic cell line; PC, pancreatic cancer.
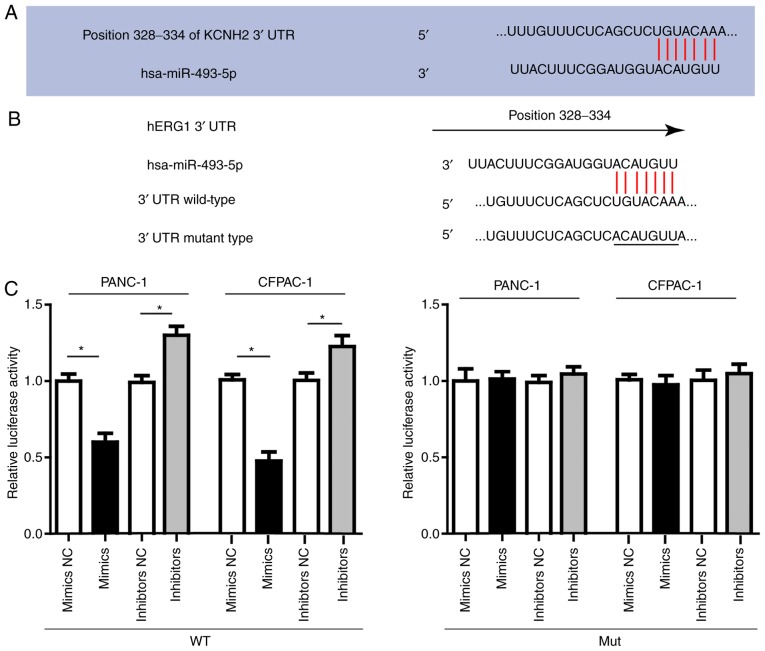
Then we conducted luciferase assay to investigate the direct interaction between miR-493 and hERG1. In order to verify this direct targeting relationship, the miR-493 binding sequence in the 3′-UTR of hERG1 and the mutated 3′-UTR sequence were inserted into the downstream of the firefly luciferase reporter gene in a pGL3-promoter vector and then co-transfected with miR-493 mimics, inhibitors or negative control, respectively into PANC-1, CFPAC-1 cells. As shown in Fig. 3, the relative luciferase activity in PANC-1 or CFPAC-1 cells co-transfected with pGL3-hERG1 and miR-493 mimics was significantly decreased by nearly 40% compared with that in the negative control (NC) group, while the relative luciferase activity in PANC-1 or CFPAC-1 cells co-transfected with mutated pGL3-hERG1 and miR-493 mimics or miRNA NC was no different. The luciferase activity showed relative increase when miR-493 inhibitors were used instead. In conclusion, our results confirmed that miR-493 could suppress hERG1 expression through direct binding sequences at the 3′UTR of hERG1 mRNA.
Figure 3.
hERG1 is a direct target of miR-493. (A) The putative biding sites of miR-493 in the hERG1 3′-UTR sequence. (B) The nucleotide sequence of miR-493 binding sites within the hERG1 3′-UTR region and the mutated nucleotides in miR-493 binding sites. (C) hERG1-WT and the hERG1-Mut reporter luciferase activity in PANC-1 and CFPAC-1 cells treated with miR-493 mimics, inhibitors or negative control, respectively. All data are shown as mean ± SD. n=3; *P<0.05. hERG1, human ether-a-go-go-related potassium channel 1; Mut, mutated; WT, wild-type; NC, negative control; KCNH2, hERG1.
miR-493 regulates hERG1 expression in PC cells
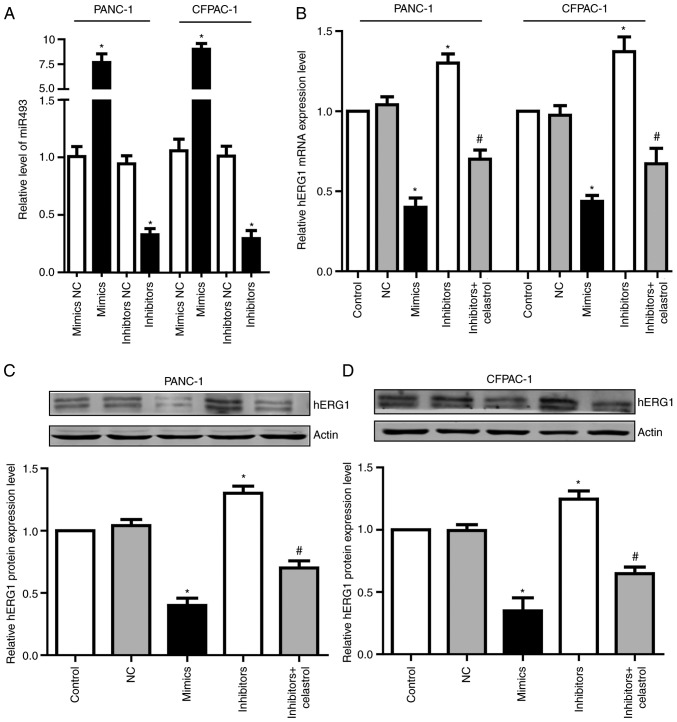
To determine the function of miR-493, we transfected miR-493 mimics or inhibitors into PANC-1 and CFPAC-1 cells. After 24 h, qRT-PCR assay was performed to detect miR-493 levels. Results showed that miR-493 mimics could significantly increase miR-493 level in both PANC-1 and CFPAC-1 cells, while miR-493 inhibitors dramatically decreased miR-493 level (Fig. 4A). To further investigate if miR-493 affected hERG1expression in PC cells, the mRNA and protein level of hERG1 were measured after the transfection of miR-493 mimics or inhibitors. As shown in Fig. 4B-D, overexpression of miR-493 led to the obvious suppression of hERG1, while knockdown of miR-493 enhances the expression of hERG1 in both mRNA and protein levels. Additionally, the elevated hERG1 level by miR-493 inhibitors could be partially reversed by celastrol, which is a well-known hERG channel inhibitor. These data demonstrated that miR-493 is an important post-transcriptional regulator of hERG1 in PC cells.
Figure 4.
hERG1 channels are inversely regulated by miR-493. (A) Relative miR-493 expression level in PC cells transfected with miR-493 mimics, inhibitors or negative control, respectively. (B-D) Relative hERG1 expression level in PC cells transfected with miR-493 mimics, inhibitors and negative control, additionally a group co-incubation miR-493 inhibitors with celastrol. All data are shown as mean ± SD. n=3; *P<0.05. hERG1, human ether-a-go-go-related potassium channel 1; PC, pancreatic cancer.
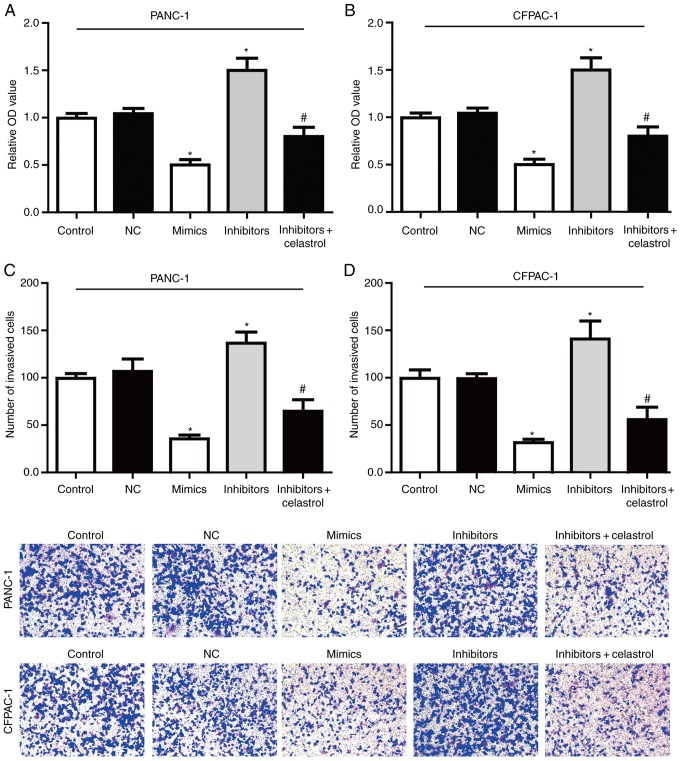
miR-493/hERG1 axis regulates PC cell proliferation and invasion
The effect of miR-493 on the proliferation of PC cells was detected in vitro by the MTT assay (Fig. 5A-B). The results showed that overexpression of miR-493 markedly reduced the cell proliferation activity in both PANC-1 and CFPAC-1 cells, while knockdown of miR-493 showed a sharp increased proliferation activity. All these data suggested that miR-493 could enhance the proliferation of PANC-1 and CFPAC-1 cells.
Figure 5.
miR-493 regulates PC cell proliferation and invasion through targeting hERG1 in vitro. (A and B) Cell proliferation was determined by MTT assay. (C and D) The invasiveness of PANC-1 and CFPAC-1 cells was assessed by Transwell assay. All data are shown as mean ± SD. n=3; *P<0.05; #P<0.05. NC, negative control; hERG1, human ether-a-go-go-related potassium channel 1; PC, pancreatic cancer.
In order to determine whether miR-493 affected cell invasion, Transwell assay were performed after transfection with miR-493 mimics, inhibitors or NC in PC cell lines. As shown in Fig. 5C-D, PANC-1 and CFPAC-1 cells transfected with miR-493 mimics showed a higher ratio in invasion. On the contrary, knockdown of miR-493 strongly inhibited the cell invasive ability. Furthermore, the enhanced invasive ability by miR-493 inhibitors could be largely reversed by co-incubation with celastrol. Therefore, our data suggested that miR-493 acts as a tumor suppressor in PC that could inhibit proliferation and invasion potentially through targeting hERG1, and plays a key role in the tumorigenesis in PC cells.
Discussion
Despite great efforts made in the treatment of PC, the prognosis of PC is still very poor with an extremely low 5-year survival rate. Clinically, PC still lacks tumor markers with enough specificity and sensitivity. Thus, it's urgently needed to explore new molecular markers to improve the early diagnosis and accurate therapy for PC.
The potassium channels have long been suggested to be related to the regulation of a variety of biological activities such as the control of cell excitability and the regulation of cell proliferation and invasion ability (22,23). Indeed, accumulating evidences have demonstrated that hERG1 channel, a human rapid delayed rectifier in the voltage-gated potassium channel family, is often aberrantly expressed in numerous carcinomas including PC. Previous study by Feng J has demonstrated that hERG1 functions as an oncogene in PC and is downregulated by miR-96 (23). Since the miRNAs expression profile is complicated in the progression of diseases, multiply miRNAs except miR-96, may involved in the process. Therefore, we tried to explore whether there is another miRNA involved in the development and progression of PC. In the present study, we provide evidence that miR-493 inhibits proliferation and invasion in PC cells and inversely regulated hERG1 expression. These results implied that multi-target therapy may offer the best hope for developing an effective PC treatment strategy.
It is becoming increasingly evident that miRNAs are important epigenetic regulators in human cancers acting as oncogenes or tumor suppressors to regulate cell proliferation, apoptosis, migration and invasion (24,25). For these reasons, we explored the potential miRNAs involved in hERG1 regulation. Based on the complementary pairing prediction by the bioinformatics tools and the expression profile of miRNA from previous study in PC by miRNA expression arrays (21), we chose miR-493 for further analysis, which has putative biding sites in the hERG1 3′-UTR sequence. Indeed, our in vitro luciferase assay study also confirmed that miR-493 was a potent regulator of the hERG1 gene. More so, upregulation of miR-493 in low-expressing PC cells decreases their malignant potential. These results indicate that miR-493 is an important tumor suppressor in PC.
Overall, our study shows that miR-493 is an important oncogene in PC development and progression. Additionally, we demonstrated that human hERG1 gene is the direct downstream target of miR-493, with its expression inversely regulated by miR-493, thus presented a novel epigenetic regulator of miR-493 in PC. Furthermore, we have shown that overexpression of miR-493 can inhibit the malignant capacity of PC cells via the hERG1 regulation, while knockdown of miR-493 promotes proliferation and invasion ability. Based on these results, we proposed the hypothesis that restoration of miR-493 expression or inhibition of hERG1 in PC might provide a novel therapeutic strategy for PC.
Acknowledgements
The present study was supported by the Project funded by China Postdoctoral Science Foundation (grant no. 2015M581491); the Scientific Fund of Heilongjiang Province for Youth (grant no. QC2015099); the Haiyan Foundation from Harbin Medical University Cancer Hospital (grant no. JJQN2017-12); and the Harbin Medical University scientific research innovation fund (grant no. 2016JCZX59).
References
- 1.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou L, Yao LT, Liang ZY, Zhou WX, You L, Shao QQ, Huang S, Guo JC, Zhao YP. Nuclear translocation of fibroblast growth factor receptor 3 and its significance in pancreatic cancer. Int J Clin Exp Pathol. 2015;8:14640–14648. [PMC free article] [PubMed] [Google Scholar]
- 3.Onyeaghala G, Nelson HH, Thyagarajan B, Linabery AM, Panoskaltsis-Mortari A, Gross M, Anderson KE, Prizment AE. Soluble MICA is elevated in pancreatic cancer: Results from a population based case-control study. Mol Carcinog. 2017;56:2158–2164. doi: 10.1002/mc.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Y, Liu L, Ma R, Gong H, Xu P, Wang C. MicroRNA-127 is aberrantly downregulated and acted as a functional tumor suppressor in human pancreatic cancer. Tumour Biol. 2016;37:14249–14257. doi: 10.1007/s13277-016-5270-0. [DOI] [PubMed] [Google Scholar]
- 5.Yang XP, Liu SL, Xu JF, Cao SG, Li Y, Zhou YB. Pancreatic stellate cells increase pancreatic cancer cells invasion through the hepatocyte growth factor/c-Met/survivin regulated by P53/P21. Exp Cell Res. 2017;357:79–87. doi: 10.1016/j.yexcr.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 6.Jixiang C, Shengchun D, Jianguo Q, Zhengfa M, Xin F, Xuqing W, Jianxin Z, Lei C. YEATS4 promotes the tumorigenesis of pancreatic cancer by activating beta-catenin/TCF signaling. Oncotarget. 2017;8:25200–25210. doi: 10.18632/oncotarget.15633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverman BR, Shi J. Alterations of epigenetic regulators in pancreatic cancer and their clinical implications. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17122138. pii: E2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao Y, Shen J, Lu Y, Lin K, Wang H, Li Y, Chang P, Walker MG, Li D. RNA sequencing analyses reveal novel differentially expressed genes and pathways in pancreatic cancer. Oncotarget. 2017;8:42537–42547. doi: 10.18632/oncotarget.16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Guo XJ, Ding YM, Jiang JX. miR-1181 inhibits invasion and proliferation via STAT3 in pancreatic cancer. World J Gastroenterol. 2017;23:1594–1601. doi: 10.3748/wjg.v23.i9.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandenberg JI, Perry MD, Perrin MJ, Mann SA, Ke Y, Hill AP. hERG K(+) channels: Structure, function, and clinical significance. Physiol Rev. 2012;92:1393–1478. doi: 10.1152/physrev.00036.2011. [DOI] [PubMed] [Google Scholar]
- 11.Villoutreix BO, Taboureau O. Computational investigations of hERG channel blockers: New insights and current predictive models. Adv Drug Deliv Rev. 2015;86:72–82. doi: 10.1016/j.addr.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Cubeddu LX. Drug-induced inhibition and trafficking disruption of ion channels: Pathogenesis of QT abnormalities and drug-induced fatal arrhythmias. Curr Cardiol Rev. 2016;12:141–154. doi: 10.2174/1573403X12666160301120217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Neut M, Haar L, Rao V, Santha S, Lansu K, Rana B, Jones WK, Gentile S. Activation of hERG3 channel stimulates autophagy and promotes cellular senescence in melanoma. Oncotarget. 2016;7:21991–22004. doi: 10.18632/oncotarget.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becchetti A, Crescioli S, Zanieri F, Petroni G, Mercatelli R, Coppola S, Gasparoli L, D'Amico M, Pillozzi S, Crociani O, et al. The conformational state of hERG1 channels determines integrin association, downstream signaling, and cancer progression. Sci Signal. 2017;10 doi: 10.1126/scisignal.aaf3236. pii: eaaf3236. [DOI] [PubMed] [Google Scholar]
- 15.Arcangeli A, Becchetti A. hERG channels: From antitargets to novel targets for cancer therapy. Clin Cancer Res. 2017;23:3–5. doi: 10.1158/1078-0432.CCR-16-2322. [DOI] [PubMed] [Google Scholar]
- 16.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 17.Rachagani S, Macha MA, Heimann N, Seshacharyulu P, Haridas D, Chugh S, Batra SK. Clinical implications of miRNAs in the pathogenesis, diagnosis and therapy of pancreatic cancer. Adv Drug Deliv Rev. 2015;81:16–33. doi: 10.1016/j.addr.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun B, Liu X, Gao Y, Li L, Dong Z. Downregulation of miR-124 predicts poor prognosis in pancreatic ductal adenocarcinoma patients. Br J Biomed Sci. 2016;73:152–157. doi: 10.1080/09674845.2016.1220706. [DOI] [PubMed] [Google Scholar]
- 19.Cai H, An Y, Chen X, Sun D, Chen T, Peng Y, Zhu F, Jiang Y, He X. Epigenetic inhibition of miR-663b by long non-coding RNA HOTAIR promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget. 2016;7:86857–86870. doi: 10.18632/oncotarget.13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Holländer NH, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 21.Kato K, Iwama H, Yamashita T, Kobayashi K, Fujihara S, Fujimori T, Kamada H, Kobara H, Masaki T. The anti-diabetic drug metformin inhibits pancreatic cancer cell proliferation in vitro and in vivo: Study of the microRNAs associated with the antitumor effect of metformin. Oncol Rep. 2016;35:1582–1592. doi: 10.3892/or.2015.4496. [DOI] [PubMed] [Google Scholar]
- 22.Debska G, Kicinska A, Skalska J, Szewczyk A. Intracellular potassium and chloride channels: An update. Acta Biochim Pol. 2001;48:137–144. [PubMed] [Google Scholar]
- 23.Feng J, Yu J, Pan X, Li Z, Chen Z, Zhang W, Wang B, Yang L, Xu H, Zhang G, Xu Z. HERG1 functions as an oncogene in pancreatic cancer and is downregulated by miR-96. Oncotarget. 2014;5:5832–5844. doi: 10.18632/oncotarget.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pengcheng S, Ziqi W, Luyao Y, Xiangwei Z, Liang L, Yuwei L, Lechen L, Wanhai X. MicroRNA-497 suppress renal cell carcinoma by targeting VEGFR-2 in ACHN cells. Biosci Rep. 2017;37 doi: 10.1042/BSR20170270. pii: BSR20170270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K, Zhang C, Liu L, Zhou J. A key role of microRNA-29b in suppression of osteosarcoma cell proliferation and migration via modulation of VEGF. Int J Clin Exp Pathol. 2014;7:5701–5708. [PMC free article] [PubMed] [Google Scholar]