Abstract
The development of a minimally invasive test, such as liquid biopsy, for early lung cancer detection in its preclinical phase is crucial to improve the outcome of this deadly disease. MicroRNAs (miRNAs) are tissue specific, small, non-coding RNAs regulating gene expression, which may act as extracellular messengers of biological signals derived from the cross-talk between the tumor and its surrounding microenvironment. They could thus represent ideal candidates for early detection of lung cancer. In this work, a methodological workflow for the prospective validation of a circulating miRNA test using custom made microfluidic cards and quantitative Real-Time PCR in plasma samples of volunteers enrolled in a lung cancer screening trial is proposed. In addition, since the release of hemolysis-related miRNAs and more general technical issues may affect the analysis, the quality control steps included in the standard operating procedures are also presented. The protocol is reproducible and gives reliable quantitative results; however, when using large clinical series, both pre-analytical and analytical features should be cautiously evaluated.
Keywords: Cancer Research, Issue 128, Lung cancer, early detection, circulating microRNAs, biomarkers, liquid biopsy, Real-Time PCR.
Introduction
Lung cancer is the most commonly diagnosed cancer worldwide, accounting for about 25% of all cancer diagnoses1. Despite the steady reduction in lung cancer mortality rate over the past 2 decades, mainly due to reduced tobacco use, lung cancer remains the leading cause of cancer death in both men and women. In 2017, it is predicted to account for approximately 25% and 20% of the total cancer deaths in the United States and Europe, respectively1,2.
Since the symptoms do not usually occur in early disease, the majority of lung cancers are diagnosed at late stages. This leads to a limited possibility for therapeutic intervention and poor efficacy of the treatments3. Therefore, many scientific efforts are aimed at identifying an effective test for early lung cancer detection.
A variety of screening trials shows that chest radiography has no value in lung cancer screening, whereas low-dose computed tomography (LDCT) is a better tool compared to radiography for detecting early stage lung cancer4. Moreover, it has been shown thatscreening with the use of LDCT reduced lung cancer mortality up to 20% among current and former heavy smokers5. These data support the use of lung cancer screening in a high risk population as defined in the guidelines by several professional societies4,6.
Of note, among the major concerns about LDCT screening, there is the high rate of false-positive results and over-diagnosis. That being so, the scientific community is looking for a strategy to overcome these issues and increase the specificity of the LDCT screening test. The development of non-invasive complementary biomarkers could be useful here. To date, there is an extensive list of candidate biomarkers under investigation, such as miRNAs, cell-free DNA, gene methylation, small proteins, and others7.
Blood-based biomarkers, including immune-response biomarkers and circulating miRNAs, are those that reach the more advanced validation phase8. The immune-response biomarkers are based on the knowledge that immune responses against both intracellular and surface tumor antigens are built up in lung cancer patients9. For instance, Ajona et al. evaluated the role of C4d, a degradation product of the classical complement pathway, in the diagnosis and prognosis of lung cancer10. Otherwise, a commercially available autoantibodies serum test examining seven tumor-related antigens has been validated in non-small cell lung cancer patients and showed 36% of sensibility and 91% of specificity11.Immune-response biomarkers are interesting, but further validation studies are needed for a potential clinical application.
miRNAs are endogenous small non-coding RNAs with a conserved mechanism and large functional significance all over the animal and plant kingdoms. The role of miRNAs is to regulate protein translation: they repress the expression of a large set of target genes12. It has been well documented that alterations ofmiRNAs' expression contribute to the pathogenesis of most human cancer12,13,14. In addition, both tumor and stromal cells release miRNAs, stabilized by their incorporation into microvesicles or through association to RNA-binding proteins, into body fluids such as plasma and serum15,16, urine17,and saliva18. Circulating miRNAs may thus reflect the host response and the dynamic interaction between the tumor and its microenvironment19. Taken together these observations have made circulating miRNAs a promising class of biomarkers for the detection of human cancer.
Circulating miRNAs can be detected by using different methods: microarray platforms20, quantitative reverse transcription PCR (RT-qPCR)21, and next generation sequencing (NGS)14,22. Various expression profile studies based on the aforementioned technologies have been developed in the last years. Microarray is a high-throughput technology, able to analyze up to thousands of miRNAs in one single assay. However, it has lower dynamic range and specificity compared to RT-qPCR or NGS. In addition, microarray is a non-quantitative method, therefore further experimental validation is required. Sequence-based methods can allow the identification of unknown miRNAs and are suitable especially in a discovery phase. On the other hand, NGS methods are still expensive and necessitate special equipment and expert bioinformaticians, thus limiting their use in large validation cohorts and in clinical settings23. At the moment, RT-qPCR is the most adopted platform for circulating miRNA detection in a diagnostic/clinical context24. There are different RT-qPCR technologies available for miRNAs analysis, but the stem-loop RT followed by TaqMan PCR is the most widely used25. This technique is extremely sensitive and accurate (single nucleotide discrimination) and has a high dynamic range; however, it allows the detection of known miRNAs only.
The microRNA quality control study (miRQC study) by Mestdagh et al. extensively investigated the characteristics of some commercially available platforms for miRNAs profiling based on the three aforesaid technologies26. By analyzing 196 miRNAs in tissues and serum samples, the performance of the different platforms was assessed in terms of reproducibility, sensitivity, accuracy, specificity, and concordance of differential expression. The results have shown that platforms based on NGS and microarray technologies had a higher reproducibility and specificity, but RT-qPCR platforms were more accurate, sensitive, and had a higher detection rate, especially for low input RNA samples, such as body fluids. Thus RT-qPCR seems to be the most suitable method for a potential application in the routine clinical diagnostics.
In 2011, Sozzi et al. analyzed the miRNA profile of plasma samples collected from volunteers enrolled in a first LDCT screeningtrial (INT-IEO trial)27 and four miRNA signatures composed by reciprocal ratios among 24 circulating miRNAs were generated. These signatures were able to discriminate disease free individuals from patients with any or lethal lung cancer at the time or up to 2 years before LDCT disease detection28. In a further paper, the same group described for the first time the miRNA signature classifier (MSC), obtained by combining the four miRNA signatures in order to stratify patients in three levels of risk: high, intermediate, and low. Its clinical utility was validated in a large retrospective set of more than 1,000 individuals enrolled in the MILD screening trial, a randomized study comparing annual or biennial LDCT arms with an observation arm29. Indeed, the combination of MSC and LDCT reduced the LDCT false-positive rate by approximately 5 times and the three MSC risk groups were associated with overall survival.
Later on, in 2013 at the "Fondazione IRCCS Istituto Nazionale dei Tumori" (Milan, IT) a prospective screening trial (BioMILD) implementing LDCT with the plasma based MSC test was launched. Volunteers enrolled in the BioMILD trial undergo LDCT and blood withdrawal that is immediately processed to separate plasma for miRNA profiling using custom made microfluidic cards. The combination of the LDCT and MSC results defines the specific screening algorithm24.
In the present work, the entire methodological protocol used in the BioMILD trial is described, starting from blood sample collection, to plasma isolation, RNA extraction, and the 24 miRNAs expression profiling using custom made RT-qPCR microfluidic cards. Given the high consistency and reproducibility, these procedures can also be used for the development of miRNA-based liquid biopsies in other diseases.
Protocol
The protocol was approved by the research ethics committee of our institution.
1. Plasma Sample Collection
Collect 10 mL of whole blood sample in Vacutainer tubes with spray-coated K2EDTA and store at room temperature. Note: To minimize the hemolysis, separate plasma within 2 h. Do not store the whole blood at low temperature (i.e., 4 °C) to avoid thermal shock and cell lysis that lead to an unspecific miRNA release.
Within 1 h, separate the plasma by a first centrifugation step at 1,258 x g and 4 °C for 10 min.
Transfer the plasma supernatant into a 15 mL tube, carefully avoiding contact with the lymphocytic ring.
Centrifuge the plasma a second time at 1,258 x g and 4 °C for 10 min.
Aliquot 1 mL of plasma into 1.5 mL cryovials, avoiding collecting the plasma fraction at the base of the tube.
Store all the aliquots at −80 °C, except one for the evaluation of the hemolysis. The molecular analysis should be performed within 5 weeks.
2. Evaluation of Hemolysis by Spectrophotometric Measurement
Immediately after the plasma separation step, in a cuvette for spectrophotometry, make a 1:10 dilution of the plasma sample in 1X PBS (e.g., 100 µL of plasma in 900 µL of 1X PBS) and mix to homogenize it. Note: The plasma sample should not be frozen before the spectrophotometric measurements, as the freeze-thawing of the sample (and in particular in lipemic samples), could form flocculates that interfere with the spectrophotometric analysis.
Read the absorbance at 375 nm, 414 nm, 541 nm, and 576 nm using a UV-Vis spectrophotometer, with a baseline correction settled at 750 nm and a path length of 10 mm. Perform a blank measurement using 1 mL of 1x PBS.
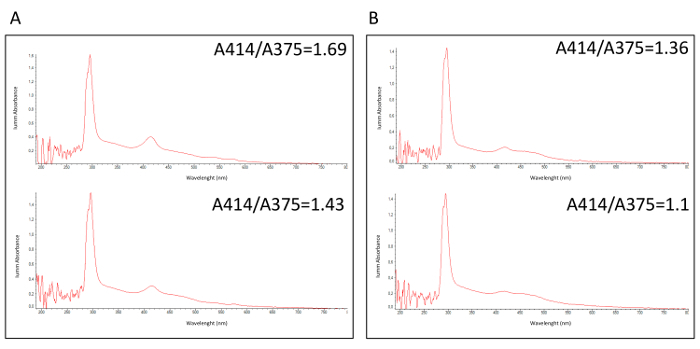
Calculate the ratio between the absorbance at 414 nm and 375 nm; if higher than 1.4, consider the sample hemolyzed and repeat the blood withdrawal (QC1 in Table 1).
3. Plasma Total RNA Extraction
Prepare a 1-Thioglycerol/Homogenization (OMG) solution with 20 µL of 1-Thioglycerol per mL of Homogenization Solution. Since 1-Thioglycerol is viscous, carefully pipette for accurate measurement. Chill the OMG Solution on ice or at 2-10 °C before using it. NOTE: The working solution is stable at 2-10 °C for 1 month.
Suspend the lyophilized DNase I by adding 275 µL of nuclease-free water into the vial and gently mix (do not vortex). As a visual aid, add 5 µL of blue dye to the reconstituted DNase I and dispense the solution into single-use aliquots in nuclease-free tubes. Store reconstituted DNase I at -30 °C to -10 °C.
Starting from 200 µL of plasma, add 200 µL of the chilled OMG solution.
Vortex 15-30 s to ensure a complete homogenization. If foaming occurs, let the sample settle on ice.
Add 200 µL of Lysis Buffer and 25 µL of Proteinase K to the homogenized sample and vortex for 20 s.
Incubate the samples for 15 min in a thermomixer preheated at 37 °C.
Load a cartridge for each sample on the deck tray of the instrument and place the plugger in the proper position.
Transfer the lysate to the appropriate position in the instrument cartridge.
Add 5 µL of DNase I solution to the proper position in the cartridge.
Add 60 µL of nuclease-free water to the base of each elution tube.
Select the "RSC miRNA Tissue" method and begin the automated purification run. The total RNA samples can be stored at −80 °C.
4. Taq-based RT
Use the Taq RT Primer Pool with the miRNAs of interest to convert the RNA to cDNA with the Taq MicroRNA Reverse Transcription Kit.
Prepare 12 µL of RT reaction mix on ice in a 1.5 mL microcentrifuge tube according to the kit instructions, using 6 µL of the custom Taq RT Primer Pool.
Add 3 µL of total RNA for a final volume of 15 µL and incubate in ice for 5 min.
Load the RT reaction on a thermo-cycler configured as follows: 16 °C for 30 min, 42 °C for 30 min, 85 °C for 5 min, and hold at 4 °C. Note: The cDNA can be store at -15 to -20 °C for at least one week.
5. Pre-amplification
Mix 2.5 µL of each RT product with 12.5 µL of Taq Master Mix 2x, 3.75 µL Custom Taq pool, and 6.25 µL nuclease-free water, for a total volume of 25 µL.
Perform the pre-amplification reaction using a thermo-cycler according to the following thermal profile: 95 °C for 10 min, 55 °C for 2 min, 72 °C for 2 min, 12 cycles of 95 °C for 15 s, and 60 °C for 4 min, then hold at 99.9 °C for 10 min and at 4 °C.
Dilute the product by adding 175 µL of 0.1X TE, pH 8.0. Note: The diluted product can be store at -15 to -20 °C for at least one week.
6. RT-qPCR Reaction on Custom Taq Array MicroRNA Cards
Use the 384-well microfluidicCustom Taq Array MicroRNA Card to measure plasma levels of the 24 specific miRNAs (spotted in duplicate) on 8 samples simultaneously. MiRBase ID (v21) for the 24 miRNAs are: hsa-miR-101-3p, hsa-miR-106a-5p, hsa-miR-126-3p, hsa-miR-133a-3p, hsa-miR-140-3p, hsa-miR-140-5p, hsa-miR-142-3p, hsa-miR-145-5p, hsa-miR-148b-3p, hsa-miR-15b-5p, hsa-miR-16-5p, hsa-miR-17-5p, hsa-miR-197-3p, hsa-miR-19b-3p, hsa-miR-21-5p, hsa-miR-221-3p, hsa-miR-28-3p, hsa-miR-30b-5p, hsa-miR-30c-5p, hsa-miR-320a, hsa-miR-451a, hsa-miR-486-5p, hsa-miR-660-5p, and hsa-miR-92a-3p.
Mix 1.13 µL of the diluted PreAmp sample with 56.25 µL of 2X Taq Universal Master Mix and 55.69 µL of nuclease-free water in a 0.5 mL tube.
Load up to 8 PCR reaction mix on the Custom Taq Array MicroRNA Cards.
Centrifuge at 311 x g for 2 min.
Seal the custom card with the array card sealer.
Run the Real-Time reaction using a Real-Time PCR System modifying the cycling parameters as followed: 94.5 °C for 10 min, 40 cycles of 97 °C for 30 s, and 59.7 °C for 60 s.
7. Data Extrapolation and Ratios Generation
Use the software to obtain raw Ct values, setting an automatic baseline to remove the background signals and a fixed threshold of 0.15 for all assays and samples. Remove spots with poor amplification curves and/or poor passive reference signals.
Export raw Ct values in ".xls" format and use the Ct Mean value of the two duplicates for further analysis.
Correlate miRNAs' expression levels to historic measurements on clinical study samples (Table 2). If at least 50% of samples on each custom made microfluidic card result in a Pearson's correlation < 97.5, repeat the card (QC2 in Table 1).
Calculate the −ΔCts between all miRNAs, equivalent to the log2 values of the miRNA ratios.
8. Evaluation of Hemolysis by the Related miRNA Signature
Generate the hemolysis-related miRNA signature using the following miRNA ratios with respective cut-offs (log2 values) for short storage plasma samples (1-5 weeks at −80 °C): miR-126/451 < −0.07, 15b/451 < −3.67, 221/451 < −3.18, 30b/451 < −1.1, 126/486-5p < −0.33, 15b/486-5p < −3.86, 221/486-5 p < −3.17, 30b/486-5p < −1.42, 126/92a < 1.8, 15b/92a < −1.8, 221/92a < −1.04, 30b/92a < 0.87, 126/16 < −2.85, 15b/16 < −6.33, 221/16 < −5.9, 30b/16 < −3.68.
Classify as hemolyzed samples where at least 50% of ratios (8 out of 16) exceed respective cut-off (QC3 in Table 1).
9. Definition of the Level of Risk: High, Intermediate, and Low
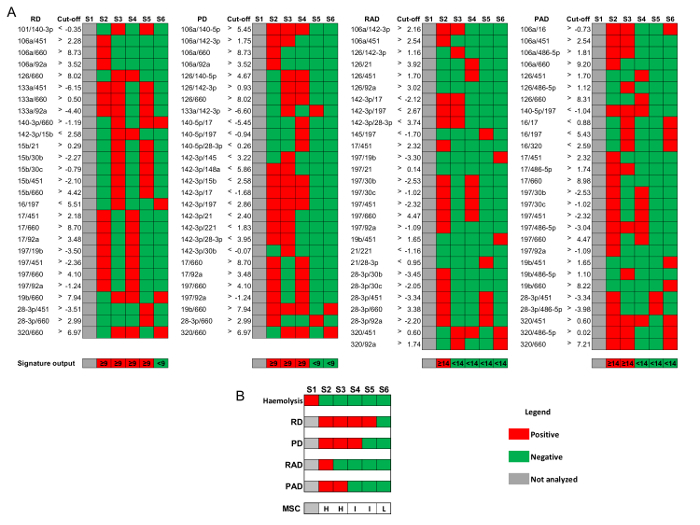
Define the four signatures of miRNA ratios composing the MSC as reported in Figure 3: risk of disease (RD), risk of aggressive disease (RAD), presence of disease (PD), and presence of aggressive disease (PAD).
For each signature, define the ratios exceeding the respective cut-off value for short storage plasma samples (1-5 weeks at −80 °C). The number of exceeding ratios needed to be considered positive is 9 out of 27 for RD and PD, and 14 out of 28 for RAD and PAD (Figure 3A).
Attribute to each sample the respective MSC risk level as follows: low risk if RDneg ∩ PDneg ∩ RADneg ∩ PADneg; intermediate risk if RDpos ∪PDpos ∩ RADneg ∩ PADneg; or high risk if RADpos ∪ PADpos (Figure 3B).
Representative Results
The BioMILD trial is a prospective study for early detection of lung cancer with the purpose of testing the efficiency of a combined LDCT-MSC approach as first-line screening tests in a large cohort of heavy smoker individuals. A class of risk is attributed to each volunteer on the basis of the miRNA test, which evaluates ratios between 24 plasma circulating miRNAs. As largely reported30,31,32, hemolysis is a critical issue for the analysis of circulating miRNA, since false results could be obtained due to a nonspecific release of miRNAs by blood cells. In the proposed methodological workflow, a pre-analytical quality control step (QC1 in Table 1) to identify hemolyzed, and thus non-analyzable, plasma samples was described. Figure 1 reports a spectrophotometric analysis of hemolyzed (Figure 1A) and non-hemolyzed (Figure 1B) plasma samples with respective A414/A375 values. In a clinical context were the result of a molecular test is crucial for volunteer management, this QC1 allows repeating the blood sampling and, in most cases, restoring the sample.
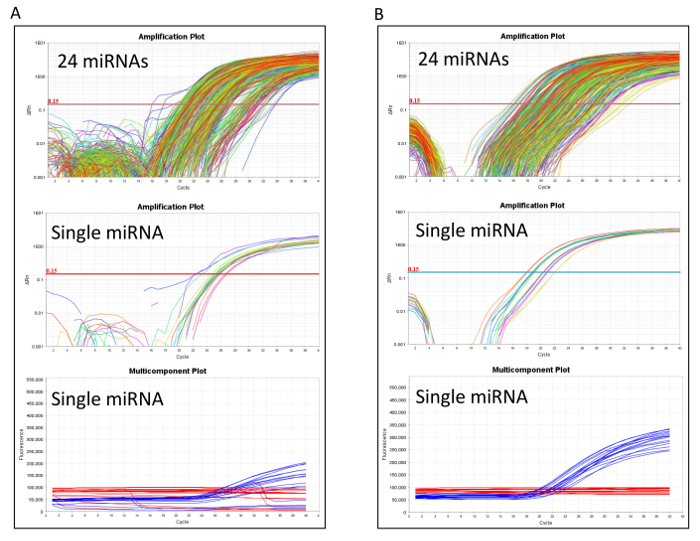
After molecular processing, the RT-qPCR performance should be accurately evaluated to identify any technical issues. Figure 2A shows a RT-qPCR with a low performance due to a poor passive reference signal. In this case reloading the pre-amplification product to a new microfluidic card is recommended. When the RT-qPCR performs well, as in Figure 2B, the card proceeds to the first post analytical quality control step to ensure that the miRNA expression values are comparable with the historical measurement (QC2 in Table 1). If the QC2 step fails, a technical problem may have occurred during reverse transcription or in the pre-amplification reactions, and it is convenient to repeat the entire molecular processing starting from reverse transcription.
The last quality control step (QC3 in Table 1) evaluates the hemolysis also at the molecular level. Expression levels of 4 erythrocyte-specific miRNAs (mir-16, mir-451, mir-486-5p, and mir-92a) were compared to those of 4 non-hemolysis related miRNAs (mir-126, mir-15b, mir-221, and mir-30b), generating the hemolysis-related miRNA signature composed by the 16 (4 x 4) miRNA ratios. Positive samples are classified as hemolyzed, while negative samples proceed to the final MSC risk level classification as described in Protocol Section 9 and Figure 3.
Figure 1:Spectrophotometric analysis of fresh plasma samples. Spectrophotometric profiles of (A) 2 hemolyzed and (B) 2 non-hemolyzed plasma samples, measuring the absorbance ratio between wavelengths A414 and A375. Please click here to view a larger version of this figure.
Figure 2: RT-qPCR performance in microfluidic cards analysis. Amplification and multicomponent plots comparing (A) poor and (B) good quality microfluidic cards. All 24 miRNAs and a single exemplifying miRNA are reported in each panel. The multicomponent plots illustrate the passive reference signals occurring in these RT-qPCR reactions. Please click here to view a larger version of this figure.
Figure 3: MSC algorithm for risk stratification. Representative results considering 6 plasma samples from volunteers enrolled in BioMILD screening trial.(A) miRNA-ratios' signatures of risk of disease (RD), risk of aggressive disease (RAD), presence of disease (PD), presence of aggressive disease (PAD), and corresponding cut-off values (log2) for plasma samples stored at -80 °C for at least 1 week and up to 5 weeks. (B) Combination of the four signatures to stratify analyzable samples in high (H), intermediate (I), and low (L) MSC risk level. Please click here to view a larger version of this figure.
Discussion
Pitfalls and Challenges of the Methodological Workflow As BioMILD is the first prospective study analyzing circulating miRNAs as a diagnostic tool, thresholds and cut-offs utilized for data-analysis were generated from retrospective series. In order to overcome the issue of the different storage interval of plasma samples, the parameters can be refined to possibly obtain an increase of the MSC performance. Nevertheless, before starting miRNAs analysis, several aspects of the overall process need to be carefully evaluated, such as the inclusion criteria, the sample collection and processing, and the bioinformatics algorithm adopted for data analysis. The following sections will focus on the most critical points of the workflow described in the present paper, addressing the major issues of each single step and then proposing strategies to overcome them.
Study Population Enrollment criteria for the BioMILD screening trial are: 50-75 years old, heavy current or former (less than 10 years) smokers with at least 30 pack-years and no prior history of cancer within 5 years. In this population, the estimated lung cancer prevalence and incidence per year were about 1% and 0.7%, respectively. In order to apply the MSC test as described in the proposed protocol, the population analyzed should maintain similar characteristics. In particular, the QC2 step described above involves comparison with the given historical data obtained from a similar population. While a correlation below 97.5 could be indicative of technical problems, on the other hand, most individuals with altered circulating miRNA levels reflecting the risk of having lung cancer are actually below that threshold. In other words, if the QC2 step with the parameters reported here is applied to a population at higher risk of developing lung cancer, it may exclude acceptable samples. New, proper reference parameters should then be defined that are specific for each population analyzed according to expected results.
Blood Collection and Hemolysis The collection of biological samples is a crucial pre-analytical step, since it may impair sample quality and characteristics, and thus modify the final results of analyses. Regarding blood samples, it is important to collect fresh blood and process the specimen as quickly as possible. Samples should neither be stored at 4 °C nor frozen after blood withdrawal, since freezing and thawing passages may induce cell lysis (hemolysis) and RNA degradation. To minimize hemolysis process, it is suggested to separate plasma within 2 h from blood collection31 .
Hemolysis is largely due to breakdown of red blood cells (RBCs) and consequent release of their content in the surrounding environment. This could be a problem for circulating miRNAs testing because release of RBCs intracellular content dramatically alters the microRNA profile in blood, potentially affecting the accuracy of plasma-based analysis30.
Being able to identify also minimal levels of hemolysis in plasma samples is crucial for studies based on circulating miRNAs as biomarkers. Hemolysis can initially be assessed by visual inspection, since after the two centrifugation steps the color of plasma can range from yellow to red for highly hemolyzed samples. However, mere visual inspection may not be sensitive enough in the context of molecular biomarker research.
A simple, pre-analytical method to identify hemolyzed plasma samples is the spectrophotometric measurement at wavelength (λ) = 414 nm of the main oxyhemoglobin peak absorbance, after appropriate adjustment for any other source of interference. For that reason, we normalized the 414 nm peak to the absorbance value at 375 nm, indicative of the lipidic content33. Alternatively, or better, in combination, a lipemia-independenthemolysis score (HS) can be used34. This spectrophotometrically-based procedure can identify at least 6.1 mg/dL of free hemoglobin. Otherwise, hemolyzed specimens can be identified through the detection of erythrocyte-specific miRNAs. Since even a very low percentage of hemolysis can elicit a considerable increase in erythrocyte-specific miRNA levels32, Fortunato et al. identified a specific hemolysis-related signature based on 16 miRNAs-ratios that can efficiently classify hemolysis in plasma samples33. All the mentioned assays may be used in combination since the absorbance ratio 414/375 and/or the HS are adopted in a pre-analytical phase saving further costs, whereas measuring hemolysis-related miRNAs can be used in a post-analytical phase as a quality control step.
Circulating miRNAs Isolation and RNA Purity The extraction RNA procedure is a very influential step for the success of the subsequent analyses. Starting from a low input sample, such as plasma, extraction needs to be as efficient as possible to ensure an accurate quantification of miRNAs. Plasma is characterized by high concentrations of many proteins, including nucleases, and other components which can interfere with downstream enzymatic reactions of RT-qPCR and could affect the circulating-miRNAs detection. Several extraction methods have been used in these types of studies, but in general, organic extraction followed by silica-based RNA enrichment allows the removal of plasma inhibitors better then TRIzol alone35. Nevertheless, in the last few years, manual extraction has been replaced by automated extraction. The minor risk of contamination, the higher reproducibility, and the economic time use are the main advantages of automated systems compared to manual isolation protocols. Moreover, automated extraction methods yielded the highest miRNA amounts from blood samples and the highest efficiency compared to manual protocols36.
Normalization Issue Normalization of plasma miRNA levels is still a controversial issue. Indeed, differences in sample quality or quantity can interfere with the identification of true changes in miRNAs expression levels caused by the presence or risk of diseases, and thus leading to ambiguous data interpretations and misleading conclusions. So far, the lack of consensus has resulted in various normalization strategies37.
Total RNA extracted from plasma samples is usually below the threshold of standard quantification methods such as UV spectrophotometry or fluorescence-based spectrophotometry, thus precluding RNA mass standardization38. Consequently, a fixed volume rather than a fixed amount of eluted RNA should be chosen as input for the RT reaction39.
Housekeeping transcripts used for miRNA analysis in tissue samples, like RNU48 and RNU6, are not suitable for normalizing extracellular miRNA measurements, as they both are not always detectable in circulation, due to RNase-mediated degradation40,and are also deregulated in a disease-specific manner37. It is unclear if any circulating miRNAs could effectively be used as reference controls. For example, miR-16 is frequently proposed for housekeeping, but different studies have shown that it is deregulated in plasma samples of cancer patients28,37. Moreover, miR-16 levels are highly affected by hemolysis30.
While for high-throughput assays measuring several miRNAs and normalizing on the mean expression value is generally accepted41, when a limited number of miRNAs needs to be analyzed,this normalization approach cannot be applied. In order to bypass the normalization issue, Boeri et al. previously established an algorithm based on reciprocal ratios among 24 miRNAs28. Such an approach, despite being very useful for the development of the diagnostic tool, does not easily allow distinguishing effectively altered miRNAs that possibly having a role in tumor development, and thus identifying miRNAs as functioning calibrators. Another effective strategy, adopted by the group of Bianchi et al. that developed a serum miR-Test for lung cancer early detection, was based on normalizing on the geometric mean of a group of 6 "housekeeping serum-miRNAs" varying less among samples and classes of patients42.
Features of miRNA-based Liquid Biopsy and Potential Application Plasma-based MSC is a molecular non-invasive test studied for early diagnosis of lung cancer in heavy smokers. The MSC test can identify intermediate or high-risk subjects before lung cancer detection by LDCT, with high sensibility (SE, 87%) and negative predictive value (NPV, 99%). Specifically, MSC could implement LDCT screening by reducing false-positive results to 3.7%, from 19.7% for LDCT alone29. The MSC test has also shown to have a good prognostic performance when considering only lung cancer patients, and could be used as a monitoring tool alone or in combination with other clinical pathological parameters43,44. Potentially, the MSC test could enable greater consistency of lung cancer diagnostic algorithms, thus decreasing screening cost-effectiveness.
Besides MSC, other non-invasive tests for early lung cancer detection have been reported. Bianchi et al. proposed a circulating miR-Test based on a 34-miRNA signature45, then reduced it to a 13 miRNA signature42. Serum samples fordiscovery and validation were obtained from the Continuous Observation of Smoking Subjects (COSMOS) LDCT screening trial, performed at the European Institution of Oncology (IEO, Milan, Italy). The miR-Test showed 78% accuracy for lung cancer detection in high-risk volunteers. Between the 13 serum-miRNAs and the 24 composing the MSC, only 5 miRNAs were overlapping (miR-92a, miR-30b, miR-30c, miR-148a, and miR-140-5p).This could be due to the different type of starting samples (serum vs. plasma) and to data elaboration. In fact, for the miR-Test, the authors selected 6 miRNAs behaving as housekeeping in serum, 3 of which were included also among the MSC miRNA ratios (miR-15b, miR-19b, and miR-197). Nevertheless, the MSC in plasma and the miR-Test in serum samples more deeply validate the use of miRNA-based liquid biopsy for detection of lung cancer.
MiRNAs profiling for the identification of lung cancer biomarkers has been performed also on sputum samples46. Sputum can be easily collected and the miRNA expression levels appear to be extremely stable in it. However, lung cancer screening volunteers are older than 50-55 years and are heavy smokers with co-morbidities such as COPD, thus making sputum difficult to obtain. Indeed, among the different body fluids, plasma is one of the best alternatives to investigate the potential role of miRNAs as (but not limited to) lung cancer biomarkers.
Overall, the procedure described in the present paper can be relevant in the context of various diseases, with no restriction in the type of the pathology investigated (from cancer, to nervous system and cardiovascular disorders, or diabetes) and in the purpose of clinical use (a biomarker for diagnosis, prognosis, or even response to treatment).
Disclosures
Gabriella Sozzi, Mattia Boeri, and Ugo Pastorino are co-inventors of three patent applications licensed to Gensignia Life Sciences, regarding the miRNA signature disclosed in this article. All the remaining authors declare no conflicts of interest in relation to the work submitted.
Acknowledgments
The authors thank Paola Suatoni, Elena Bertocchi, Carolina Ninni, Annamaria Calanca, and Chiara Banfi for handling volunteers in the trials and for administrative assistance; and Claudio Jacomelli and Claudio Citterio for data management. The work was supported by the Italian Association for Cancer Research [Investigator Grants No. 15928 to UP, 14318 to GS, and 12162 (Special Program "Innovative Tools for Cancer Risk Assessment and early Diagnosis", 5x1000)]; the Italian Ministry of Health [Grant No. RF-2010]; Grant UO1 CA166905 from the National Cancer Institute (USA). MB was supported by a Fondazione Pezcoller Fellowship.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- Malvezzi M, et al. European cancer mortality predictions for the year 2017, with focus on lung cancer. Ann Oncol. 2017. p. 10. [DOI] [PubMed]
- Miller KD, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- Kanne JP. Screening for lung cancer: what have we learned. AJR Am. J Roentgenol. 2014;202(3):530–535. doi: 10.2214/AJR.13.11540. [DOI] [PubMed] [Google Scholar]
- Aberle DR, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender R, et al. American Cancer Society lung cancer screening guidelines. CA Cancer J Clin. 2013;63(2):107–117. doi: 10.3322/caac.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Vila MA, et al. Liquid Biopsy in Non-Small Cell Lung Cancer. Front Med (Lausanne) 2016. p. 69. [DOI] [PMC free article] [PubMed]
- Sozzi G, Boeri M. Potential biomarkers for lung cancer screening. Transl Lung Cancer Res. 2014;3(3):139–148. doi: 10.3978/j.issn.2218-6751.2014.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd FA, Douillard JY, Blumenschein GR. Immunotherapy for non-small cell lung cancer: novel approaches to improve patient outcome. J Thorac Oncol. 2011;6(10):1763–1773. doi: 10.1097/JTO.0b013e31822e28fc. [DOI] [PubMed] [Google Scholar]
- Ajona D, et al. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst. 2013;105(18):1385–1393. doi: 10.1093/jnci/djt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, et al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol. 2011;22(2):383–389. doi: 10.1093/annonc/mdq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014. [DOI] [PubMed]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke M, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28(6):655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Park NJ, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15(17):5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012;18(3):268–274. doi: 10.1097/PPO.0b013e318258b743. [DOI] [PubMed] [Google Scholar]
- Castoldi M, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12(5):913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, et al. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res. 2010;38(17):5919–5928. doi: 10.1093/nar/gkq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberio P, Callari M, Angeloni V, Daidone MG, Appierto V. Challenges in using circulating miRNAs as cancer biomarkers. Biomed Res Int. 2015. p. 731479. [DOI] [PMC free article] [PubMed]
- Boeri M, et al. Recent advances of microRNA-based molecular diagnostics to reduce false-positive lung cancer imaging. Expert Rev Mol Diagn. 2015;15(6):801–813. doi: 10.1586/14737159.2015.1041377. [DOI] [PubMed] [Google Scholar]
- Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50(4):244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- Mestdagh P, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- Pastorino U, et al. Early lung-cancer detection with spiral CT and positron emission tomography in heavy smokers: 2-year results. Lancet. 2003;362(9384):593–597. doi: 10.1016/S0140-6736(03)14188-8. [DOI] [PubMed] [Google Scholar]
- Boeri M, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108(9):3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzi G, et al. Clinical Utility of a Plasma-Based miRNA Signature Classifier Within Computed Tomography Lung Cancer Screening: A Correlative MILD Trial Study. J Clin Oncol. 2014;32(8):768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5(3):492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6(9):24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, et al. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front Genet. 2013;4(94) doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato O, et al. Assessment of circulating microRNAs in plasma of lung cancer patients. Molecules. 2014;19(3):3038–3054. doi: 10.3390/molecules19033038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appierto V, et al. A lipemia-independent NanoDrop((R))-based score to identify hemolysis in plasma and serum samples. Bioanalysis. 2014;6(9):1215–1226. doi: 10.4155/bio.13.344. [DOI] [PubMed] [Google Scholar]
- Kim DJ, et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn. 2012;14(1):71–80. doi: 10.1016/j.jmoldx.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulstein G, Marienfeld R, Miltner E, Wiegand P. Automation of DNA and miRNA co-extraction for miRNA-based identification of human body fluids and tissues. Electrophoresis. 2016;37(21):2742–2750. doi: 10.1002/elps.201600365. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data Normalization Strategies for MicroRNA Quantification. Clin Chem. 2015;61(11):1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser K, McIntyre LA, White RJ, Stewart DJ. Customized Internal Reference Controls for Improved Assessment of Circulating MicroRNAs in Disease. PLoS ONE. 2015;10(5):0127443. doi: 10.1371/journal.pone.0127443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, et al. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7(7):41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montani F, et al. miR-Test: A Blood Test for Lung Cancer Early Detection. J Natl Cancer Inst. 2015;19(6):063. doi: 10.1093/jnci/djv063. [DOI] [PubMed] [Google Scholar]
- Sestini S, et al. Circulating microRNA signature as liquid-biopsy to monitor lung cancer in low-dose computed tomography screening. Oncotarget. 2015;20(32):32868–32877. doi: 10.18632/oncotarget.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verri C, et al. Mutational Profile from Targeted NGS Predicts Survival in LDCT Screening-Detected Lung Cancers. J Thorac. Oncol. 2017. p. 10. [DOI] [PMC free article] [PubMed]
- Bianchi F, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3(8):495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyoba J, Shan S, Roa W, Bedard EL. Diagnosing Lung Cancers through Examination of Micro-RNA Biomarkers in Blood, Plasma, Serum and Sputum: A Review and Summary of Current Literature. Int J Mol Sci. 2016;17(4):494. doi: 10.3390/ijms17040494. [DOI] [PMC free article] [PubMed] [Google Scholar]


