Abstract
Altered expression of microRNAs (miRNAs or miRs) contributes to lung carcinogenesis. The present study performed an in silico analysis of differentially expressed miRNAs in different peripheral blood samples from patients with various diseases vs. controls using the Gene Expression Omnibus (GEO) database data, and assessed miR-105-1 expression in 32 normal lung and 142 non-small cell lung cancer (NSCLC) tissue samples using reverse transcription-quantitative polymerase chain reaction. Survival data were calculated using Kaplan-Meier curves and a log-rank test. The stepwise forward Cox regression model was performed for univariate and multivariate analyses of independent predictor of overall survival (OS) of patients. The data on in silico and tissue microarray analyses of miRNA expression revealed reduced miR-105-1 expression in different types of human cancer, particularly in NSCLC. The level of miR-105-1 expression was confirmed to be downregulated in NSCLC tissues compared with that in normal lung tissues. Reduced miR-105-1 expression was associated with larger tumor size as well as poor OS and disease-free survival (DFS) of patients. Multivariate survival analysis demonstrated that reduced miR-105-1 expression and tumor size were independent predictors for OS of NSCLC patients. In conclusion, reduced miR-105-1 expression in NSCLC tissues is associated with poor OS and DFS of NSCLC patients.
Keywords: microRNA, microRNA-105-1, non-small cell lung cancer, biomarker, survival
Introduction
Lung cancer is one of the most frequently diagnosed types of human malignancy and the leading cause of cancer mortality globally, particularly in developed countries (1,2). In China, lung cancer morbidity and mortality have increased in recent decades (1). Pathologically, lung cancer is classified as non-small cell lung cancer (NSCLC) and SCLC, and NSCLC accounts for 75–80% of all lung cancer cases (3). Surgery remains the most curative treatment option for patients with NSCLC and prolongs survival rate of such patients; however, a great number of patients are diagnosed at the late stages of the disease, leading to surgery not being an option (4). Despite advances in surgery, chemotherapy, radiotherapy and targeting therapy, the prognosis of patients with advanced NSCLC remains poor (5) which leads to substantial financial and psychological burdens for patients and their families associated with NSCLC diagnosis and therapy (6–9). In the early stages, NSCLC rarely exhibits noticeable symptoms (8,9); thus, the development and identification of biomarkers for early detection and prediction of prognosis may aid medical oncologists to detect the disease early or to provide more aggressive treatment to effectively control the disease.
The dysfunctional regulation of microRNAs (miRNAs or miRs) has been associated with human disease, including various types of cancer (10,11). miRNAs are a class of naturally-occurring small non-coding RNAs of 18–22 nucleotides in length, which are able to post-transcriptionally inhibit the expression of their targeting messenger RNAs (mRNAs). A single miRNA may regulate hundreds of targeting mRNAs by specifically binding to the 3′-untranslated region of mRNA, while one type of mRNA may also be regulated by different miRNAs (12). miRNAs are usually localized in the exosome of cells (13). Cancer-secreted exosomes/miRNAs can be detected in tissue samples or other samples such as blood (13–15). Thus, miRNAs may be ideal markers to foster predictive or early diagnostic tumor markers or for proficient therapeutic decisions.
miR-105 has been reported to be altered in human cancer and lost miR-105 expression was associated with poor patient survival (16). However, little is known about the association between miR-105-1 expression and the survival of patients with NSCLC. The present study performed in silico analysis of differentially expressed miRNAs in different peripheral blood samples from patients with various diseases vs. controls using Gene Expression Omnibus (GEO) database data, and assessed miR-105-1 expression in 32 normal lung and 142 non-small cell lung cancer (NSCLC) tissue samples using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The aim of the present study was to evaluate and identify the utility of miR-105-1 as a prognostic marker for patients with NSCLC.
Materials and methods
Acquisition of GEO database data
The present study downloaded the raw data on GSE61741 and GSE24709 from the GEO database (17,18). GEO is the repository database to store all high-throughput gene expression data of hybridization arrays, chips or microarrays. The GSE61741 dataset includes 1,049 miRNA expression data in normal controls, chronic obstructive pulmonary disease (COPD), lung cancer and other types of cancer, while the GSE24709 dataset includes 47 miRNA expression data from 19 normal control and 28 lung cancer patients. The present study utilized these two sets of differentially expressed miRNA data to identify differentially expressed miRNAs for NSCLC. Hierarchical clustering was performed using the multiple experiment viewer (MeV) 4.7.1 software programs (http://www.tm4.org/).
Patients and tissue samples
The present study collected tumor and matched non-tumorous tissue specimens from patients with NSCLC who underwent surgery in Shanghai Tenth People's Hospital, Tongji University School of Medicine (Shanghai, China) between January 2008 and December 2014. Two experienced pathologists confirmed NSCLC diagnosis independently according to the World Health Organization criteria (19) and tumor staging according to the Seventh Edition of the American Joint Committee on Cancer tumor-node-metastasis (TNM) staging system for NSCLC (20). Specimens included 32 paired cancer and adjacent non-cancerous tissues, and 110 cases of NSCLC tumor tissues. The present study was approved by the Ethics Committee of Shanghai Tenth People's Hospital, Tongji University School of Medicine (approval no. SHSY-IEC-PAP-15-18). Patients and/or their legal surrogates provided written informed consent to the surgical procedures and participation in the present study by donating the tissue specimens.
The demographic and clinicopathological characteristics of the patients were documented, including the clinical information, sex, age, smoking history, tumor characteristics, lymph-node metastasis, tumor differentiation, histological subtype, TNM stage, invasion of lung membrane, vascular invasion diameter, overall survival (OS) and disease-free survival (DFS). The last follow-up was conducted on July 30, 2015 through direct correspondence or phone interview. Mortality or tumor relapse was verified by patients or their relatives, or from medical or social security records. OS was computed in months from the date of diagnosis to the time of mortality, regardless of the cause. DFS was defined as the period from the initial date of diagnosis to the time of tumor progression by computed tomography scan, or to the time of mortality due to the disease.
RNA isolation and RT-qPCR
Total RNA from NSCLC and normal tissues was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the protocol of the manufacturer. RNA concentration was measured using NanoDrop ND-1000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and the quality was assessed using electrophoresis in 1.5% denaturing agarose gels and viewed on a Kodak Gel Logic 2200 imaging system (Kodak, Rochester, NY, USA). TaqMan probe-based qPCR was carried out using a commercial kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the protocol of the manufacturer (21). RT reactions were performed using AMV Reverse Transcriptase (Takara Biotechnology Co., Ltd., Dalian, China) and qPCR was performed using a standard TaqMan PCR kit protocol on the Applied Biosystems 7900HT Sequence Detection system (Thermo Fisher Scientific, Inc.). A specific primer (sequence, 5′-AGGACUCAAAUGCUCAG-3′) and TaqMan probe (sequence, 5′-UCAAAUGCUCAGACUCCUGU-3′) were used to quantify the expression of hsa-miR-105-1 (PN: 000441). Thermocycling conditions were as follows: Initial denaturation at 94°C for 10 min, followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, with a final extension at 72°C for 10 min. Each reaction was independently tested in duplicate a minimum of three times. U6 was used as the internal control and miR-105-1 levels were quantified using the 2−ΔΔCq method (22). miR-105-1 level was summarized and recorded as high vs. low levels of expression based on the median value.
Statistical analysis
Levels of miR-105-1 were summarized and recorded as the mean ± standard deviation. The independent t-test was used to calculate the difference between two groups of data. The χ2 test was used to evaluate the difference between the groups. Kaplan-Meier curves and log-rank tests were used to analyze the OS or DFS of patients with NSCLC. Multivariate Cox proportional hazards regression models were performed to explore the character of multiple characteristics in the prognosis of patients with NSCLC. All statistical analyses were performed using the SPSS 20.0 software program (IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Reduced miR-105-1 expression using miRNA microarray profiling data
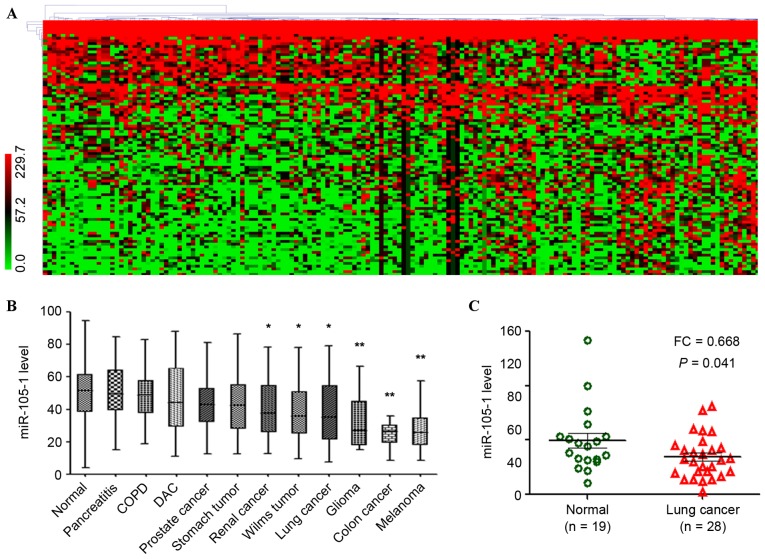
The present study first performed an in silico analysis using GEO database data. GSE61741 included peripheral blood profiles of patients with various diseases and controls. There were 93 differentially expressed miRNAs between normal controls and lung cancer, with the cut-off point of ≥2-fold changes for upregulated miRNAs and ≤0.5-fold changes for downregulated miRNAs (Fig. 1A). This included certain upregulated miRNAs such as miR-130b-3p, miR-135b and miR-92 (23,24), and certain downregulated miRNAs such as miR-144-3p, miR-30 and miR-486 (25–27), which were reported previously in lung cancer.
Figure 1.
Reduced expression of miR-105-1 in lung cancer using the GEO datasets. (A) Data on GEO GSE61741 dataset, including 1,049 miR expression data in normal controls, COPD, lung cancer and other types of cancer, were clustered using MeV 4.7.1 software. The color ratio bar (range, 0–299.7) indicates intensity of gene upregulation (red), downregulation (green) and no change (black). (B) Relative expression levels of miR-105-1 in different types of cancer vs. normal controls (GSE61741). (C) Relative expression levels of miR-105-1 in non-small cell lung cancer, adjacent normal tissues and non-cancerous lung diseases from GEO GSE24709 datasets. *P<0.05 and **P<0.01 vs. normal. GEO, Gene Expression Omnibus; COPD, chronic obstructive pulmonary disease; DAC, ductal adenocarcinoma; FC, fold change; miR, microRNA.
The usefulness of other newfound miRNAs as prognostic markers for NSCLC patients was then analyzed, including miR-105-1 [fold change (FC)=0.43, P=0.044; Fig. 1A]. The different expression levels of the dysregulated miRNAs in a control, lung cancer, COPD and pancreatitis with different types of tumor were investigated, and the results revealed that miR-105-1 was significantly downregulated in the majority of tumor types, particularly in lung cancer (P<0.05; Fig. 1B).
The present study then validated the aforementioned findings in another set of NSCLC vs. non-cancerous tissue samples. GSE24709 data also demonstrated that the miR-105-1 level was significantly reduced in lung cancer compared with that in normal controls (FC=0.668, P=0.041; Fig. 1C).
Validation of the reduced miR-105-1 level in NSCLC tissue samples
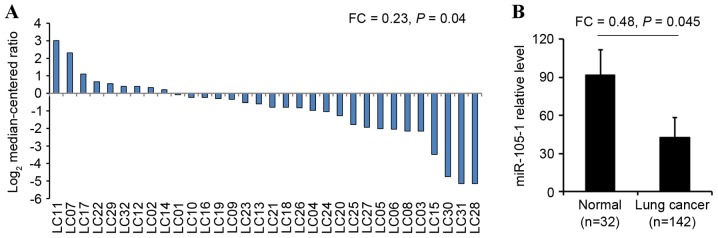
The present study next performed RT-qPCR to quantify the miR-105-1 level in 142 (90 I–II and 52 III–IV grades) NSCLC specimens and 32 non-cancerous tissues from patients with NSCLC. miR-105-1 levels were observed to be significantly lower in NSCLC tissues (21.36±3.89) compared with those in paired adjacent non-cancerous tissues (92.89±10.75). The difference between tumor and normal tissues was statistically significant (FC=0.23, P=0.04; Fig. 2A). In addition, the level of miR-105-1 expression was also lower in all NSCLC tumor biopsies (44.59±5.93) compared with the expression in normal lung tissues (92.89±10.75) and the differences were statistically significant (FC=0.48, P=0.045; Fig. 2B).
Figure 2.
Validation of the reduced miR105-1 levels in NSCLC tissue vs. normal lungs. (A) RT-qPCR. miR-105-1 levels in NSCLC (n=142) vs. adjacent non-tumor tissue (n=32). Tissues below the line have significantly lower miR105-1 expression levels compared with the normal tissues. (B) RT-qPCR. miR-105-1 levels in NSCLC tissue samples (n=32) and paired adjacent non-tumor tissues (n=32). NSCLC, non-small cell lung cancer; LC, lung cancer; FC, fold change; miR, microRNA.
Association of miR-105-1 expression with clinicopathological data from patients with NSCLC
The present study next associated miR-105-1 expression with clinicopathological data from patients with NSCLC and observed that miR-105-1 expression levels were inversely associated with tumor size (P=0.013; Table I), but there was no association of miR-105-1 expression with other clinicopathological data from NSCLC patients, including sex, age, smoking history, tumor differentiation, histology, vascular invasion, lymph-node metastasis, TNM stage or invasion of the lung membrane (P>0.05; Table I).
Table I.
Univariate analysis of overall survival based on patients with non-small cell lung cancer stratified by clinical characteristics.
| Overall survival | ||||||
|---|---|---|---|---|---|---|
| Variable | N | miR-105-1 expression, mean ± SD | P-value | Mean months | 95% CI (mean) | P-value (log-rank test) |
| Age, years | 0.39 | 0.74 | ||||
| ≥60 | 90 | 39.76±3.83 | 31.46 | 29.38–32.14 | ||
| <60 | 52 | 46.35±4.38 | 33.52 | 30.51–35.69 | ||
| Sex | 0.89 | 0.90 | ||||
| Male | 86 | 45.39±3.47 | 31.97 | 28.36–32.34 | ||
| Female | 56 | 44.38±6.54 | 32.42 | 29.38–33.13 | ||
| Tobacco smoking | 0.74 | 0.32 | ||||
| Never | 35 | 41.35±2.85 | 32.33 | 28.74–29.16 | ||
| Ever | 41 | 40.29±6.58 | 30.26 | 27.43–35.06 | ||
| Unknown | 66 | 42.36±6.89 | 31.29 | 27.04–33.65 | ||
| Lymph node metastasis | 0.07 | 0.03 | ||||
| Negative | 48 | 46.75±8.95 | 33.69 | 32.05–34.73 | ||
| Positive | 82 | 39.87±15.74 | 30.83 | 28.95–32.22 | ||
| Unknown | 12 | 40.39±10.88 | 30.26 | 28.34–33.16 | ||
| Tumor differentiation | 0.10 | 0.12 | ||||
| Poorly | 51 | 40.23±5.96 | 28.74 | 26.47–30.96 | ||
| Moderately | 85 | 41.05±6.25 | 30.51 | 28.48–32.52 | ||
| Well | 6 | 43.47±4.89 | 31.26 | 29.56–33.03 | ||
| Histology | 0.89 | 0.22 | ||||
| Adenocarcinoma | 90 | 42.68±9.87 | 28.99 | 27.06–31.13 | ||
| Squamous cell carcinoma | 52 | 43.15±6.63 | 32.04 | 29.37–34.59 | ||
| TNM stage | 0.06 | 0.011 | ||||
| I–II | 92 | 42.67±5.24 | 32.48 | 30.56–34.33 | ||
| III–IV | 50 | 40.38±6.69 | 27.59 | 24.69–29.38 | ||
| Invasion of lung membrane | 0.08 | 0.08 | ||||
| Negative | 28 | 40.56±2.04 | 31.69 | 28.47–32.64 | ||
| Positive | 102 | 41.36±6.67 | 29.08 | 27.95–31.02 | ||
| Unknown | 12 | 43.25±5.26 | 29.31 | 27.65–35.43 | ||
| Vascular invasion | 0.87 | 0.67 | ||||
| Negative | 130 | 41.26±5.03 | 30.59 | 28.74–32.69 | ||
| Positive | 8 | 42.65±3.38 | 31.26 | 29.38–33.15 | ||
| Unknown | 4 | 41.96±5.74 | 29.63 | 27.31–31.31 | ||
| Tumor size, cm | 0.01 | 0.00 | ||||
| ≥5 | 35 | 35.26±6.67 | 26.15 | 24.18–29.34 | ||
| <5 | 107 | 42.37±5.89 | 33.06 | 31.25–34.31 | ||
SD, standard deviation; miR, microRNA; TNM, tumor node metastasis; CI, confidence interval.
Association of miR-105-1 levels and clinicopathological data with survival of NSCLC patients
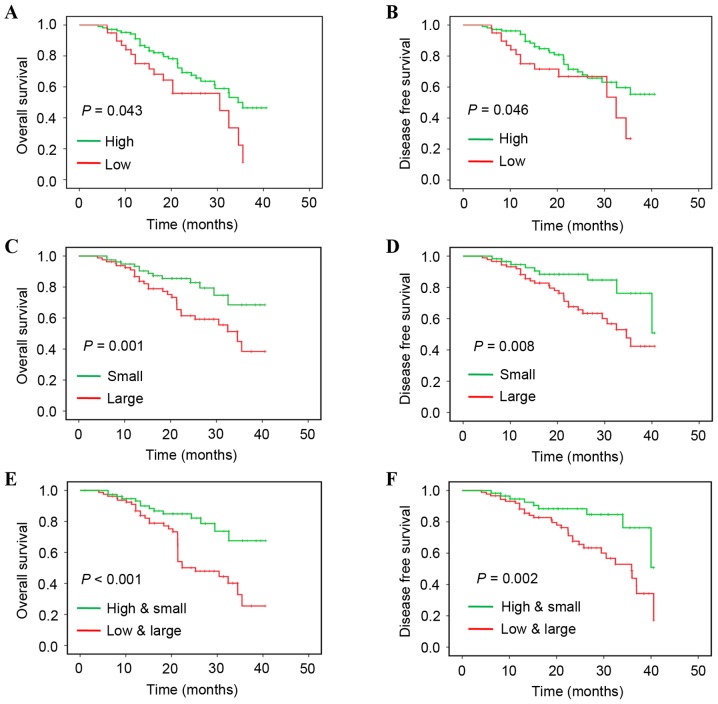
The present study additionally associated miR-105-1 levels and clinicopathological data with the survival of patients with NSCLC. miR-105-1 levels were recorded as high vs. low using the median value as the cut-off point. Kaplan-Meier survival curves were plotted to estimate the prognostic value of the clinicopathological characteristics for OS. The data of the present study revealed that a reduced miR-105-1 level was associated with a shorter OS (P=0.043; Fig. 3A) and DFS (P=0.046; Fig. 3B), while lymph-node metastasis (P=0.031), TNM stage (P=0.011) and tumor size (≥5 cm; P=0.001) were all associated with shorter OS of patients with NSCLC (Fig. 3 and Table II).
Figure 3.
Kaplan-Meier curve analysis of overall survival and disease-free survival. (A) Overall survival stratified by miR-105-1 level. (B) Disease-free survival stratified by miR-105-1 level. (C) Overall survival stratified by tumor size. (D) Disease-free survival stratified by tumor size. (E) Overall survival stratified by miR-105-1 level and tumor size. (F) Disease-free survival stratified by miR-105-1 level and tumor size.
Table II.
Univariate and multivariate analyses of overall survival of patients with non-small cell lung cancer.
| miR-105-1 multivariate analysis | ||||||
|---|---|---|---|---|---|---|
| Factor | HR | 95% CI (univariate) | P-value | HR | 95% CI (multivariate) | P-value |
| Age | 1.24 | 1.11–1.35 | 0.68 | 1.31 | 1.15–1.38 | 0.55 |
| Sex | 0.89 | 0.74–1.02 | 0.66 | 0.85 | 0.77–0.89 | 0.61 |
| Smoking history | 1.31 | 1.24–1.65 | 0.23 | 1.48 | 1.35–1.74 | 0.09 |
| Lymph-node metastasis | 1.89 | 1.56–2.21 | 0.04 | 1.95 | 1.66–2.32 | 0.04 |
| Tumor differentiation | 1.33 | 0.94–1.52 | 0.12 | 1.31 | 0.92–1.46 | 0.14 |
| Histology | 1.06 | 0.84–1.52 | 0.32 | 1.08 | 0.89–1.63 | 0.31 |
| TNM stage | 2.31 | 2.04–2.66 | 0.01 | 2.47 | 2.25–2.89 | 0.01 |
| Invasion of lung membrane | 1.62 | 1.44–1.81 | 0.09 | 1.68 | 1.49–1.84 | 0.07 |
| Vascular invasion | 0.92 | 0.74–1.02 | 0.77 | 0.93 | 0.75–1.08 | 0.75 |
| Tumor size | 2.26 | 2.08–2.69 | <0.01 | 3.01 | 2.65–3.33 | <0.01 |
| miR-105-1 expression | 0.66 | 0.55–0.71 | 0.03 | 0.64 | 0.51–0.70 | 0.02 |
HR, hazard ratio; CI, confidence interval; miR, microRNA; TNM, tumor node metastasis.
Univariate and multivariate analyses with Cox proportional hazards regression model revealed that lymph node metastasis [P=0.04; hazard ratio (HR)=1.89; 95% confidence interval (CI), 1.56, 2.21], advanced TNM stage [P=0.01; HR=2.31 (95% CI, 2.04, 2.66)], larger tumor size [P=0.001; HR=2.26 (95% CI, 2.08, 2.69)] and reduced miR-105-1 levels [P=0.034; HR=0.66 (95% CI, 0.55, 0.71)] were all predictors for poorer NSCLC prognosis (Table II). When combining the reduced miR-105-1 level with tumor size, multivariate analysis demonstrated that the aforementioned variables were independent predictors for poor OS (P<0.001; Fig. 3E) and DFS (P=0.002; Fig. 3F).
Discussion
Sensitive and specific tumor-markers for early NSCLC detection may be useful to decrease lung cancer morbidity and mortality (28,29). In the present study, we evaluated and identified miR-105-1 as a prognostic marker able to predict OS and DFS of patients with NSCLC. An in silico analysis of miR-105-1 was performed, together with other differentially expressed miRNAs in lung cancer vs. other conditions using GEO database data. The present study then validated the aforementioned findings in another set of NSCLC vs. non-cancerous tissue samples. The data of the present study revealed that a downregulated miR-105-1 level was associated with tumor size and poor OS and DFS of patients with NSCLC.
The findings of the present study are consistent with those from previous studies on various types of tumor. For example, Guan et al (16) revealed that the miR-105 level was markedly decreased in gliomas compared with that in non-neoplastic brain tissues and that the reduced miR-105 expression was statistically associated with advanced tumor grades, such as grade IV glioma. The authors speculated that downregulation of miR-105 expression was associated with poor clinical outcome, and was involved in glioma tumorigenesis and progression; thus, the detection of miR-105 levels may be a novel biomarker in the prediction of glioma prognosis. Shen et al (30) reported that miR-105 expression was prominently low in hepatocellular carcinoma (HCC) cell lines and tissues compared with that in normal human hepatocyte and adjacent non-cancerous tissues, respectively. The authors also observed that miR-105 functioned as a potential tumor suppressor to inhibit expression of the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway genes and may represent a potential therapeutic target for patients with HCC. Another previous study demonstrated that miR-105 inhibited prostate tumor growth via the inhibition of cyclin-dependent kinase 6 (31). However, not all published data supported this association or the tumor-suppressive activity of miR-105-1. For example, Honeywell et al (32) reported that the levels of miR-105-1 were upregulated in cancerous tissue analyzed by RT-qPCR. This inconsistent finding may be due to the tumor type or contamination of normal tissue in the tumor samples. However, another in vitro study revealed that cancer-secreted miR-105 destroyed vascular endothelial barriers and promoted cancer metastasis (14). Thus, additional studies are required to clarify the role of miR-105 in NSCLC.
To date, the precise role and potential patho-physiological mechanism responsible for or involving the antitumor activity of miR-105-1 require additional investigation, including NSCLC. The next step will be to predict and identify the targeting genes of miR-105-1 in NSCLC and, based on that data, the role of miR-105-1 in lung tissue may be speculated. Thus, targeting miR-105-1 may be used as a therapeutic strategy for NSCLC patients.
Although the present study revealed reduced miR-105-1 levels in NSCLC tissue, which was associated with poor OS and DFS of patients with NSCLC, there are several limitations. Primarily, the miR-105-1 levels in tissues were detected using RT-qPCR, which was not microdissected and included a mixture of normal and tumor tissues. Furthermore, the data of the present study are from an association study, and little is known about the molecular and patho-physiological mechanisms of miR-105-1 in NSCLC.
In conclusion, the data of the present revealed that a reduced level of miR-105-1 in NSCLC tissues was associated with a shorter OS and DFS of patients with NSCLC, and that miR-105-1 was a predictor for shorter OS of patients with NSCLC.
Acknowledgements
We appreciate the experimental support of Central Laboratory for Medical Research, Shanghai Tenth People's Hospital (Shanghai, China). The present study was supported by grants from the National Natural Science Foundation of China (Beijing, China; grant nos. 81472501, 81201535, 81301993, 81372175, 81472202 and 81302065) and Health and Family Planning Commission projects (Shanghai, China; grant nos. 201540228 and 201440398).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Hill A, Fisher P, Yeomanson D. Non-small cell lung cancer. BMJ. 2012;345:e6443. doi: 10.1136/bmj.e6443. [DOI] [PubMed] [Google Scholar]
- 4.Cykert S, Dilworth-Anderson P, Monroe MH, Walker P, McGuire FR, Corbie-Smith G, Edwards LJ, Bunton AJ. Factors associated with decisions to undergo surgery among patients with newly diagnosed early-stage lung cancer. JAMA. 2010;303:2368–2376. doi: 10.1001/jama.2010.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fervers B. Chemotherapy in elderly patients with resected stage II–IIIA lung cancer. BMJ. 2011;343:d4104. doi: 10.1136/bmj.d4104. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan R, Alatise OI, Anderson BO, Audisio R, Autier P, Aggarwal A, Balch C, Brennan MF, Dare A, D'Cruz A, et al. Global cancer surgery: Delivering safe, affordable, and timely cancer surgery. Lancet Oncol. 2015;16:1193–1224. doi: 10.1016/S1470-2045(15)00223-5. [DOI] [PubMed] [Google Scholar]
- 7.Global Burden of Disease Cancer Collaboration, Fitzmaurice C, corp-author. Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et al. The Global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Lung Screening Trial Research Team, corp-author. Church TR, Black WC, Aberle DR, Berg CD, Clingan KL, Duan F, Fagerstrom RM, Gareen IF, Gierada DS, et al. Results of initial low-dose computed tomographic screening for lung cancer. N Engl J Med. 2013;368:1980–1991. doi: 10.1056/NEJMoa1209120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasparini P, Cascione L, Landi L, Carasi S, Lovat F, Tibaldi C, Alì G, D'Incecco A, Minuti G, Chella A, et al. microRNA classifiers are powerful diagnostic/prognostic tools in ALK-, EGFR-, and KRAS-driven lung cancers. Proc Natl Acad Sci USA. 2015;112:14924–14929. doi: 10.1073/pnas.1520329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 13.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 16.Guan Y, Chen L, Bao Y, Li Z, Cui R, Li G, Wang Y. Identification of low miR-105 expression as a novel poor prognostic predictor for human glioma. Int J Clin Exp Med. 2015;8:10855–10864. [PMC free article] [PubMed] [Google Scholar]
- 17.NCBI Resource Coordinators: Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lantuejoul S, Rouquette I, Brambilla E, Travis WD. New WHO classification of lung adenocarcinoma and preneoplasia. Ann Pathol. 2016;36:5–14. doi: 10.1016/j.annpat.2015.11.010. (In French) [DOI] [PubMed] [Google Scholar]
- 20.Edge SB, Compton CC. The American Joint Committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Schaap-Oziemlak AM, Raymakers RA, Bergevoet SM, Gilissen C, Jansen BJ, Adema GJ, Kögler G, le Sage C, Agami R, van der Reijden BA, Jansen JH. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010;19:877–885. doi: 10.1089/scd.2009.0112. [DOI] [PubMed] [Google Scholar]
- 24.Zhao B, Zhu Y, Cui K, Gao J, Yu F, Chen L, Li S. Expression and significance of PTEN and miR-92 in hepatocellular carcinoma. Mol Med Rep. 2013;7:1413–1416. doi: 10.3892/mmr.2013.1406. [DOI] [PubMed] [Google Scholar]
- 25.Gao F, Wang T, Zhang Z, Wang R, Guo Y, Liu J. Regulation of activating protein-4-associated metastases of non-small cell lung cancer cells by miR-144. Tumour Biol. 2015 Aug 8; doi: 10.1007/s13277-015-3866-4. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 26.Qi F, He T, Jia L, Song N, Guo L, Ma X, Wang C, Xu M, Fu Y, Li L, Luo Y. The miR-30 family inhibits pulmonary vascular hyperpermeability in the premetastatic phase by direct targeting of Skp2. Clin Cancer Res. 2015;21:3071–3080. doi: 10.1158/1078-0432.CCR-14-2785. [DOI] [PubMed] [Google Scholar]
- 27.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 28.Liu D, Hu X, Zhou H, Shi G, Wu J. Identification of aberrantly expressed miRNAs in gastric cancer. Gastroenterol Res Pract. 2014;2014:473817. doi: 10.1155/2014/473817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zen K, Zhang CY. Circulating microRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 30.Shen G, Rong X, Zhao J, Yang X, Li H, Jiang H, Zhou Q, Ji T, Huang S, Zhang J, Jia H. MicroRNA-105 suppresses cell proliferation and inhibits PI3K/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. 2014;35:2748–2755. doi: 10.1093/carcin/bgu208. [DOI] [PubMed] [Google Scholar]
- 31.Han Y, Jia C, Cong X, Yu F, Cai H, Fang S, Cai L, Yang H, Sun Y, Li D, et al. Increased expression of TGFβR2 is associated with the clinical outcome of non-small cell lung cancer patients treated with chemotherapy. PLoS One. 2015;10:e0134682. doi: 10.1371/journal.pone.0134682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honeywell DR, Cabrita MA, Zhao H, Dimitroulakos J, Addison CL. miR-105 inhibits prostate tumour growth by suppressing CDK6 levels. PLoS One. 2013;8:e70515. doi: 10.1371/journal.pone.0070515. [DOI] [PMC free article] [PubMed] [Google Scholar]