Abstract
Cellular senescence is a stress response that is characterized by a stable cellular growth arrest, which is important for many physiological and pathological processes, such as cancer and ageing. Recently, senescence has also been implicated in tissue repair and regeneration. Therefore, it has become increasingly critical to identify senescent cells in vivo. Senescence-associated β-galactosidase (SA-β-Gal) assay is the most widely used assay to detect senescent cells both in culture and in vivo. This assay is based on the increased lysosomal contents in the senescent cells, which allows the histochemical detection of lysosomal β-galactosidase activity at suboptimum pH (6 or 5.5). In comparison with other assays, such as flow cytometry, this allows the identification of senescent cells in their resident environment, which offers valuable information such as the location relating to the tissue architecture, the morphology, and the possibility of coupling with other markers via immunohistochemistry (IHC). The major limitation of the SA-β-Gal assay is the requirement of fresh or frozen samples.
Here, we present a detailed protocol to understand how cellular senescence promotes cellular plasticity and tissue regeneration in vivo. We use SA-β-Gal to detect senescent cells in the skeletal muscle upon injury, which is a well-established system to study tissue regeneration. Moreover, we use IHC to detect Nanog, a marker of pluripotent stem cells, in a transgenic mouse model. This protocol enables us to examine and quantify cellular senescence in the context of induced cellular plasticity and in vivo reprogramming.
Keywords: Developmental Biology, Issue 128, Cellular senescence, in vivo reprogramming, pluripotent stem cells, reprogrammable mouse model, muscle injury, cardiotoxin, regeneration, senescence-associated β-galactosidase staining
Introduction
Cellular senescence is a form of stress response characterized by a stable cell-cycle arrest. In the last decade, research has firmly established that senescence is associated with various biological and pathological processes including embryonic development, fibrosis, and organism ageing1,2. Cellular senescence was first identified in human fibroblasts at the end of their replicative lifespan triggered by telomere shortening3. Besides replicative stress, there are many other stimuli that can induce senescence, including DNA damage, oxidative stress, oncogenic signals, and genomic/epigenomic alterations, any of which may eventually activate the p53/p21 and/or pRB pathways to establish and reinforce the permanent growth arrest1. One of the important characteristics of senescent cells is that they remain metabolically active and robustly express a senescence-associated secretory phenotype (SASP): secretion of many inflammatory cytokines, growth factors, and extracellular matrix factors4. SASP factors have been proposed to play an important role in mediating and amplifying the senescence effect, due to their potent effects on attracting immune cells and altering local and systemic tissue milieus1. Interestingly, senescence has been recently proposed to be important for tissue repair and regeneration5,6. In addition, data from several labs, including ours, has suggested that tissue damage-induced senescence might enhance cellular plasticity, via SASPs, to promote regeneration7-9. Therefore, all the emerging data highlight the importance of studying senescence in vivo.
In the post induced pluripotent stem cell (iPSC) era, cellular plasticity is the capacity of a cell to acquire a new identity and to adopt an alternative fate when exposed to different stimuli both in culture and in vivo10. It is known that full reprogramming can be achieved in vivo11,12, where the expression of the the cassette containing four Yamanaka factors: Oct4, Sox2, Klf4, and c-Myc (OSKM) can be induced in vivo to promote teratomas formation in multiple organs. Therefore, a reprogrammable mouse model (i4F) can be used as a powerful system to identify critical regulators and pathways that are important for cellular plasticity11.
A suitable and sensitive in vivo system is essential to understand how cellular senescence regulates cellular plasticity in the context of tissue regeneration. Here, we present a robust system and a detailed protocol to evaluate the link between senescence and cellular plasticity in the context of tissue regeneration. The combination of cardiotoxin (CTX) induced muscle damage in the Tibialis Anterior (TA) muscle group, a well-established system to study tissue regeneration, and the i4F mouse model, allows the detection of both cellular senescence and in vivo reprogramming during muscle regeneration.
To evaluate the link between cellular plasticity and senescence, i4F mice are injured with CTX to induce acute muscle damage and treated with doxycycline (0.2 mg/mL) over 7 days to induce in vivo reprogramming. While a CTX induced acute muscle damage and regeneration protocol has been recently published13, for ethical reasons, this procedure will be omitted in the current protocol. TA muscle samples will be collected at 10 days post injury13, when the peak of senescent cells have been previously observed14. Here, this detailed protocol describes all the steps required to evaluate the level of senescence (via SA-β-Gal) and reprogramming (via IHC staining of Nanog).
Senescence-associated beta-galactosidase (SA-β-Gal) assay is the most commonly used assay to detect senescent cells both in culture and in vivo15. Compared to other assays, the SA-β-Gal assay allows the identification of the senescent cells in their native environment with intact tissue architecture, which is particularly important for in vivo study. Moreover, it is possible to couple the SA-β-Gal assay with other markers using IHC. However, the SA-β-Gal assay does require fresh or frozen samples, which remains a major limitation. When fresh or frozen tissues are routinely available, such as frozen TA muscle samples, SA-β-Gal is obviously the most suitable assay to detect senescent cells. Nanog is the marker used to detect reprogramed cells for two reasons: 1) it is an essential marker for pluripotency; 2) more importantly, its expression is not driven by doxycycline (dox), therefore it detects induced pluripotency rather than the forced expression of the Yamanaka cassette.
It is important to note, the staining protocols presented in this study can be conducted separately to simplify the quantification procedure, but can also be done in a co-staining procedure to visualize both senescent and pluripotent stem cells on the same section.
Protocol
Animals were handled as per European Community guidelines and the ethics committee of the Institut Pasteur (CETEA) approved protocols.
1. Preparations of the Stock Solutions
Prepare the materials for muscle sample fixation.Dissolve 0.5 g of tragacanth gum with 20 mL water at RT to make the freezing-embedding medium for muscle fixation.
- Prepare the solutions for SA-β-Gal staining.
- Prepare the stock solutions of K3Fe(CN)6 (100 mM), K4Fe(CN)6 (100 mM), MgCl2 (1M) by dissolving the respective powders in distilled water.
- Prepare the stock solution of 0.2% C20H6Br4Na2O5 (Eosin) by diluting it in water.
- Prepare the stock solution of C14H15BrClNO6(X-gal) (50 mg/mL) by dissolving the X-gal powder in C3H7NO (dimethylformamide, DMF).
- Store K3Fe(CN)6 and K4Fe(CN)6 solutions at 4 °C, and MgCl2 at RT. NOTE: X-gal can be stored in aliquot at -20 °C, up to 6 months. Eosin solution can be kept at RT and reused after filtering if necessary. K3Fe(CN)6 and K4Fe(CN)6 solutions are protectedfromthe light. X-gal solution is not stable in water and has to be protected from light.
Preparation of the solutions for Nanog staining: the permeabilization solution contains 0.1% Na3C6H5O7 (trisodium citrate), 0.1% C14H22O(C2H4O)n(n = 9-10) (Triton X-100) in distilled water, which should be stored at 4 °C. Prepare the blocking solution containing 5% fetal bovine serum (FBS) in phosphate-buffered saline (PBS), which should be stored at RT.
2. SA-β-Gal Staining on Frozen TA Muscle Section
Prepare the fixation material for the TA muscle by placing a small amount of the tragacanth gum on a slice of cork.
- Injured mice of both sexes (2 month old, C57/B6) were injured 10 days before with cardiotoxin (CDX) as described previously13. Use non-injured (PBS injection) TA from the same mouse as the negative control. If in vivo reprogramming is desired, treat each mouse with Dox (1 mg/mL) in the drinking water on the same day (protected from the light), right after CTX injury. NOTE: The Dox solution needs to be changed every 3 days for a total treatment duration of 7 days.
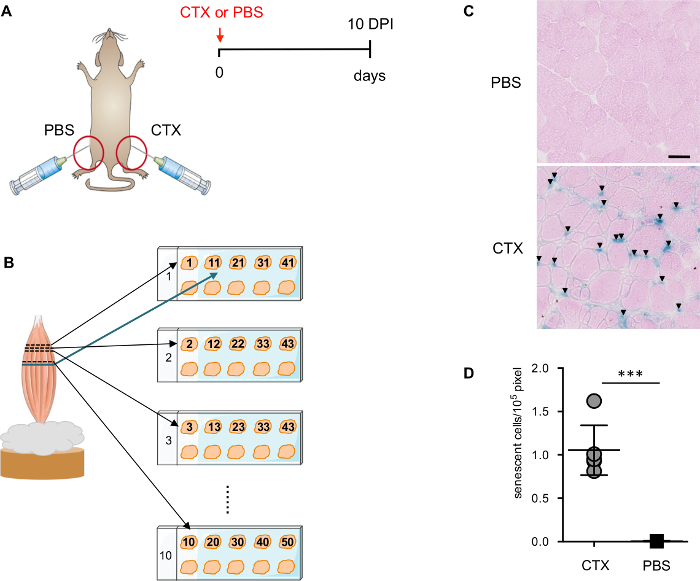
- Isolate both TA muscles (injured and control) from mice as described previously (Figure 1A)13. To ensure the transverse sections, insert the distal tendon of the TA muscle into the tragacanth gum and leave roughly ¾ part of the muscle outside (Figure 1B) and freeze directly in liquid nitrogen cooled isopentane for <1 min. NOTE: Make sure the TA muscle is in a perpendicular position and in the center of the cork. Samples can be stored at -80°C or directly cryosectioned in 10 µm sections.
- Process the TA muscles as described in the Figure 1B.
- Distribute the 1st-10th sections in the correct order onto ten different slides at the upper left position of each slide. Place the 11th section adjacent to the 1st section on the first slide; the 12th section will follow the same order to be placed right besides the 2nd section on the second slide. Repeat this process till 10 sections/slide are obtained for ten slides (100 sections in total) to ensure minimum 1 mm distance between the first section and the last section.
Fix the sections for 4 min in PBS containing 1% paraformaldehyde and 0.2% glutaraldehyde. Wash with PBS, 2x 10 min. Next, incubate the sections in PBS (pH = 5.5) for 30 min. Perform all the steps at RT. NOTE: The fixation has to be mild to maintain enzymatic activity. Perform this step under the hood. The pH of the PBS is critical and the X-gal solution must be protected from light.
Incubate sections in the X-gal solution containing: 4 mM K3Fe(CN)6, 4 mM K4Fe(CN)6, 2 mM MgCl2, and 400 µg/mL X-Gal in PBS, pH = 5.5. Incubate in the dark at 37 °C for a minimum of 24 h. Wash the slides with PBS, 3x 10 min, at RT. NOTE: The incubation requires a minimum of 24 hours and can last for 48 hours to maximize the SA-β-Gal signal. The solution needs to be changed after 24 hours of incubation. The slides should be protected from light. If only SA-β-Gal staining is desired, continue with step 2.5. If co-staining with Nanog is desired, please skip forward to step 3.1.
Fix slides in 1% paraformaldehyde in PBS for 30 min. Wash slides with PBS, 3x 10 min. Counterstain with 0.2% eosin. Immerse the slides in the eosin solution for 1 min and rinse them with distilled water briefly. Finally, mount the slides with aqueous non-fluorescing mounting medium (see Table of Materials). NOTE: It is essential to perform the post-fixation. Perform this step under the hood. All steps are performed at RT.
3. Immunohistochemistry Using Anti-Nanog Antibody
- Fix the slides with PBS containing 4% paraformaldehyde for 10 min. Wash with PBS, 2x 10 min. Add 200 µL of the permeabilization solution directly onto the slides and incubate for 5 min.
- Wash slides with PBS, 2x 5 min and in the last wash, use 200 µL PBS containing 0.25% BSA directly on the slides. Perform all these steps at RT. Caution: Perform the fixation step under the hood.
Incubate slides with the primary anti-Nanog antibody (final concentration: 1.25 ug/mL) overnight at 4 °C in PBS containing 5% FBS. Wash slides with PBS, 2x 10 min and in the last wash, use 200 µL PBS containing 0.25% BSA for 5 min. Perform all the washing steps at RT. NOTE: Incubate slides in a box with wet paper towel to prevent evaporation.
Incubate slides with 100 µL of rAb-HRP secondary antibody from a ready to use kit (see Table of Materials) for 45 min. Wash slides with PBS, 3x 5 min, to remove secondary antibody. Perform all the steps at RT with protection from light.
- Visualization
- First, dilute the 3,3'-diaminobenzidine (C12H14N4C12H18Cl4N4(4HCl), DAB) in the substrate buffer provided by the ready to use kit (20 µL of DAB for 1 mL of substrate buffer solution). Add 100 µL of DAB solution directly onto each slide and incubate for a maximum of 10 min at RT. NOTE: The diluted DAB solution should be prepared freshly and can be stored for up to one week at 4 °C. The incubation time can be adjusted to minimize the background signal but must be kept the same for all slides.
- Remove the DAB solution by rinsing with water. Counterstain the slides with fast red solution (ready to use, see the Table of Materials) for 20 min. Wash the slides with water again briefly.
- Dehydrate them with 95% ethanol for 5 min followed by 100% ethanol, 2x 5 min. Finally, mount the slides with quick-hardening mounting medium (see Table of Materials).
- Observe the slides under the microscope in bright field at 20X to avoid background. NOTE: Fast red solution can be kept at RT and re-used after filtering.
4. Analysis and Quantification
- SA-β-Gal quantification
- Scan the slides and choose the best sections on each slide based on the acquired images. Use at least 2 sections/TA for the quantification. Judge the section's quality based on tissue integrity, quality of the staining, and counterstaining. For the quantification, choose the two highest quality sections with the maximum possible distance in between samples, which allows better representation of SA-β-Gal quantification throughout the whole TA muscle.
- Quantification of SA-β-Gal positive cells (Figure 2).
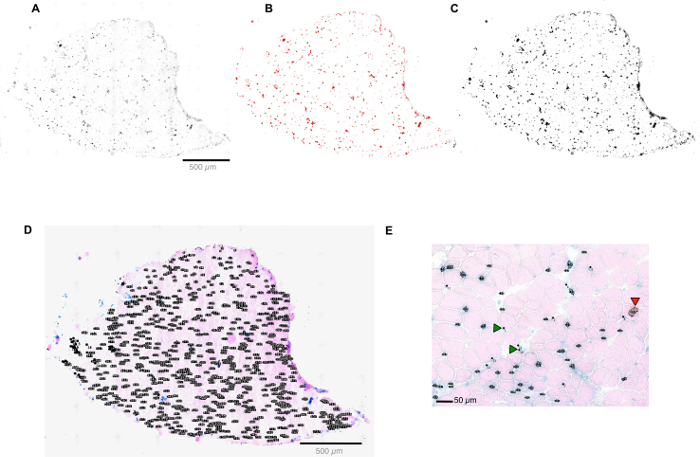
- Determine the rank of the pixel size by manually selecting the smallest and the largest positive SA-β-Gal cells.
- Open the digital image of the scanned slide using ImageJ software.
- In the interface, click 'Analyze' > 'Tools' > 'ROI Manager' > 'Analyze' > 'set measurement' > 'select Area'. Use the selection tool and surround the smallest and biggest positive SA-β-Gal cell and add it to the ROI manager by clicking on 'Add'. Measure the sizes using the 'Measure' button and save the values for later use.
- Adjust the threshold parameter to ensure all the visible positive cells are selected.
- Analyze particles by clicking 'Analyze' > 'Analyze Particles' > 'Size (pixel^2)', and apply the value defined in step 4.1.2.1, enter 'Circularity': '0.00-1.00', click 'ok' ; a summary of all the counted particles is shown in the ROI manager (Figure 2D).
- Transfer all the selected particles to the original image. Adjust the selection manually to ensure accurate quantification. Add positive cells (green arrow, Figure 2E) and/or remove false positive cells (red arrow, Figure 2E). Finally, measure the area by outlining the section (Figure 2D) using the 'selection tool'. Click 'ROI manager' > 'Add' > 'Measure'.
- Importantly, normalize the number of positive cells by the area of the section.
- Nanog quantification
- Count the Nanog positive cells under the microscope in the bright field manually at 20X magnification. NOTE: The fast red counterstaining allows good evaluation of the morphology of the TA muscle. However, the staining is very light and lacks a clear outline of the section. The scanner often fails to identify the boundary of the sections and cannot focus correctly. It is possible to use marker to circle the edges of the sections before scanning or use a more sensitive scanner to detect the section. For the current protocol, it is most efficient to count the Nanog positive cells manually.
Representative Results
Detecting muscle injury-induced cellular senescence
It has been recently demonstrated that muscle injury induces transient cellular senescence14. At 10 days post-injury (DPI), the majority of the damaged myofibers are undergoing regeneration with centrally located nuclei, a hallmark of regenerating myofibers, and the architecture of the muscle is re-established. The infiltrating inflammatory cells are dramatically reduced while remaining visible in certain regions. 10 DPI is a good time point to detect senescent cells by SA-β-Gal, since there are fewer necrotic and inflammatory cells present in the muscle to interfere with the staining. To determine the specificity of the staining, TA injected with PBS from the same mouse (Figure 1A) is used as a critical negative control.
To ensure a better and more precise evaluation of the SA-β-Gal positive cells, sections from different planes of the TA muscle are placed in the same slide (Figure 1B). Counter staining with eosin is important for the automatic quantification of the SA-β-Gal positive cells by ImageJ software. Eosin counter staining outlines the section, which allows the digital scanner to detect the sections with the correct focus. It is important to carefully define the range and the threshold of the detection (Figure 2A-D). In addition, a manually curated process is essential to permit more accurate detection and quantification (Figure 2E).
Cellular senescence facilitates in vivo reprogramming in muscle
Reprogrammable mouse model (i4F) provides an ideal system to evaluate the impact of senescence on cellular plasticity and regeneration. Upon muscle injury, i4F mice are treated with dox to induce reprogramming in vivo. 7 days dox (1 mg/mL) treatment is sufficient to induce reprogramming on the cellular level, while still being well tolerated by mice (Figure 3A). Therefore, we harvest injured muscles at 10 DPIs from i4F mice treated with dox for 7 days.
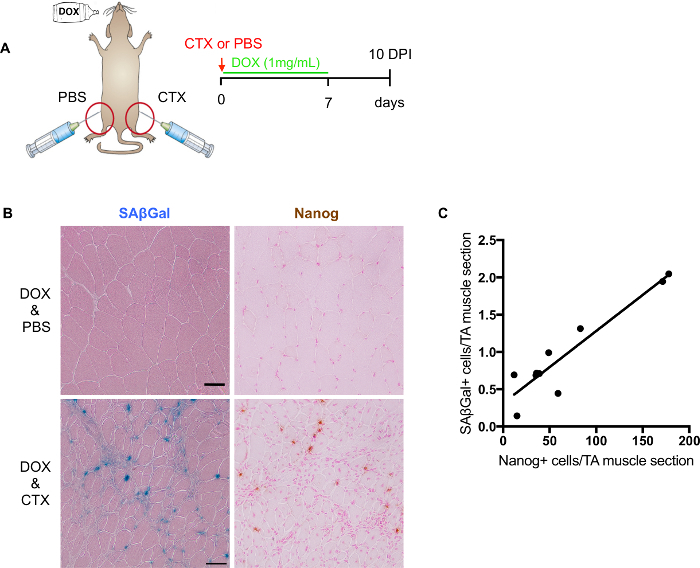
Although it is possible to perform co-staining of SA-β-Gal with Nanog, it is not recommended for quantification due to potential interfering staining. As mentioned above, counter staining is essential for digital scanner detection. The best counter staining for the SA-β-Gal together with Nanog is fast red orhematoxylin. However, counter staining might mask over either the SA-β-Gal or Nanog signal. Therefore, for more accurate quantification, it is better to perform them separately on consecutive slides (Figure 3B). By quantifying SA-β-Gal positive and Nanog positive cells from adjacent slides, we established a positive correlation between them (Figure 3C), suggesting a potential involvement of senescence on cellular plasticity and regeneration.
Figure 1: Evaluate senescence level after muscle injury. (A) Schematic representation of the muscle injury strategy used to induce senescence. (B) Schematic representation of the muscle section preparation. (C) Representative images of SAβGal staining counterstained with eosin. Arrows point to the SA-β-Gal+ cells. Scale bars = 50 µm. (D) Quantification of SA-β-Gal+ cells in injured and non-injured TA-muscle. Each dot corresponds to an individual animal. Statistical significance was assessed by the two-tailed Student´s t-test: ***p <0.001. Please click here to view a larger version of this figure.
Figure 2: Quantification of SA-β-Gal+ cells by ImageJ software. (A-D) Screen shots of a muscle section in the ImageJ software interface. Screen shot of converting a muscle section image to gray scale (A); Selecting all the SA-β-Gal+ cells in the section (B); Analyzing the selected particles (C); Summary of all the counted particals in the ROI manager (D). (E) Screen shot of the manual curation process. Please click here to view a larger version of this figure.
Figure 3: Evaluation of in vivo reprogramming after muscle injury. (A) Schematic representation to evaluate in vivo reprogramming and senescence level after muscle injury. (B) Representative images of SA-β-Gal and Nanog staining on frozen sections of damaged skeletal muscle. SAβGal staining counterstained with eosin (left); immunohistochemical staining of Nanog counterstained with fast red (right). Non-injured muscles are shown on the top and injured muscle below. Scale bars = 50 µm. (C) Quantification and correlation of SA-β-Gal + and Nanog+ cells in consecutive sections (n = 9 mice, value represents the average of 2 sections per mouse). Please click here to view a larger version of this figure.
| Volume for 50 mL | |
| 100 mM stock K3Fe(CN)6 solution | 2 mL |
| 100 mM stock K4Fe(CN)6 solution | 2 mL |
| 1 M MgCl2 | 100 μL |
| 50 mg/mL X-Gal | 400 μL |
| PBS (pH 5.5) | 45.50 mL |
Table 1: Composition of 50 mL X-gal solution.
Discussion
Here, we present a method to detect both senescent and pluripotent stem cells in the skeletal muscle of reprogrammable mice. This method could be used to evaluate and quantify both senescence and induce cellular plasticity in vivo, and examine the role of senescence in tissue repair and regeneration.
In the current protocol, the senescence-associated β-galactosidase (SA-β-Gal) assay is used to detect in vivo senescent cells in the skeletal muscle. This assay detects the increased lysosomal β-galactosidase activity at suboptimum pH (6.0 or 5.5), associated specifically with senescent cells, while this enzyme's activity is typically measured at acidic pH 4.516,17. Therefore, it is important to adjust the pH (pH = 6 or 5.5) to ensure a specific detection of senescence-associated activity. In addition, counter staining with eosin is essential for the automatic quantification of SA-β-Gal+ cells, where the weak and diffuse signals are not counted. To avoid potential variability, it is preferable that the same person performs the entire counting procedure.
Although the SA-β-Gal assay is the most widely used and accepted biomarker for senescent cells, it is not an exclusive marker for senescence. It has been suggested that over-confluent cells in culture might cause false positivity for SA-β-Gal18. The sensitivity of the assay can be cell type and tissue type dependent in vivo19. Therefore, it is necessary to use other independent canonical markers, such as lack of proliferation, increased expression of senescence mediators (p16, ARF, p53, p21, and p27), and the secretion of various SASP factors, to further confirm and characterize senescence in vivo. Moreover, proper negative controls are indispensable for interpreting results, especially for in vivo study.
Despite the fact that SA-β-Gal assay is not perfect, it does provide particularly valuable information for in vivo study. It permits the detection of senescent cells in their resident environment with intact tissue architecture, providing critical information facilitating the further understanding of the senescent cells' role in different physiological and pathological contexts. Moreover, it can be coupled with the immunostaining of other markers, such as cell surface markers to determine the cellular identity of senescent cells; or stemness markers to examine the potential involvement of senescence in regeneration and tumorigenesis. Previously, we performed the SA-β-Gal assay with immunohistochemical staining of Nanog in the same section or in close proximity to investigate the potential link between senescence and in vivo reprogramming8. While this protocol is focused on skeletal muscle, it can be certainly extended to other tissues.
Recently, cellular senescence has been implicated in tissue repair and regeneration, most likely via SASPs7,8,9. Understanding the mechanisms of how senescence contributes to tissue repair and regeneration will certainly have a tremendous impact on regenerative medicine. This assay provides an important and valuable tool to facilitate the identification and quantification of senescent cells in vivo.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We are indebted to Clemire Cimper for her excellent technical support. Work in the laboratory of H.L. was funded by Institut Pasteur, Centre National pour la Recherche Scientific, and the Agence Nationale de la Recherche (Laboratoire d'Excellence Revive, Investissement d'Avenir; ANR-10-LABX- 73), the Agence Nationale de la Recherche (ANR-16-CE13-0017-01) and Fondation ARC (PJA 20161205028). C.C. and A.C. are funded by the Ph.D. and postdoctoral fellowships from the Revive Consortium.
References
- Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15(7):482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Davaapil H, Brockes JP. Recurrent turnover of senescent cells during regeneration of a complex structure. Elife. 2015;4 doi: 10.7554/eLife.05505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteiro L, et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354(6315) doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- Chiche A, et al. Injury-Induced Senescence Enables In Vivo Reprogramming in Skeletal Muscle. Cell Stem Cell. 2016. [DOI] [PubMed]
- Ritschka B, et al. The senescence-associated secretory phenotype induces cellular plasticity and tissue regeneration. Genes Dev. 2017;31(2):172–183. doi: 10.1101/gad.290635.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17(3):183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Abad M, et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502(7471):340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- Ohnishi K, et al. Premature Termination of Reprogramming In Vivo Leads to Cancer Development through Altered Epigenetic Regulation. Cell. 2014;156(4):663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Guardiola O, et al. Induction of Acute Skeletal Muscle Regeneration by Cardiotoxin Injection. J Vis Exp. 2017. [DOI] [PMC free article] [PubMed]
- Le Roux I, Konge J, Le Cam L, Flamant P, Tajbakhsh S. Numb is required to prevent p53-dependent senescence following skeletal muscle injury. Nat Commun. 2015;6:8528. doi: 10.1038/ncomms9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4(12):1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5(2):187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- Krishna DR, Sperker B, Fritz P, Klotz U. Does pH 6 beta-galactosidase activity indicate cell senescence? Mech Ageing Dev. 1999;109(2):113–123. doi: 10.1016/s0047-6374(99)00031-7. [DOI] [PubMed] [Google Scholar]
- Cristofalo VJ. SA beta Gal staining: biomarker or delusion. Exp Gerontol. 2005;40(10):836–838. doi: 10.1016/j.exger.2005.08.005. [DOI] [PubMed] [Google Scholar]


