Abstract
This release of the FunCoup database (http://funcoup.sbc.su.se) is the fourth generation of one of the most comprehensive databases for genome-wide functional association networks. These functional associations are inferred via integrating various data types using a naive Bayesian algorithm and orthology based information transfer across different species. This approach provides high coverage of the included genomes as well as high quality of inferred interactions. In this update of FunCoup we introduce four new eukaryotic species: Schizosaccharomyces pombe, Plasmodium falciparum, Bos taurus, Oryza sativa and open the database to the prokaryotic domain by including networks for Escherichia coli and Bacillus subtilis. The latter allows us to also introduce a new class of functional association between genes - co-occurrence in the same operon. We also supplemented the existing classes of functional association: metabolic, signaling, complex and physical protein interaction with up-to-date information. In this release we switched to InParanoid v8 as the source of orthology and base for calculation of phylogenetic profiles. While populating all other evidence types with new data we introduce a new evidence type based on quantitative mass spectrometry data. Finally, the new JavaScript based network viewer provides the user an intuitive and responsive platform to further evaluate the results.
INTRODUCTION
Advances in high-throughput biology are generating vast amounts of data for determining the function and interaction patterns proteins use to create complex biological processes in a cell. The results from the current efforts directed at determining protein functions and interactions are spread across different databases, for instance DIP (1), IntAct (2), GEO (3), Encode (4) and BioGrid (5), are specialized on different types of experimental techniques. On their own, these data sources provide a rather incomplete picture of the interactional landscape responsible for the complex biology observed in a cell. Fortunately, these data can be converted and combined into networks of gene/protein associations, where genes/proteins are represented by nodes and the associations are depicted by links. Such networks appear to be scale-free, i.e. with a node degree distribution that follows a power law. Despite that this network property is not uncontroversial (6), the majority of nodes in such a network do have only a few links, except the so-called hubs, that interact with many partners (7). This indicates that gene/protein networks capture some fundamental properties of complex biological systems, albeit far from complete and with false positives. Despite these shortcomings, gene/protein networks have become indispensable for applications such as functional annotation of proteins (8,9), understanding of cellular regulatory mechanisms (10), pathway annotation (11), gene prioritization, and disease gene discovery (12).
Several integrated global networks exist, including FunCoup (13–15), STRING (16), GeneMANIA (17) and GIANT (18). Although other ways to integrate data from various data sources are available, many networks use Bayesian techniques. FunCoup uses a unique redundancy weighted Bayesian integration (15) to combine functional association data of currently 10 different types (15). These data types are mRNA co-expression (MEX), phylogenetic profile similarity (PHP), protein interaction (PIN), subcellular co-localization (SCL), co-miRNA regulation by shared miRNA targeting (MIR), domain-domain interaction (DOM), protein co-expression (PEX), protein abundance profile similarity from quantitative mass spectrometry (QMS), shared transcription factor binding (TFB), and genetic interaction profile similarity (GIN). FunCoup relies on transfer of orthology information between the included proteomes, using the comprehensive InParanoid (19) database, to increase the quality and the coverage of the inference of functional association between genes/proteins. FunCoup employs unique scoring functions for each type of data (e.g. Pearson linear correlation for mRNA co-expression, PPI scoring for PPI). These scores are then turned into a Bayesian score for each network link using a set of known functional associations, i.e. the Gold standard, contrasted with a set of randomly generated associations.
The new FunCoup release includes a complete overhaul of the underlying data, including an update of the existing data sources, addition of new data sources where new experimental data has become available, and addition of a new informative data type, Quantitative Mass Spectrometry. The update also contains purified Gold standards that improve the quality of inferred associations, and six added model species, including two prokaryotes. Visualization of the more comprehensive, higher quality networks is provided through a new network viewer, which is a huge improvement over the old java viewer.
We strive to make FunCoup a tool for discovery of novel functional associations. Therefore, we are avoiding the use of curated interaction data as evidence and focus on high-throughput machine-generated data. Interaction information can also be obtained using text-mining of biomedical literature. However, this may potentially add additional sources of error, e.g. in identifier mapping, distinguishment between positive and negative interactions, or species identification, and is therefore prone to spurious associations (20). By using 10 different evidence types, FunCoup is able to capture functional associations of a wide range and provide high coverage without the use of text mining.
FunCoup is now equipped with an intuitive and user-friendly web interface including a lightweight, interactive network viewer designed to handle large networks.
MATERIALS AND METHODS
Proteomes
The proteomes of the species present in the previous release of FunCoup have been updated using the latest available release of the Quest for Orthologs (QFO) (RELEASE 2016_04, http://www.ebi.ac.uk/reference_proteomes) database. We have also introduced six new species; four eukaryotic: Plasmodium falciparum, Schizosaccharomyces pombe, Bos taurus, Oryza sativa and two prokaryotic: Escherichia coli and Bacillus subtilis, gathered from QFO except for Oryza sativa, which was obtained from UniProt (release March 2017) (21). FunCoup extensively transfers information between orthologous genes across different species. To avoid duplicity, this is however not done for evidences that are similarly derived in all species. These include Phylogenetic Profiles, Domain Interactions, and Sub-cellular Co-localization. The newly added species were selected both due to their potential for transfer of orthology information to the organisms already available in FunCoup, and the amount and quality of publicly accessible data, including orthology information in the latest release of InParanoid (version 8) orthology database (19).
Data sets
Data for almost all evidence types were updated with new available datasets. For PIN, the latest version of iRefIndex (version 14) (22) was used. For PEX the latest version of the Human Protein Atlas (HPA) v.15 (23) was used. The latest version of the Cellular Component ontology from the Gene Ontology (GO) (downloaded in June 2016) (24) was used to update the SCL evidence type, and InParanoid v.8 served as new source for phylogenetic profiles (PHP). For MEX and TFB, the data used in FunCoup 3.0 was supplemented with new available datasets. MEX now includes 64 new data sets from the Gene Expression Omnibus (downloaded in June 2016) (3). In case of GIN, we added the recent follow up study by Costanzo et al. (25), providing a larger coverage of the genetic interaction landscape of S. cerevisiae, and for E. coli we included the comprehensive study by Babu et al. (26) (Supplementary Table S1).
Quantitative mass spectrometry
In addition to the existing evidence types we introduce quantitative mass spectrometry data (QMS) as new source of evidence in this release of FunCoup. QMS was not included in previous releases due to poor coverage of open access data, but this has changed in recent years. QMS data sets for Homo sapiens, Mus musculus, Arabidopsis thaliana and Danio rerio were obtained via PaxDB (v. 4.0) (27). PaxDB is a database hosting a collection of standardized mass spectrometry datasets across different species and conditions/tissues. In a preprocessing step the 25% most abundant proteins per condition were extracted and labeled accordingly. These profiles were further evaluated using an adapted jaccard index (15). Here two proteins that are highly abundant across the same tissues achieve a high score.
Gold standards
In FunCoup, the gold standards are used to assign a log likelihood score to a bin representing a window of raw evidence score values, e.g. correlations. All the interactions that fall into that bin inherit the gold standard-derived score (13). The new FunCoup networks were inferred using five different gold standards, derived from KEGG metabolic and signaling pathways (see Supplementary Table S2), protein-protein interactions (PPIs), shared protein complexes, and shared operons (see Table 1). The quality of the gold standards is one of the key elements for inferring accurate networks. Therefore we updated the signaling and metabolic gold standards using KEGG v. 79 (28), increasing the number of pathways by 48% for signaling and 34% for metabolic pathways. A novelty in release 4 is that we extracted complex data from iRefIndex v14 and added them to the previously used curated complex data (15). This increased the complex gold standards by a factor 12 on average. iRefIndex v14 was also used for the PPI gold standard, filtering as before for interactions that are also present in the other gold standards or are reported in at least two experiments. Finally we introduced a new type of gold standard for prokaryotic organisms, shared operon. The underlying assumption is that genes organized in an operon participate in the same or similar functions (29,30). We obtained the data sets from OperonDB v.3 (31).
Table 1. Amount of links used for the positive gold standards, in total for all species: shared protein–protein interaction(PPI), KEGG signaling pathway (Signaling), KEGG metabolic pathway (Metabolic), shared protein complex (Complex), and organization in same operon (Operon).
| Gold standard | FunCoup 4 |
|---|---|
| PPI | 115 799 |
| Signaling | 4 805 854 |
| Metabolic | 2 248 802 |
| Complex | 1 854 271 |
| Operon | 5895 |
RESULTS
Networks
The updated database contains comprehensive networks for H. sapiens and 16 model organisms with 49 122 943 links between 200 100 genes in total (see Table 2; Supplementary Figure S1). Most species have a relatively high gene coverage between 70 and 90%, with a few exceptions. For C. intestinalis the coverage is 37%, due to that most of the links were inferred via orthology transfer. The other two species with low coverage are P. falciparum (42%) where coverage is low probably due to that most of the studies in this model organism are focusing on host-parasite interacting genes, and O. sativa (28%) where the reason may be attributed to a relatively recent whole genome duplication.
Table 2. Comparison of number of links and genome sizes between Funcoup 3 and Funcoup 4.
| Species | Genes (% genome coverage) | Links | ||
|---|---|---|---|---|
| FunCoup 3 | FunCoup 4 | FunCoup 3 | FunCoup 4 | |
| Arabidopsis thaliana | 16375 (60) | 19461 (71) | 5106648 | 5597050 |
| Caenorhabditis elegans | 12389 (61) | 13942 (69) | 3206664 | 3618485 |
| Canis familiaris | 17239 (89) | 17742 (89) | 3537089 | 3853720 |
| Ciona intestinalis | 5642 (40) | 6098 (37) | 1137425 | 1373106 |
| Drosophila melanogaster | 11398 (83) | 9768 (73) | 1987503 | 2174621 |
| Danio rerio | 15003 (57) | 16612 (73) | 4168563 | 3938535 |
| Gallus gallus | 12317 (74) | 12289 (79) | 2037840 | 1608939 |
| Homo sapiens | 18113 (84) | 18355 (82) | 4477041 | 6403719 |
| Mus musculus | 19226 (83) | 17708 (79) | 5314496 | 6157297 |
| Rattus norvegicus | 18562 (81) | 18322 (82) | 5460769 | 5560189 |
| Saccharomyces cerevisiae S288c | 5766 (86) | 6234 (90) | 1353169 | 806515 |
| Total | 152030 (72) | 156531 (74) | 3435200 | 3735652 |
| New in FunCoup 4 | ||||
| Bacillus subtilis strain 168 | - | 3856 (92) | - | 60553 |
| Bos taurus | - | 17906 (90) | - | 4551013 |
| Escherichia coli K-12 | - | 3624 (88) | - | 111500 |
| Oryza sativa | - | 12184 (28) | - | 2996703 |
| Plasmodium falciparum | - | 2273 (43) | - | 133158 |
| Schizosaccharomyces pombe | - | 3726 (73) | - | 277840 |
| Total | 152030 (72) | 200100 (68) | 34603907 | 49122943 |
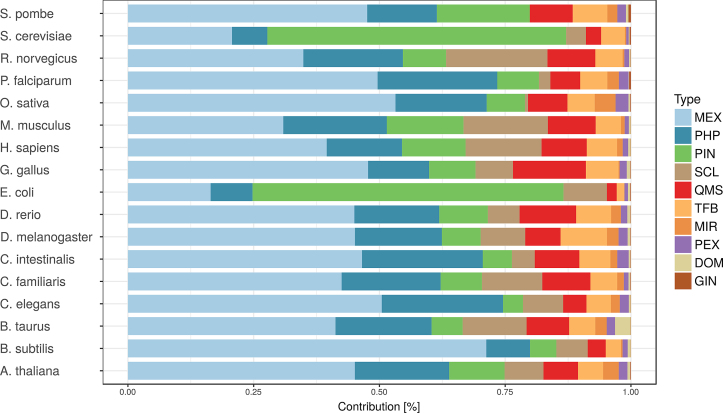
On average we gained 10% more functional associations than in the previous release, for the species present in both releases. In particular the H. sapiens network increased by 43% (see Table 2). This increase is primarily attributed to addition of new data covering a greater range of tissues and experimental conditions as well as bigger parts of the genome. A direct comparison of the data amount used in FunCoup 3 and FunCoup 4 is shown in Table 3. The largest increase is for MEX followed by GIN and SCL. Using InParanoid v8 for inferring PHP almost tripled the amount of species used for inferring PHP profiles, from 93 to 273. The three evidence types contributing the most to FunCoup's networks are MEX, PHP and PIN, while PEX, DOM and GIN are contributing the least (see Figure 1). Their modest contribution is not related to low quality but rather to the low amount of publicly available data (Supplementary Table S1).
Table 3. Comparisons of number of datapoints used for Funcoup 3 and FunCoup 4 for each evidence type.
| Evidence type | ||
|---|---|---|
| FunCoup 3 | FunCoup 4 | |
| PIN | 53886 | 70878 |
| MEX | 920690 | 2807555 |
| DOM | 144826 | 223822 |
| GIN | 288287 | 904740 |
| MIR | 62304 | 62304 |
| PEX | 12238 | 14578 |
| PHP | 188068 | 266236 |
| SCL | 151439 | 307578 |
| TFB | 70975 | 77703 |
| QMS | - | 99239 |
| Total | 1892713 | 4834633 |
Protein interaction (PIN), mRNA co-expression (MEX), domain-interaction (DOM), protein co-expression (PEX), genetic interaction profile similarity (GIN), co-miRNA regulation by shared miRNA targeting (MIR), protein co-expression (PEX), phylogenetic profile similarity (PHP), sub-cellular co-localization (SCL), shared transcription factor binding (TFB) and quantitative mass spectrometry(QMS).
Figure 1.
Evidence contribution per species. Evidence data types are: MEX: mRNA co-expression; PHP: phylogenetic profile similarity; PIN: protein interaction networks; SCL: sub-cellular co-localization; MIR: comiRNA regulation by shared miRNA targeting; DOM: domain interactions; PEX: protein co-expression; TFB: shared transcription factor binding; GIN: genetic interaction profile similarity and QMS: quantitative mass spectrometry data. The total contribution (LLRs) is normalized such that for each species it sums up to 1.
Additional factors responsible for the improved networks are the bigger gold standard sets (Table 1) and the introduction of new species. Larger gold standards allow the LRR scores to be better tuned and more accurately assigned, producing more reliable networks. Compared to the previous release of FunCoup, 73% more gold standard links were used on average for the species present in both releases. This increase is primarily driven by the purification of the links in the complex gold standard class which yielded a 6-fold increase.
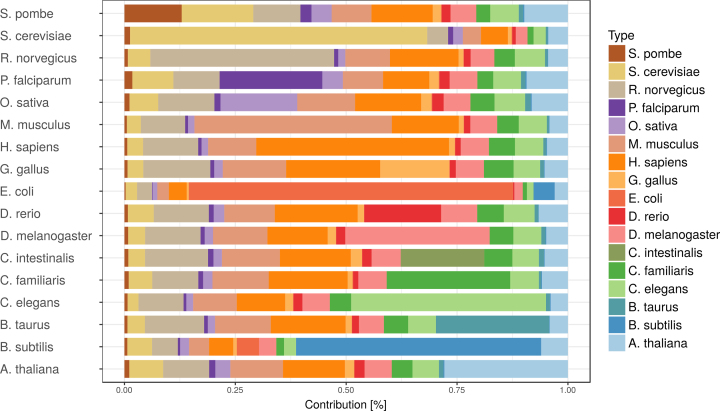
Including more species gives more opportunities for orthology based evidence transfer, which increases coverage of the networks. The level of orthology transfer between species is shown in Figure 2. For most species, the majority of the network support comes from other other species, even though the data from the species itself is the largest single contributor. Some exceptions to this rule exist. For S. cerevisiae, E. coli and B. subtilis, most of the support comes from the species itself. In the case of S. cerevisiae this can be explained by the large amount of experimental PPI S. cerevisiae data, while for the two prokaryotes the explanation is that they belong to a different phylogenetic domain than the other species. For S. pombe, G. gallus, and D. rerio, the species itself is not even the largest single contributor. These species come with relatively less own data, yet are well placed for orthology transfer.
Figure 2.
Evidence source species contributions for all evidences. The total contribution (LLRs) is normalized such that for each species it sums up to 1.
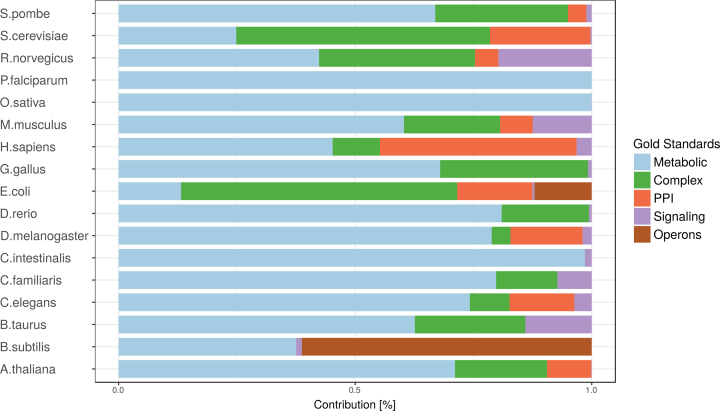
Each gold standard gives rise to a network; these are merged into the summary network by taking the maximum link support in any of the gold standard networks. The frequency that each gold standard network has the highest link support is shown in Figure 3. The distribution is dominated by the KEGG metabolic pathways for all species except for S. cerevisiae and E. coli, where protein complexes play a more prominent role, and B. subtilis which is dominated by links from the shared operon class.
Figure 3.
Distributions of gold standard contributions, showing the fraction of links where a given gold standard has the highest LLR score.
New network viewer
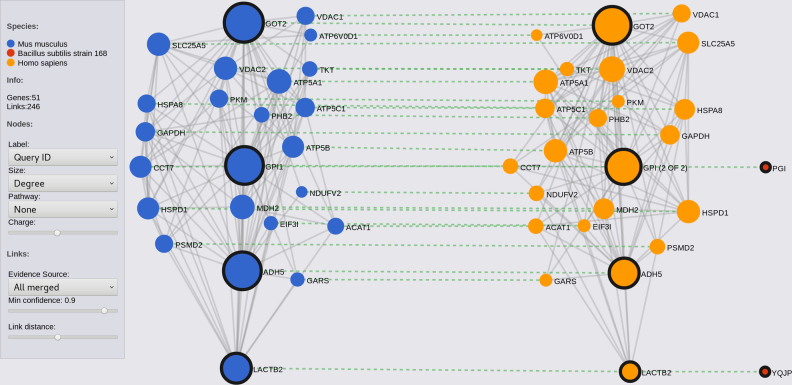
We have implemented a new dynamic network viewer for FunCoup 4, see Figure 4. The new viewer is based on the javascript library D3 v4 (32) replacing the previously available java applet (33) and the static picture of the network. In the new implementation, the nodes (colored circles) represent genes while edges (gray lines) depict their functional associations. The genes submitted in the network query are highlighted by a bold black border. For a comparative interactomics query, the black border highlights also the genes orthologous to the query, while the ortholog relation between the species are visualized by dashed green edges and node colors emphasize the different species.
Figure 4.
The new FunCoup network viewer, showing the comparative interactomics feature. The network of the query in H. sapiens (orange circles) is linked to orthologous networks in M. musculus (blue circles) and B. subtilis (red circles). As query we used the 4 human genes, LACTB2, ADH5, GOT2 and GPI, which have been identified as an evolutionarily conserved ancient metazoan protein complex. The query genes and their orthologs are highlighted with bold black border, and the orthology relation between genes is represented using green dashed lines whereas gray solid lines are functional associations within a species.
All nodes can be dragged and dropped to different positions. Hovering over a node or a link makes the elements of the network which are not connected to the highlighted object fade out into the background. Other intuitive applications, e.g. the mouse wheel or double click can be used for zooming and a click outside the network elements can be used to move the whole graph.
The menu box on the left is grouped in three section; Info, Nodes and Links. The sections Nodes and Links have various options to manipulate the network. The Info section displays additional information about a node or a link when the user hovers over it, otherwise the total number of genes and links within the subnetwork are shown. Within the Nodes section the user can vary node Label and node Size, highlight a Pathway or manipulate a node Charge. Label: the default node label refers to the query identifier, but can be set to UniProt, Ensembl or NCBI ID. Additionally the label can also display species name, node degree or, if set to none, hide all the labels. Size: Node sizes scale with node degrees to emphasize gene importance. This can be adapted to scale depending on the number of participated pathways or not scale at all if set to none. Pathway: This option is disabled per default. If a pathway is chosen the viewer highlights participating nodes in black. Charge: This slider alters the tension between the nodes.
The Link section contains three options, Evidence source, Min confidence and Link distance. Evidence source: Per default, a link represents the functional association inferred using all gold standards. Setting this option restricts the underlying data representing a link. One can restrict it to either one of the gold standards, species, evidence sources or known links. Min confidence: This option can be used to alter the minimum confidence score for the displayed links. Link distance: Here one can manipulate the distance of the links within the subnetwork.
Comparative interactomics example
To demonstrate the power of the latest FunCoup release we selected 4 human genes, LACTB2, ADH5, GOT2 and GPI, which have been identified by Wan et al. (34) as an evolutionarily conserved, ancient protein complex (see Figure 4). A standard FunCoup web query on the human network reveals a densely connected subnetwork including the 30 highest ranked neighbouring genes. To see if this complex also exists in other organisms we use the advanced feature of the web search called ‘comparative interactomics’ by unfolding the ‘advanced’ field underneath the query box, selecting the ‘interactomics’ tab and then the species of interest. For this example we use mouse and to test the definition of ancient we also try to find this complex within the prokaryotic organism B. subtilis. As a result we obtain a subnetwork for each species where ortholog genes are connected via green lines between the networks. To investigate this even further we use the tab ‘Interactions’. Here all the evidence sources, scores and ortholog transfers are visualized as boxes for each link. The last box indicates if the link has been experimentally verified.
Overall the query produces a dense network in the queried species H. sapiens and between the orthologous genes in M. musculus. Comparing the H. sapiens subnetwork to the prokaryote B. subtilis gives a completely different picture as we can only find two orthologous genes in B. subtilis which have no functional association. The lack of network conservation in prokaryotes suggests that this complex arose in the eukaryotic lineage.
DISCUSSION AND OUTLOOK
We have described the fourth release of the FunCoup database of functional association networks. After a complete overhaul of data sources and addition of new sources where appropriate, FunCoup 4 surpasses FunCoup 3 in terms of network sizes for most species, in particular for H. sapiens. A large part of the increase was due to orthology transferred data, which gained a lot from the addition of six new species, which has also enabled the database to open up to the prokaryotic domain.
The prokaryotic networks, despite having most of their interactions inferred from species-specific data sets, received substantial contributions from eukaryotic species, on par with e.g. S. cerevisiae, which indicates a successful integration in the database. Successful use of the new type of evidence, i.e. QMS, is witnessed by its relatively large contribution to the resulting networks, being the fifth (out of 10) biggest contributor for most of the species.
The challenge of mapping identifiers between different data sources included in the final database is something we have struggled with previously, and this release is no different. Databases can sometimes change their primary identifiers e.g. InParanoid switched to UniProt IDs from Ensembl IDs. This makes a fully automated update of data sources impossible. The absence of a universal identifier system often leads to many-to-many mappings or secondary mappings for some data sources, which may result in loss of data or ambiguous mappings for some genes/proteins. This remains a challenge we will continue to address in future releases.
To investigate the robustness of the FunCoup framework, we split each gold standard randomly into a test set with 20% of the links and a training set with the remaining 80%, and measured how much of the test set links could be recovered (Supplementary Figure S2). Overall, the recovery rate of the held out gold standard was far higher than the false positive rate, indicating that the gold standards have good coverage. The recovery rate, which reached 0.7 for S. cerevisiae, varies considerably between species however, indicating which gold standards should be prioritized for improvement in the future.
FunCoup contains some of the most comprehensive functional association networks that are available. With 10 evidence types and five gold standards, it is able to capture a broader range of interactions and functional associations than most other available networks. This diversity of data produces high coverage, yet FunCoup refrains from using some evidence types, such as text-mining, which often has a high error rate, and curated data. The reason for the latter is that we do not want to replicate other secondary databases, but want to focus FunCoup on novel interactions that can be used for discovery of new interaction partners and mechanisms.
Supplementary Material
ACKNOWLEDGEMENTS
The computations were performed on the Tegner resource provided by the Swedish National Infrastructure for Computing (SNIC) at PDC. SNIC support at PDC-HPC is acknowledged for assistance concerning technical aspects of making the code to run on the PDC-HPC resources. We also acknowledge the SciLifeLab IT department for hosting and helping to maintain the web server.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council grant [2015-05342]. Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D.. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004; 32:D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N. et al. . The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014; 42:D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chatr-aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A. et al. . The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017; 45:D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khanin R., Wit E.. How scale-free are biological networks. J. Comput. Biol. 2006; 13:810–818. [DOI] [PubMed] [Google Scholar]
- 7. Wang X., Gulbahce N., Yu H.. Network-based methods for human disease gene prediction. Brief. Funct. Genomics. 2011; 10:280–293. [DOI] [PubMed] [Google Scholar]
- 8. Vazquez A., Flammini A., Maritan A., Vespignani A.. Global protein function prediction from protein-protein interaction networks. Nat. Biotech. 2003; 21:697–700. [DOI] [PubMed] [Google Scholar]
- 9. Beiki H., Nejati-Javaremi A., Pakdel A., Masoudi-Nejad A., Hu Z.-L., Reecy J.M.. Large-scale gene co-expression network as a source of functional annotation for cattle genes. BMC Genomics. 2016; 17:846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Studham M.E., Tjärnberg A., Nordling T.E.M., Nelander S., Sonnhammer E.L.L.. Functional association networks as priors for gene regulatory network inference. Bioinformatics. 2014; 30:i130–i138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogris C., Guala D., Helleday T., Sonnhammer E.L.L.. A novel method for crosstalk analysis of biological networks: improving accuracy of pathway annotation. Nucleic Acids Res. 2017; 45:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guala D., Sjölund E., Sonnhammer E.L.L.. MaxLink: network-based prioritization of genes tightly linked to a disease seed set. Bioinformatics. 2014; 30:2689–2690. [DOI] [PubMed] [Google Scholar]
- 13. Alexeyenko A., Sonnhammer E.L.. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009; 19:1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alexeyenko A., Schmitt T., Tjärnberg A., Guala D., Frings O., Sonnhammer E.L.L.. Comparative interactomics with Funcoup 2.0. Nucleic Acids Res. 2012; 40:D821–D828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmitt T., Ogris C., Sonnhammer E.L.L.. FunCoup 3.0: database of genome-wide functional coupling networks. Nucleic Acids Res. 2014; 42:D380–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. et al. . The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zuberi K., Franz M., Rodriguez H., Montojo J., Lopes C.T., Bader G.D., Morris Q.. GeneMANIA Prediction Server 2013 Update. Nucleic Acids Res. 2013; 41:W115–W122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C. et al. . Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015; 47:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sonnhammer E.L., Östlund G.. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015; 43:D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harmston N., Filsell W., Stumpf M.P.. What the papers say: Text mining for genomics and systems biology. Hum. Genomics. 2010; 5:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Razick S., Magklaras G., Donaldson I.M.. iRefIndex: A consolidated protein interaction database with provenance. BMC Bioinformatics. 2008; 9:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. et al. . Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 24. Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015; 43:D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D. et al. . A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016; 353:aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Babu M., Arnold R., Bundalovic-Torma C., Gagarinova A., Wong K.S., Kumar A., Stewart G., Samanfar B., Aoki H., Wagih O. et al. . Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli. PLOS Genet. 2014; 10:e1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang M., Weiss M., Simonovic M., Haertinger G., Schrimpf S.P., Hengartner M.O., von Mering C.. PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell. Proteomics. 2012; 11:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M.. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016; 44:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dandekar T., Snel B., Huynen M., Bork P.. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998; 23:324–328. [DOI] [PubMed] [Google Scholar]
- 30. Snel B., Lehmann G., Bork P., Huynen M.A.. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000; 28:3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pertea M., Ayanbule K., Smedinghoff M., Salzberg S.L.. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009; 37:D479–D482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bostock M., Ogievetsky V., Heer J.. D3 - Data-Driven Documents. IEEE Trans. Vis. Comput. Graph. 2011; 17:2301–2309. [DOI] [PubMed] [Google Scholar]
- 33. Klammer M., Roopra S., Sonnhammer E.L.L.. jSquid: a Java applet for graphical on-line network exploration. Bioinformatics. 2008; 24:1467–1468. [DOI] [PubMed] [Google Scholar]
- 34. Wan C., Borgeson B., Phanse S., Tu F., Drew K., Clark G., Xiong X., Kagan O., Kwan J., Bezginov A. et al. . Panorama of ancient metazoan macromolecular complexes. Nature. 2015; 525:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.