Abstract
The peroxisome was the last organelle to be discovered and five decades later it is still the Cinderella of eukaryotic compartments. Peroxisomes have a crucial role in the detoxification of reactive oxygen species, the beta-oxidation of fatty acids, and the biosynthesis of etherphospholipids, and they are assumed to be present in virtually all aerobic eukaryotes. Apicomplexan parasites including the malaria and toxoplasmosis agents were described as the first group of mitochondriate protists devoid of peroxisomes. This study was initiated to reassess the distribution and evolution of peroxisomes in the superensemble Alveolata (apicomplexans, dinoflagellates, ciliates). We established transcriptome data from two chromerid algae (Chromera velia, Vitrella brassicaformis), and two dinoflagellates (Prorocentrum minimum, Perkinsus olseni) and identified the complete set of essential peroxins in all four reference species. Our comparative genome analysis provides unequivocal evidence for the presence of peroxisomes in Toxoplasma gondii and related genera. Our working hypothesis of a common peroxisomal origin of all alveolates is supported by phylogenetic analyses of essential markers such as the import receptor Pex5. Vitrella harbors the most comprehensive set of peroxisomal proteins including the catalase and the glyoxylate cycle and it is thus a promising model organism to investigate the functional role of this organelle in Apicomplexa.
Keywords: Vitrella brassicaformis, Toxoplasma gondii, glyoxylate cycle, diagnostic peroxins, catalase, phylogeny
Introduction
Peroxisomes, also designated as “microbodies,” are organelles of eukaryotic cells that are surrounded by a single membrane and required, for example, for the breakdown of long-chained fatty acids (Rhodin 1954; de Duve and Baudhuin 1966; Gabaldón 2010). Compartmentalization prevents oxidative damage due to concomitantly formed highly reactive hydrogen peroxide (H2O2) that is rapidly eliminated by oxidoreductases such as the catalase (Reddy and Mannaerts 1994). Peroxisomes with specific cellular functions or metabolic capacities are known as “Woronin bodies” in filamentous fungi (Markham and Collinge 1987; Pieuchot and Jedd 2012), “glycosomes” in kinetoplastids (Opperdoes and Borst 1977) as well as diplonemids (Morales et al. 2016), and “glyoxysomes” in land plants, fungi and some protists (Breidenbach and Beevers 1967; Tolbert and Essner 1981; Kunze and Hartig 2013).
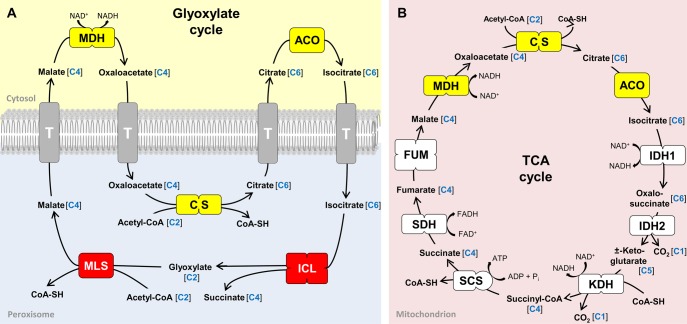
Glyoxysomes harbor the eponymous glyoxylate cycle (Kornberg and Krebs 1957) that uses the C2 unit acetyl-CoA as carbon source for the synthesis of succinate (C4 unit, fig. 1A;Schnarrenberger and Martin 2002). This anabolic bypass of the mitochondrial TCA cycle is required for the gluconeogenetic formation of carbohydrates and frequently linked to the catabolism of fatty acids (McCammon et al. 1990). Accordingly, germination of oilseed plants like castor beans essentially depends on a functional glyoxylate cycle (Breidenbach and Beevers 1967; Eastmond and Graham 2001; Yang et al. 2009), whereas humans are generally not able to convert lipids into sugar because they are lacking this pathway (de Figueiredo et al. 2009). The glyoxylate cycle is composed of five enzymes, the aconitase (ACO), isocitrate lyase (ICL), malate synthase (MLS), malate dehydrogenase (MDH), and citrate synthase (CS; fig. 1A). Their differential subcellular localization in both the peroxisome and the cytosol challenged the traditional view that all enzymes of a metabolic pathway are located in the same compartment (Kunze et al. 2006). In land plants, three of the enzymes (ICL, MLS, CS) are imported into the peroxisomal matrix, whereas the two other ones (MDH, ACO), which catalyze the oxidation of malate to oxaloacetate and the conversion of citrate to isocitrate, reside in the cytosol (fig. 1A). The unexpected localization of the MDH and ACO is associated with a rapid ping-pong-like exchange of the four carbohydrate intermediates through the peroxisomal membrane (Kunze and Hartig 2013). Homologous isoforms of three enzymes of the glyoxylate cycle (MDH, CS, ACO) are also essential for the mitochondrial TCA cycle (highlighted in yellow, fig. 1B;Schnarrenberger and Martin 2002) and additional MDH isoenzymes are required for the plastid energy metabolism (Scheibe 2004). In contrast, the ICL and MLS are specific for the glyoxylate cycle and were used in the past as marker enzymes for the identification of peroxisomes (highlighted in red, fig. 1A; Breidenbach and Beevers 1967; Tolbert and Essner 1981; Lazarow and Fujiki 1985; Kunze et al. 2006).
Fig. 1.
Peroxisomal glyoxylate cycle (A) and the mitochondrial TCA cycle (B) in Vitrella brassicaformis CCMP3155. A comparable localization of the pathways is found in Chromera velia, dinoflagellates, land plants, and some fungi (Kunze et al. 2006). Single boxes and multiple boxes symbolize the stoichiometry of the enzyme, for example, monomers, homodimers, heterodimers, and homotetramers, respectively. Enzymes highlighted in yellow are integrated in both pathways with two of them located in the cytosol concerning the glyoxylate cycle in plants and fungi. Enzymes highlighted in red are peroxisome-specific and are located in the peroxisomal matrix. The amount of carbon atoms in each reaction intermediate is highlighted in blue. The enzymes involved in translocation of reaction intermediates across the peroxisomal membrane are currently unknown. Possible transmembrane transporters are marked by a “T.”
Different other metabolic pathways including the beta-oxidation of fatty acids, the catabolism of purines, the biosynthesis of amino acids and of etherphospholipids as well as the cellular antioxidant system for the detoxification of reactive oxygen species (ROS) are located in peroxisomes (Reddy and Mannaerts 1994; Schrader and Fahimi 2008). The composition of metabolic pathways within these organelles can even vary in different cellular stages and tissue types, which is induced by the adaptation of peroxisomes to the specific needs of the cells (Gabaldón 2010; Hayashi et al. 2000; Smith and Aitchison 2009). Photorespiration serves as a carbon recovery system in land plants that results from the oxygenase reaction of the ribulose-1, 5-bisphosphate carboxylase/oxygenase and comprises distinct enzymatic reactions in three organelles, that is, chloroplasts, peroxisomes, and mitochondria (Maurino and Peterhansel 2010). The pathway, which is also known as the oxidative photosynthetic carbon cycle, requires a comprehensive set of transporters that mediate the metabolite exchange between the compartments. The “peroxins” represent a constant protein pool for the maintenance of peroxisomes. Peroxins are also responsible for peroxisome biogenesis, protein import, translocation and recycling as well as peroxisome proliferation (Smith and Aitchison 2013). Mutations in these proteins were shown to cause fatal neurological disorders in humans (Schlüter et al. 2006; Smith and Aitchison 2013).
The evolutionary origin of the tiny peroxisome, the last eukaryotic organelle that was discovered (Rhodin 1954; de Duve and Baudhuin 1966), has been controversially discussed for decades. Peroxisomes proliferate analogous to mitochondria and plastids by fission (Fagarasanu et al. 2007), but they do not contain DNA. Christian de Duve favored an endosymbiotic descent over the alternative explanation that peroxisomes originated as offshoots of the endoplasmatic reticulum (ER; de Duve 1982, 2007). The former idea was supported by Cavalier-Smith, who moreover proposed a simultaneous symbiotic origin of mitochondria, chloroplasts and microbodies (Cavalier-Smith 1987). However, this hypothesis as well as the recently proposed actinobacterial origin of peroxisomes, which probably reflects a phylogenetic artifact (Duhita et al. 2010; Gabaldón and Capella-Gutiérrez 2010), proved to be wrong. Erdmann et al. discovered in the baker yeast Saccharomyces cerevisiae that mutations of two peroxin genes (pex1, pex4) resulted in peroxisome-deficient strains (Erdmann et al. 1989) and specific mutagenesis approaches allowed to identify 18 different assembly mutants (Erdmann and Kunau 1992; van der Leij et al. 1992). The complementation with the wild type genes pex3 and pex19 resulted in the regeneration of functional peroxisomes from offshoots of the ER (Hoepfner et al. 2005). Evidence for an origin of peroxisomes as a genuine eukaryotic invention is provided by the peroxisomal protein import machinery (Schlüter et al. 2006; Gabaldón et al. 2006), which resembles a modification of the ancient ER-associated degradation (ERAD) system for ubiquitinated proteins (Bolte et al. 2011; Schliebs et al. 2010). Speijer proposed that peroxisomes emerged driven by the need for cellular detoxification in a mitochondrial eukaryote (Speijer 2011; Speijer 2015). Accordingly, early aerobic protists accumulated increasing amounts of ROS, which provided a strong selective force for subcellular compartmentalization and thus for the formation of the peroxisome. The model is supported by the observation that the beta-oxidation pathway in peroxisomes is derived from the mitochondrial one (Bolte et al. 2015) and the recent finding that the peroxisomal membrane partially derives from the mitochondrial membrane during peroxisomal biogenesis (Sugiura et al. 2017).
Protein transfer into peroxisomes is usually mediated by a specific import system that recognizes peroxisomal targeting signals (PTS; Rucktäschel et al. 2011). PTS1 with the conserved C-terminal tripeptide consensus motif (S/A/C)-(K/R/H)-(L/M) is the most frequently observed import signal. PTS1 is recognized by Pex5, a receptor that also binds peroxisomal proteins without well-defined targeting signals (van der Klei and Veenhuis 2006). Import of proteins with the N-terminal PTS2 [(R/K)-(L/V/I)-X5-(H/Q)-(L/A)] is mediated by Pex7 (Lanyon-Hogg et al. 2010).
The current study was focused on the distribution and evolution of peroxisomes in alveolates (Adl 2012), a monophyletic superensemble comprising the basal grouping ciliates, dinoflagellates, and apicomplexans. The three groups developed conspicuous lineage-specific cellular characteristics, such as the presence of two distinct nuclei in ciliates (Coyne et al. 1996), the liquid crystalline chromosomes of giant genomes in dinoflagellates (Wisecaver and Hackett 2011) and the adaptation to parasitism resulting in fast evolving apicomplexans (Swapna and Parkinson 2017). According to their parasitic life style most researchers thought for decades that peroxisomes are missing in the latter group (Schlüter et al. 2006; Gabaldón 2010). The ciliate Tetrahymena pyriformis, which served as a reference species for the introduction of the term “peroxisome” (de Duve and Baudhuin 1966), harbors an archetypal glyoxylate cycle including the ICL and MLS (fig. 1A; Hogg and Kornberg 1963). Transcripts of these key enzymes were also found in dinoflagellates (Butterfield et al. 2013), thus providing independent support for the ultrastructure-based prediction that peroxisomes are abundant in this class of alveolates (Bibby and Dodge 1973). The genomes of completely sequenced apicomplexan parasites such as Cryptosporidium, Theileria, Eimeria, Babesia and the malaria agent Plasmodium falciparum are lacking ICL and MLS genes thus documenting the absence of the glyoxylate cycle. Furthermore, Schlüter et al. (2006) concluded based on their comparative in silico analyses that Apicomplexa are devoid of peroxisomes. However, the genome of the feline pathogen Toxoplasma gondii harbors a catalase (EC 1.11.1.6; XP_002368095), which is regarded as a marker enzyme of peroxisomes. Two contradictory studies suggested either a cytosolic or a peroxisomal localization (Ding et al. 2000; Kaasch and Joiner 2000). Accordingly, and based on the detection of some additional maker proteins, the presence of peroxisomes in apicomplexans is still a matter of debate (Ding et al. 2004; Gabaldón et al. 2016).
The surprising discovery of the apicomplexan algae Chromera velia and Vitrella brassicaformis (Chromeridae; Moore et al. 2008; Janouškovec et al. 2010; Oborník et al. 2012) allowed to investigate the biology of free-living relatives of malaria and toxoplasmosis agents. We established transcriptomes from both chromerids, the peridinin-containing dinoflagellate Prorocentrum minimum and the Manila clam parasite Perkinsus olseni, which represents the most basal lineage of dinoflagellates (Bachvaroff et al. 2014). The distribution of peroxisomal marker genes was investigated in these evolutionary key species and furthermore in alveolate protists whose genomes have been sequenced such as P. falciparum, T. gondii, C. parvum, and Tetrahymena thermophila. We could detect the complete set of peroxins, the five enzymes of the glyoxylate cycle and 35 additional peroxisome-specific enzymes in ciliates, dinoflagellates, and chromerids. Phylogenetic analyses of ICL, MLS, the acetyl-CoA acyltransferase (ACAA1), Pex1 and Pex5 were performed to retrace the evolutionary origin of the glyoxylate cycle, of the beta-oxidation and of peroxisomes in alveolates.
Materials and Methods
Cultivation of Protists and RNA Isolation
The apicomplexan alga V. brassicaformis CCMP3155 was cultivated in L1-medium at 22 °C. 1 l Erlenmeyer flasks were shaken permanently at 100 rpm in New Brunswick Scientific Innova 42 incubator shaker under continuous light. The heterotrophic dinoflagellate P. olseni strain PRA-181 was obtained from Chris Dungan. Cultivation was performed in 5–50 ml 850 mOsm/kg (29ppt) DME: Ham’s F-12 Perkinsus sp. propagation medium (Burreson et al. 2005) containing 3% (v/v) fetal bovine serum (FBS) and 100 U µg/ml penicillin–streptomycin for axenic growth (DME/F12-3ps) within cell culture flasks without shaking and light at 22 °C. Cell pellets were stored in liquid nitrogen and total as well as mRNA was isolated as previously described (Petersen et al. 2014).
Construction of cDNA Libraries, Illumina Sequencing, and Transcriptome Assembly
The preparation of a 300 bp paired-end (PE) Illumina RNA library from P. olseni PRA-181 was performed analogous to the proceedings for the apicomplexan alga C. velia CCAP 1602/1 and the dinoflagellate P. minimum CCMP1329 (Petersen et al. 2014). Illumina sequencing was conducted with both MiSeq and HiSeq 2000 sequencers. The sequences were converted to the FASTQ format and the fastq-mcf tool of ea-utils (Aronesty 2011) was used for a general quality control. Sequence reads of one 150 bp PE MiSeq run were de novo assembled with VELVET 1.2.07 (Zerbino and Birney 2008). The assembled contigs were extended and scaffolded with half of a 101 bp HiSeq run using SSPACE 2.0 (Boetzer et al. 2011).
The isolation of mRNA, normalization, reverse transcription and construction of two 150–350 bp PE Illumina cDNA libraries of V. brassicaformis CCMP3155 was performed by the Vertis Biotechnology AG (Freising, Germany). The normalized library was used for two 150 bp PE MiSeq runs and one 110 bp PE Genome Analyzer IIx (GA) run, whereas the nonnormalized library was used for half of a 101 bp PE HiSeq run. Different assemblies were established and their quality was checked based on the transcript length of glycolytic reference genes. First, all sequence data were de novo assembled with VELVET 1.2.07. Second, the assembled contigs were extended and scaffolded with the 101 bp HiSeq run using SSPACE 2.0, and for the final transcriptome data set both assemblies were merged with MIRA 3.4.1.1 (Chevreux et al. 2004).
Identification of Peroxisome-Specific Genes in Newly Established Transcriptomes and Genome-Sequenced Eukaryotes
The basis of the current study was the reliable identification of peroxisome-specific genes from a broad range of alveolates via BLAST analyses. This aim was challenged by the presence of paralogous proteins from other subcellular compartments (see e.g. Bolte et al. 2011). Accordingly, and in order to exclude false-positive hits, we decided against automated sequence similarity searches with predefined threshold E-values (Žárský and Tachezy 2015). Instead, we used an individual two-step BLAST strategy for the establishment of a manually curated set of peroxisomal proteins. Homologs of peroxisomal query sequences were identified with a species-specific TBLASTN (BLASTP) search and BLASTX searches with these proteins in peroxisome-containing reference species allowed us to differentiate between orthologs and paralogs.
Peroxisomal proteins for BLAST analyses were retrieved from the “peroxisome pathway” of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto 2000) and the PeroxisomeDB (Schlüter et al. 2010). Proteins of the ciliate T. thermophila were preferentially used as query sequences for local and online BLAST searches in our newly established transcriptomes and the alveolate reference genomes, respectively. The E-value threshold for the initial BLAST hits was set to ≤ 1e-04 due to the fast evolutionary rate of many apicomplexan proteins. The peroxisomal inventory of the parasitic Apicomplexa T. gondii and P. falciparum was also investigated in the species-specific genome databases ToxoDB v.12.0 and PlasmoDB v.12.0 (Gajria et al. 2007; Aurrecoechea et al. 2009). PTS sequences were identified with the PSORT II Web server (Nakai and Horton 1999) and the Target Signal Predictor Tool of the PeroxisomeDB (Schlüter et al. 2010).
Phylogenetic Analyses
The alignments generated by ClustalW (Thompson et al. 1997) were manually refined using the ED option of the MUST program package (Philippe 1993). G-blocks were used to eliminate both highly variable and/or ambiguous portions of the alignments (Talavera and Castresana 2007). Maximum likelihood (ML) analyses were performed with RAxML version 7.9.5 (Stamatakis and Alachiotis 2010) under the LG + F+Γ4 model, based on the LG-matrix of amino acid replacements (Le and Gascuel 2008) with empirical amino acid frequencies and four discrete gamma rates. Bootstrap analyses (100 replicates) were performed with RAxML under the same model with the rapid bootstrap option, to estimate the support for internal nodes.
Results
Establishment of cDNA Sequences from C. velia, V. brassicaformis, P. minimum, and P. olseni
We cultivated the apicomplexan alga V. brassicaformis CCMP3155 and the dinophycean Manila clam parasite P. olseni PRA-181, isolated polyA(+) mRNA and constructed PE cDNA libraries for Illumina sequencing. The raw data of 150-bp MiSeq, 110-bp GA, and 101-bp HiSeq runs were used for transcriptome shotgun assemblies (TSAs = cDNA contigs). The construction of the V. brassicaformis library was challenging, but the quality of both TSAs is good and allowed to frequently identify full-length cDNA sequences. The establishment of the high quality TSAs of P. minimum CCMP1329 and C. velia CCAP 1602/1 with cDNA sequences of up to 13 kb has been previously described (Petersen et al. 2014). We screened the four transcriptome data sets of alveolate key species for peroxisomal marker genes and identified 204 cDNA sequences, which have been deposited at DDBJ/EMBL/GenBank under the accession numbers KT633933 and KR704675–KR704725 (C. velia CCAP 1602/1), KR704726–KR704777 (V. brassicaformis CCMP3155), KR704778–KR704835 (P. olseni PRA-181), and KR704836–KR704877 (P. minimum CCMP1329).
Marker Genes of the Glyoxylate Cycle in Ciliates, Dinoflagellates, and Apicomplexa
We started our systematic investigation of the distribution of peroxisomes in Alveolata with TBLASTN searches for the diagnostic enzymes of the glyoxylate cycle ICL and MLS. All five enzymes of this pathway were identified as expected in the genome of the ciliate T. termophila SB210 (table 1), which confirmed the biochemical data of de Duve and Baudhuin (1966). The complete enzyme set could also be identified in the transcriptomes of the peridinin-containing dinoflagellate P. minimum and the heterotrophic clam parasite P. olseni, which represents the most basal branching lineage of dinoflagellates (Bachvaroff et al. 2014). This finding and the presence of ICL and MLS in further EST data sets (Butterfield et al. 2013) as well as the incomplete genome of Symbiodinium minutum (supplementary table S1, Supplementary Material online) is indicative of a common occurrence of the glyoxylate cycle in this group of protists. The pathway is missing in the malaria and toxoplasmosis agents P. falciparum and T. gondii (Apicomplexa) that only contain the three isoenzymes CS, ACO, and MDH of the mitochondrial TCA cycle (table 1, fig. 1B). However, we detected the complete set of enzymes for the glyoxylate cycle including ICL and MLS in our newly established transcriptome data sets of the two apicomplexan algae C. velia and V. brassicaformis. Moreover, we could identify the MLS in the partially sequenced genome of Ascogregarina taiwanensis (Apicomplexa, Conoidasida, Gregarinasina) a parasite of the Asian tiger mosquito (supplementary table S1, Supplementary Material online).
Table 1.
Distribution of Peroxisomal Marker Proteins in Eight Alveolate Key Species
| Protein | Abbr. | Size (aa) | Tt | Po | Pm | Vb | Cv | Cp | Tg | Pf |
|---|---|---|---|---|---|---|---|---|---|---|
| Glyoxylate pathway | ||||||||||
| Isocitrate lyase | ICL | 576 | ✓ | ✓ | ✓ | ✓ | ✓1 | − | − | − |
| Malate synthase | MLS | 562 | ✓ | ✓1 | ✓ | ✓ | ✓1 | − | − | − |
| Citrate synthase | CS | 480 | ✓ | ✓2 | ✓ | ✓2 | ✓ | − | ✓m | ✓m |
| Aconitase | ACO | 898 | ✓C | ✓C | ✓C | ✓C | ✓C | − | ✓C | ✓C |
| Malate dehydrogenase | MDH | 333 | ✓ | ✓ | ✓1 | ✓ | ✓ | ✓C | ✓m | ✓m |
| Peroxisome | ||||||||||
| Biogenesis factor 1 | Pex1 | 1130 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Biogenesis factor 2 | Pex2 | 333 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Biogenesis factor 3 | Pex3 | 400 | − | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Ubiquitin carrier protein | Pex4 | 157 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Biogenesis protein 5 | Pex5 | 728 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Biogenesis protein 6 | Pex6 | 941 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Biogenesis protein 7 | Pex7 | 317 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Biogenesis protein 10 | Pex10 | 381 | ✓ | − | − | ✓ | ✓ | − | ✓ | − |
| Biogenesis factor 11 | Pex11 | 248 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Biogenesis protein 12 | Pex12 | 393 | ✓ | ✓ | (✓) | ✓ | ✓ | − | ✓ | − |
| Biogenesis factor 13 | Pex13 | 304 | − | − | − | − | − | − | − | − |
| Membrane protein 14 | Pex14 | 507 | − | ✓ | (✓) | ✓ | ✓ | − | ✓ | − |
| Membrane protein 15 | Pex15Sc | 383 | − | − | − | − | − | − | − | − |
| Biogenesis factor 16 | Pex16 | 367 | ✓ | ✓ | (✓) | ✓ | ✓ | − | ✓ | − |
| Membrane protein receptor | Pex19 | 248 | ✓ | ✓ | ✓ | ✓ | − | − | − | − |
| Biogenesis protein 22 | Pex22 | 283 | − | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Biogenesis factor 26 | Pex26Hs | 305 | − | − | − | − | − | − | − | − |
| Membrane channel | PMP22 | 283 | − | ✓ | − | ✓ | ✓ | − | − | − |
| Membrane protein 4 | PMP27 | 190 | ✓ | ✓ | − | − | − | − | − | − |
| ATP/ADP-transporter | PMP34 | 331 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| Fatty acid ABC-transporter | PMP70 | 1338 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | − |
| ROS metabolisma | MPV17a | 288 | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓ | ✓ |
Note.—Presence is indicated by a check mark, absence by a dash. Subscripted characters: PTS, peroxisomal targeting sequence; 1 PTS1; 2 PTS2; m mitochondrial; c cytosolic. The protein size in amino acid (aa) positions is shown for the reference species Arabidopsis thaliana. Abbreviations for organisms: Tt, Tetrahymena thermophila; Po, Perkinsus olseni; Pm, Prorocentrum minimum; Vb, Vitrella brassicaformis; Cv, Chromera velia; Cp, Cryptosporidium parvum; Tg, Toxoplasma gondii; Pf, Plasmodium falciparum. (✓) Transcripts are missing in P. minimum, but have been identified in other dinoflagellates (Pex12: Symbiodinium sp., Oxyrrhis marina [FE865168, EG736886]; Pex14: Lingulodinium polyedrum, Alexandrium tamarense [GABP01017358.1, CK432205]; Pex16: Symbiodinium sp. [GBSC01009331.1]). Sc, Saccharomyces cerevisiae; Hs, Homo sapiens.
MPV17, which is involved in the production of reactive oxygen species (ROS), represents in contrast to former conclusions an inner mitochondrial membrane protein in mice (Spinazzola et al. 2006).
In silico prediction of intracellular protein targeting with TargetP gave a mitochondrial localization of the CS and MDH from Plasmodium and Toxoplasma (table 1), which is in agreement with their functional role within the TCA cycle (fig. 1B). Peroxisomal prediction programs allowed us to identify C-terminal and N-terminal targeting sequences for the MLS and CS in P. olseni, for the ICL and MLS in C. velia, and for the CS in V. brassicaformis (table 1). The conservation of peroxisomal targeting sites hence indicates that the intracellular localization of the glyoxylate cycle in dinoflagellates and apicomplexan algae is comparable to those of land plants (fig. 1A). The observed localization of a functional metabolic pathway in two cell compartments is analogous to the close collaboration of mitochondria and peroxisomes in the beta-oxidation of fatty acids (Speijer 2017), which seems to be widely distributed in alveolates (table 2).
Table 2.
Metabolic Inventory of Peroxisomal Pathways in Eight Alveolate Species
| Functional Category | Protein | Abbr. | Tt | Po | Pm | Vb | Cv | Cp | Tg | Pf |
|---|---|---|---|---|---|---|---|---|---|---|
| Fatty acid oxidation | ||||||||||
| α-oxidation | 2-Hydroxyacyl-CoA lyase | HPCL2 | ✓ | − | − | − | ✓1 | − | − | ✓ |
| Phytanoyl-CoA hydrolase | PHYH | ✓2 | ✓ | ✓2 | ✓2 | ✓2 | − | − | − | |
| β-oxidation | α-Methylacyl-CoA-racemase | AMACR | ✓ | ✓ | − | − | − | − | − | − |
| Acyl-CoA-oxidase | ACOXa | ✓ | − | ✓1 | ✓1 | ✓1 | − | ✓1 | − | |
| Multifunctional protein | DBP | ✓1 | ✓1 | ✓1 | ✓1 | ✓1 | − | ✓1# | − | |
| Sterole carrier protein 2 | SCPX | ✓ | ✓1 | ✓ | − | ✓1 | − | ✓# | − | |
| Multifunctional protein | PBE | ✓ | ✓1 | ✓1 | − | − | − | − | − | |
| Acetyl-CoA acyltransferase 1 | ACAA1 | ✓ | ✓2 | ✓2 | ✓2 | ✓2 | − | ✓ | − | |
| 2, 4-dienoyl-CoA reductase | PDCR | ✓ | − | ✓1 | ✓ | ✓2 | − | ✓1 | − | |
| δ(3, 5)-δ(2, 4)-dienoyl-CoA isomerase | ECH | ✓ | ✓ | ✓1 | ✓1, 2 | ✓1 | − | ✓ | − | |
| ATP-binding cassette, subfamily D | ABCD | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | − | |
| Long-chain acyl-CoA synthetase | ACSL | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Solute carrier family 27, member 2 | VLACS | − | ✓ | ✓1 | ✓ | ✓ | − | ✓ | − | |
| Other oxidation | Acyl-CoA thioesterase 8 | PTE | − | ✓1 | − | − | − | − | − | − |
| Nucleoside disphosphate-linked m. | NUDT19 | − | − | − | − | ✓ | − | − | − | |
| Amino acid metabolism | Multifunctional protein | AGT | − | ✓1 | ✓ | ✓ | ✓ | − | ✓ | − |
| d-Amino-acid oxidase | DAOa | ✓ | − | − | − | − | − | − | − | |
| Isocitrate dehydrogenase | IDH | ✓ | ✓ | ✓ | ✓ | ✓ | − | ✓1 | ✓m | |
| N1-acetylpolyamine oxidase | PAOXa | − | ✓1 | ✓ | ✓ | ✓ | − | − | − | |
| l-Pipecolate oxidase | PIPOXa | − | ✓ | ✓ | ✓1 | ✓1 | − | − | − | |
| hydroxymethylgluatryl-CoA lyase | HMGCL | ✓ | − | − | ✓ | ✓ | − | ✓ | − | |
| (S)-2-hydroxy-acid oxidase | HAOa | ✓ | − | − | ✓1 | ✓1 | − | − | − | |
| Antioxidant system | ||||||||||
| Hydrogen peroxide metabolism | Catalase | CAT | ✓1 | − | − | ✓ | − | − | ✓1 | − |
| Superoxide dismutase | SODa | ✓ | ✓ | − | − | − | ✓ | ✓ | ✓ | |
| Nitric-oxide synthase, inducible | INOS | − | − | ✓ | − | − | − | − | − | |
| Peroxiredoxin 1 | PRDX1 | − | ✓ | − | ✓ | ✓ | ✓ | ✓ | ✓1 | |
| Peroxiredoxin 5 | PRDX5 | − | ✓ | ✓ | − | − | − | − | − | |
| Glutathione metabolism | Glutathione S-transferase kappa 1 | GSTK1 | − | ✓ | ✓1 | ✓1 | ✓1 | − | − | − |
| Etherphospholipid biosynthesis | Dihydroxyacetone phosphate acyltr. | DHAPAT | ✓1, 2 | ✓1 | ✓ | ✓ | ✓ | − | ✓ | − |
| Alkyldihydroxyacetonephosphate syn. | AGPS | ✓ | − | ✓2 | ✓2 | ✓ | − | − | − | |
| Fatty acyl-CoA reductase | FAR | ✓1, 2 | − | ✓ | ✓ | ✓ | − | ✓ | − | |
| Purine metabolism | Xanthine dehydrogenase | XDHa | − | ✓ | ✓2 | ✓ | ✓ | − | − | − |
| Retinol metabolism | Dehydrogenase/reductase SDR family | DHRS4 | ✓ | ✓1 | − | ✓1 | ✓1 | − | − | − |
| Sterol precursor biosynthesis | Mevalonate kinase | MVK | ✓2 | − | − | − | − | − | − | − |
| Phosphomevalonate kinase | PMVK | − | − | ✓ | − | − | − | − | − | |
Note.—Presence of proteins is indicated by a check mark, absence by a dash. Subscripted characters show the presence of conserved peroxisomal targeting sequences (PTS1, PTS2). Abbreviations for organisms: Tt, Tetrahymena thermophila; Po, Perkinsus olseni; Pm, Prorocentrum minimum; Vb, Vitrella brassicaformis; Cv, Chromera velia; Cp, Cryptosporidium parvum; Tg, Toxoplasma gondii; Pf, Plasmodium falciparum; 1 PTS1; 2 PTS2; m mitochondrial targeting sequence in Pf.
H2O2 releasing enzymes.
Distribution of Peroxins in Alveolata
The presence of the glyoxylate cycle in alveolate key species indicates that peroxisomes are widely distributed in this superensemble. However, the apparent mitochondrial localization of the pathway in the complex green alga Euglena gracilis and the protist Acanthamoeba castellanii (Ono et al. 2003; Gawryluk et al. 2014) diminishes the predictive value of ICL and MLS for the presence of this organelle. Therefore, we also searched in our established transcriptomes and in public databases for peroxisome-specific proteins according to the respective KEGG pathway. In alveolates we could detect 14 of the 17 referred peroxins (table 1) with a diagnostic core set of eight proteins (Pex1, Pex2, Pex5, Pex6, Pex7, Pex11, Pex12, Pex16). The remaining three peroxins Pex13, Pex15, and Pex26 are generally missing in the sequenced genomes of ciliates, the early branching dinoflagellate P. olseni and Apicomplexa. Eleven peroxins could be identified in the genome of the ciliate T. thermophila that served as a reference for the current study. The absence of Pex3, Pex14, and Pex22 in the genomes of all 15 sequenced ciliates (supplementary fig. S1, Supplementary Material online) clearly documents that respective orthologs are dispensable for the formation of a functional peroxisome in this basal lineage of alveolates. Dinoflagellates comprise a comparable number of peroxins, ten were discovered in P. minimum and all but one in the early branching clam parasite P. olseni. The remaining protein Pex10 is probably not required for peroxisome biogenesis, which shows that the crucial pool of pex genes can even vary between different alveolate lineages. In contrast, we identified 13 marker genes in the transcriptomes of the apicomplexan algae C. velia and could detect the complete set of 14 peroxins in V. brassicaformis (table 1, fig. 2). This finding is contradictory to the former view that apicomplexans are lacking peroxisomes (Schlüter et al. 2006; Gabaldón 2010), but their absence might be still valid for parasitic representatives. Accordingly, we used the Pex proteins of Chromera as query sequences for comprehensive BLAST analyses in the publicly available transcriptome and genome data sets.
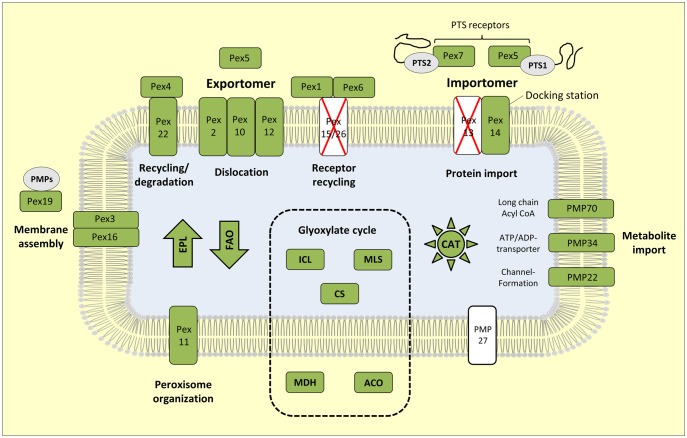
Fig. 2.
Peroxisomal marker proteins of the apicomplexan alga Vitrella brassicaformis CCMP3155. A comprehensive map including peroxisomal functions that corresponds to the KEGG pathway “Peroxisome” is shown for all eight alveolate references species in supplementary figures S1–S8, Supplementary Material online. The presence of orthologous proteins is highlighted in green, their absence is marked in white. Marker proteins of yeast, metazoan, and plants that are lacking in all alveolate species analyzed in the current study are crossed with a red X. PTS, peroxisomal targeting sequence; Pex, Peroxin; PMP, peroxisomal membrane protein; EPL, etherphospholipid biosynthesis; FAO, fatty acid oxidation; CAT, catalase of antioxidant system. The arrows for EPL and FAO reflect the anabolic and catabolic metabolism, respectively.
In P. falciparum and Cryptosporidium parvum, we failed to identify any peroxisome gene with the exception of pex4 and pex22 (table 1, supplementary table S2, Supplementary Material online). A comparable pattern has been observed for all analyzed Plasmodium, Cryptosporidium, Babesia, and Theileria strains as well as Gregarina niphandrodes (supplementary table S1, Supplementary Material online), thus providing strong evidence for the absence of peroxisomes in these genera. The Pex4/Pex22 complex, whose major physiological role is the monoubiquitination of peroxisomal import receptors (El Magraoui et al. 2014), has obviously an alternative function and might be involved in protein degradation. In contrast, we surprisingly identified 13 of 14 apicomplexan pex-genes in T. gondii (table 1, supplementary table S2, Supplementary Material online) and could also detect twelve of them in the closely related avirulent species Hammondia hammondi (supplementary table S1, Supplementary Material online; Walzer et al. 2013). The majority of peroxins is also present in other coccidian parasites such as Sarcocystis neurona, Cyclospora cayetanensis, and two Eimeria strains (Apicomplexa, Conoidasida, Coccidia). In Neospora caninum, a pathogen of dogs (Reid et al. 2012), we could also identify the complete set of 13 peroxins (supplementary table S1, Supplementary Material online). This finding unequivocally documents that peroxisomes are, in clear contradiction to previous conclusions based on 10,000 T. gondii ESTs (Ding et al. 2000), widely distributed among apicomplexan parasites of the class Coccidia. Moreover, the presence of pex1, pex5, and pex7 in the only partially sequenced genome of A. taiwanensis (6.15 of 30 MB; supplementary table S1, Supplementary Material online) strongly indicates that peroxisomes are also present in ascogregarines (Apicomplexa, Conoidasida, Gregarinasina).
Peroxisome-Specific Markers of Alveolates
Peroxisome Biogenesis
The structural components for peroxisome biogenesis include import systems for peroxisomal matrix and membrane proteins, the essential factor for peroxisome biogenesis Pex11 and specific transporters for metabolite import (fig. 2). A comprehensive overview of the distribution of these components among alveolates is provided as supporting information (supplementary S1 Text, Supplementary Material online).
Metabolic Enzymes
Peroxisomes were previously dismissed as the cellular cleaner of ROS, but they exhibit central functions far beyond H2O2 metabolism such as the degradation of fatty acids, metabolism of amino acids and biosynthesis of ether lipids (van den Bosch et al. 1992; Singh 1997; Bussell et al. 2013; Lodhi and Semenkovich 2014). We established a comprehensive inventory of 47 enzymes, which are regarded to be peroxisome-specific according to the respective KEGG map, for all eight alveolate reference species of the current study (supplementary figs. S1–S8, Supplementary Material online). Apart from the glyoxylate cycle key markers ICL and MLS (see above; table 1), we could identify 35 additional enzymes with known peroxisomal functions, but failed to find homologs of the ten remaining ones (table 2, supplementary table S3, Supplementary Material online). The explanatory power of conserved targeting signals for peroxisomal protein import is limited, but our in silico analyses revealed PTS1 and/or PTS2 for nearly half of the enzymes that are supposed to be located in the lumen of this organelle (53/109; table 2). The most common C-terminal tripeptide sequence of alveolates was “-SRL,” with the overall consensus motif (S/A/G)-(R/K/N)-(L/M/A).
The absence of peroxisomes in Plasmodium and Cryptosporidium provided the unique opportunity to calibrate the data set, that is, to differentiate between marker enzymes that are exclusively present in peroxisomal alveolates and ubiquitous equivalents. The latter might represent paralogous isoenzymes, displaying a variable localization in different species or they are per se subjected to dual targeting in different compartments of the cell (Ast et al. 2013).
We investigated the metabolic inventory of the eight alveolate genomes serving as a reference. An overview is presented in table 2 and a more detailed description of the presence of marker enzymes of the antioxidant system, anabolic and catabolic pathways is provided as supporting information (supplementary S2 Text, Supplementary Material online). Apart from sporadically occurring markers, we could identify five enzymes required for fatty acid oxidation (ACOX, DBP, ACAA1, ECH) and etherphospholipid biosynthesis (DHAPAT) that are universally present in the peroxisomal alveolates but missing in Cryptosporidium and Plasmodium (table 2, supplementary table S3, Supplementary Material online). Our analyses revealed the presence of a characteristic metabolic inventory of the peroxisome for the apicomplexan parasite T. gondii (table 2, supplementary table S3, fig. S7, Supplementary Material online), thus providing clues about the functional role of this organelle in apicomplexan parasites. The detection of genes for beta-oxidation of fatty acids and the catalase in related genera such as Ascogregarina (supplementary tables S1 and S4, Supplementary Material online) provided independent evidence, apart from the crucial set of peroxins, for the presence of a functional peroxisome.
Phylogenetic Analyses of Peroxisomal Markers
ICL and MLS
On the basis of our working hypothesis that all peroxisomes of contemporary alveolates descend from the last common ancestor of this superensemble, we investigated the applicability of diagnostic peroxins and metabolic enzymes for phylogenetic analyses. The two key enzymes of the glyoxylate cycle, ICL and MLS (fig. 1), are a priori suitable markers, because of their unique metabolic function, sequence conservation and a considerable length of more than 500 amino acid (aa) positions as well as an universal presence in ciliates, dinoflagellates and photoautotrophic Apicomplexa (table 1, supplementary table S1, Supplementary Material online). However, the phylogenetic RAxML analyses of the ICL and MLS revealed an evolutionary history that is dominated by gene duplication and horizontal gene transfer (HGT; [see supplementary S3 Text, Supplementary Material online]). The patchy distribution of these enzymes could be explained by repeated loss and gain of the glyoxylate cycle in alveolates.
Acetyl-CoA Acetyltransferase 1
Two of five additional diagnostic metabolic markers (see above), that is, DHAPAT and DBP, are occasionally part of fusion proteins (see above; XP_002368897; Lige et al. 2008) and were thus excluded from further analyses. Preliminary phylogenies of the ACOX, and ECH showed an insufficient resolution for alveolates, but the acetyl-CoA acyltransferase 1 (ACAA1; EC 2.3.1.16) for fatty acid beta-oxidation, which exhibits an N-terminal PTS2 in dinoflagellates and chromerids (table 2), was analyzed as a third marker (supplementary S3 Text, Supplementary Material online). The phylogenies document the presence of distinct subtrees comprising apicomplexans, dinoflagellates as well as ciliates, and even the presence of a common alveolate branch obtained moderate bootstrap support (supplementary figs. S9 and S15, Supplementary Material online). The considerable conservation of the ACAA1 and our phylogenetic analyses indicate that a functional beta-oxidation of fatty acids was a crucial driving force for peroxisome maintenance in alveolate evolution (Speijer 2011, 2017).
Peroxin-1 (Pex1) and Peroxin-5 (Pex5)
Our phylogenetic pre-analyses revealed that most peroxins are not suitable for phylogenetic reconstruction essentially due to their small size correlating with an insufficient number of informative sites (table 1). The sole exceptions are the ATPase Pex1 and the protein import receptor Pex5 (fig. 2).
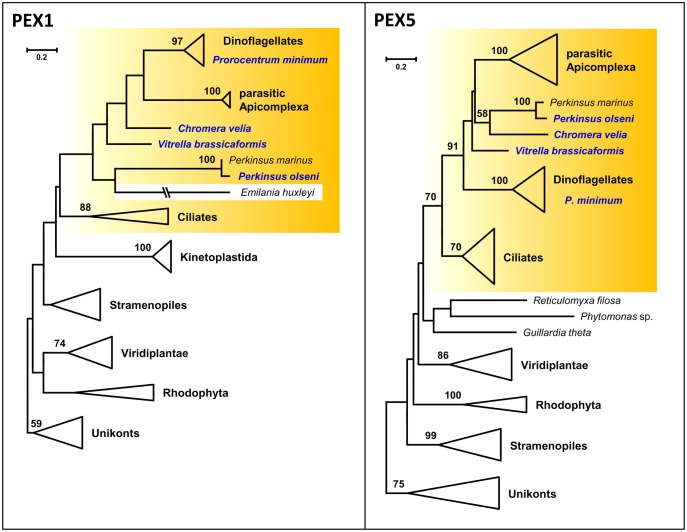
The RAxML tree of Pex1, the Cdc48-homolog of the ERAD/SELMA system that has previously been used as a phylogenetic marker (Bolte et al. 2011; Petersen et al. 2014; Kienle et al. 2016), is based on 60 sequences (fig. 3). However, alveolates exhibit in comparison to the slowly evolving unikonts highly accelerated evolutionary rates (supplementary fig. S16, Supplementary Material online) resulting in a drastic reduction of the alignment size from the large Pex1 proteins in V. brassicaformis (1508 aa; KR704726) and C. velia (1547 aa; KR704675) to only 179 comparable aa positions. The phylogenetic resolution is hence comparably low and moreover bothered by putative long branch attraction artifacts, for example, of Emiliania huxleyi (fig. 3). Nevertheless, the tree of this diagnostic ATPase is compatible with a common origin of alveolate peroxisomes.
Fig. 3.
Phylogenetic maximum likelihood RAxML trees of the peroxisomal biogenesis proteins Pex1 and Pex5. The phylogenetic analyses based on a LG + F + Γ4 model. The Pex1 tree was calculated with 60 sequences and 179 aa positions, the Pex5 tree incorporates 59 sequences with 205 aa positions. Alveolate sequences from our newly established transcriptomes are highlighted in blue. Well-defined subtrees of taxonomic units are collapsed to triangles. Alveolates are highlighted in yellow boxes. The complete phylogenetic trees and subanalyses are shown in supplementary figures S16–S18, Supplementary Material online.
Finally, the RAxML tree of the pivotal peroxisomal receptor Pex5 for peroxisomal protein import (fig. 2), which is based on 59 sequences and 205 aa positions, exhibits a high resolution combined with a moderate evolutionary rate (fig. 3, supplementary fig. S17, Supplementary Material online). Accordingly, the monophyly of alveolates is—in agreement with our working hypothesis—validated with a statistical support of 70% bootstrap proportion (BP). Moreover, the subtrees of ciliates, autotrophic dinoflagellates and parasitic apicomplexans including T. gondii are supported by 70%, 100%, and 100% BP, respectively, and the topology also provides strong support for the monophyly of Apicomplexa and dinoflagellates (Myzozoa, 91% BP; Cavalier-Smith and Chao 2004). The sole incongruence among alveolates is the association of Perkinsea with C. velia (supplementary fig. S17, Supplementary Material online). The weak boostrap support for this grouping even increased in our Pex5 subanalysis of alveolate sequences (58% vs. 84%, supplementary fig. S18, Supplementary Material online), which might either reflect an unresolved phylogenetic artifact or an authentic horizontal recruitment of the chromerid peroxin by Perkinsus. However, this exception does not diminish the value of Pex5 as a phylogenetic marker. Taken together, our comprehensive analyses of peroxisomal marker genes provided clear evidence for a common peroxisomal origin of alveolates and the vertical transmission of this tiny organelle.
Discussion
The Neglected Organelle—Presence and Function of Peroxisomes in Apicomplexa
Apicomplexa have been described as “the first group of organisms devoid of peroxisomes, in the presence of mitochondria” (Schlüter et al. 2006) and the current study was initiated to verify if peroxisomes are always missing in this lineage. We established transcriptome data sets from the algae C. velia and V. brassicaformis (Apicomplexa; Moore et al. 2008; Oborník et al. 2012; Oborník and Lukeš 2015) and detected a large set of peroxisomal markers in both free-living relatives of malaria parasites (tables 1 and 2, supplementary figs. S4 and S5, Supplementary Material online). The amount of peroxins and peroxisome-specific enzymes in Vitrella is comparable to those of land plants (fig. 2, supplementary table S1, Supplementary Material online), leading to the conclusion that their peroxisomes are functionally equivalent. To our great surprise, we could also identify eight diagnostic peroxins and two peroxisomal importers in the genera Toxoplasma, Hammondia, Eimeria, Neospora, Sarcocystis, and Cyclospora (table 1, supplementary table S1, Supplementary Material online). This set of structural proteins provides unequivocal bioinformatic evidence for the presence of a peroxisome in these apicomplexan parasites, thus disproving former conclusions about the general absence of this organelle in Toxoplasma (Ding et al. 2000; Gabaldón 2010), which was however recently questioned again (Gabaldón et al. 2016).
A comprehensive analysis of the D-bifunctional protein (DBP) from T. gondii containing inter alia two sterol carrier protein 2 (SCP-2; XP_002368897) domains for fatty acid beta-oxidation (Lige et al. 2008) is—in the light of the current study—of exceptional relevance, because its subcellular localization including immunogold labeling provided independent cell biological evidence for the presence of a peroxisome. In the current study we identified conserved DBP homologs with the three characteristic domains in Vitrella (KR704749) and several parasitic Apicomplexa including A. taiwanensis (supplementary table S4, Supplementary Material online). Lige and colleagues concluded “The characteristics of the parasite compartment containing [DBP] and cleaved products are compatible with the size of large spherical peroxisomes.” Our comparative genome analyses of Toxoplasma and related genera documented the presence of further peroxisome-specific enzymes for the oxidation of fatty acids, the metabolism of amino acids and the biosynthesis of etherphospholipids (table 2, supplementary tables S1 and S4, Supplementary Material online), thus providing first insights into the metabolic function of this organelle in apicomplexan parasites. A large set of peroxisomal enzymes including the characteristic catalase was detected in the chromerid alga V. brassicaformis CCMP3155 (fig. 2, table 2), which qualifies this free-living protist as a promising model organism to investigate the functional role of peroxisomes in Apicomplexa.
Distribution, Evolution, and Loss of Peroxisomes in Alveolates
The initial introduction of the term “peroxisome” relies on studies with mouse kidney, rat liver and the ciliate T. pyriformis (de Duve and Baudhuin 1966). The group of de Duve anticipated the evolutionary dimension of their observation and noticed: “The finding that similar particles [peroxisomes] are present in a protozoan is of great interest and suggests that they have a long evolutionary history” (Baudhuin et al. 1965). We investigated the distribution of peroxisomal key genes in the superensemble Alveolata and identified them in ciliates, various apicomplexans and dinoflagellates including the distantly-related genus Perkinsus (table 1, supplementary table S1, Supplementary Material online), which documents the abundance of peroxisomes (fig. 4). Phylogenetic analyses showed a limited phylogenetic resolution for most diagnostic genes and proposed a nonvertical evolution of the characteristic glyoxylate cycle (fig. 1, supplementary fig. S9, Supplementary Material online). Our working hypothesis of a common peroxisomal ancestry of alveolates is however supported by three markers including the essential peroxisome receptor Pex5 (fig. 3, supplementary fig. S9, Supplementary Material online). The phylogeny of the acetyl-CoA acetyltransferase 1 (ACAA1; supplementary fig. S9, Supplementary Material online) and the wealth of peroxisomal enzymes for beta-oxidation in Toxoplasma (table 1) confirm that the breakdown of fatty acids is one of the most important functions of peroxisomes that probably originated to lessen mitochondrial ROS formation (Speijer 2011, 2017). The absence of the smallest eukaryotic organelle in malaria and cryptosporidiosis parasites is indicative of two independent losses in the evolution of apicomplexans (Morrison 2009; Wasmuth et al. 2009), that is, one in the common ancestor of Plasmodium, Theileria, and Babesia (Aconoidasida) and another one in the genus Cryptosporidium (Conoidasida; table 1, fig. 4). Peroxisome-loss in alveolates might correlate with the degree of parasitic adaptation to the respective host. A plausible explanation for peroxisome-maintenance in the clam parasite Perkinsus is the protection against ROS stress in free-living zoospores (Sunila et al. 2001). Toxoplasma might require its peroxisome for the host change between cat and men (Wasmuth et al. 2009), whereas the specific adaptation of Cryptosporidium to a single host is accompanied by peroxisome loss and the absence of a reduced plastid (apicoplast).
Fig. 4.
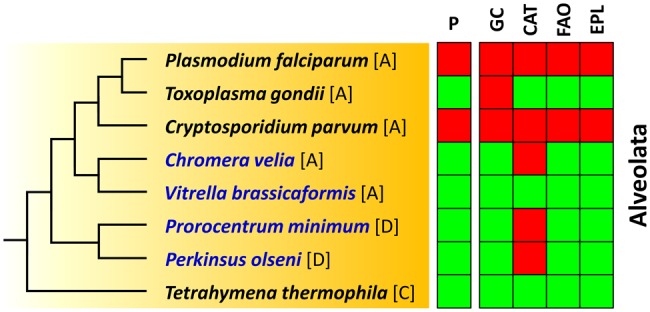
Distribution of peroxisomes and their characteristic metabolic capacities among alveolates. Key species of the current study are shown in blue. The presence and absence of the peroxisome and the most important metabolic traits of this organelle is indicated by green and red boxes, respectively. [A], Apicomplexa; [D], dinoflagellate; [C], ciliate; P, peroxisome; GC, glyoxylate cycle; CAT, catalase; FAO, fatty acid oxidation; EPL, etherphospholipid biosynthesis.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Chris Dungan for hosting V.M. and his great practical tutorial in the cultivation of Perkinsea. Furthermore, we would like to thank Boyke Bunk for his advice regarding sequence submission at GenBank, Richard Hahnke for helpful support, Hervé Philippe for constructive criticism on the manuscript, and two anonymous reviewers for their valuable suggestions. This work including the Ph.D. stipend for A.-K. L.-K. and the position of V.M. was supported by the Deutsche Forschungsgemeinschaft in the project “Von der Malaria bis zum Meeresleuchten—Evolution der Plastiden in Alveolata” (PE 894/2-1).
Literature Cited
- Adl SM. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59(5):429–493.http://dx.doi.org/10.1111/j.1550-7408.2012.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. 2011. Command-line Tools for Processing Biological Sequencing Data. Available from: https://github.com/ExpressionAnalysis/ea-utils.
- Ast J, Stiebler AC, Freitag J, Bölker M.. 2013. Dual targeting of peroxisomal proteins. Front Physiol. 4:297.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, et al. 2009. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37(Database):539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvaroff TR, et al. 2014. Dinoflagellate phylogeny revisited: using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol Phylogenet Evol. 70:314–322.http://dx.doi.org/10.1016/j.ympev.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudhuin P, Müller M, Poole B, de Duve C.. 1965. Non-mitochondrial oxidizing particles (microbodies) in rat liver and kidney an in Tetrahymena pyriformis. Biochem Biophys Res Commun. 20(1):53–59. [DOI] [PubMed] [Google Scholar]
- Bibby BT, Dodge JD.. 1973. The ultrastructure and cytochemistry of microbodies in dinoflagellates. Planta 112(1):7–16.http://dx.doi.org/10.1007/BF00386026 [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Bolte K, et al. 2011. Making new out of old: recycling and modification of an ancient protein translocation system during eukaryotic evolution. BioEssays 33(5):368–376.http://dx.doi.org/10.1002/bies.201100007 [DOI] [PubMed] [Google Scholar]
- Bolte K, Rensing SA, Maier U-G.. 2015. The evolution of eukaryotic cells from the perspective of peroxisomes. BioEssays 37(2):195–203.http://dx.doi.org/10.1002/bies.201400151 [DOI] [PubMed] [Google Scholar]
- Breidenbach RW, Beevers H.. 1967. Association of the glyoxylate cycle enzymes in a novel subcellular particle from castor bean endosperm. Biochem Biophys Res Commun. 27(4):462–469. [pii]. [DOI] [PubMed] [Google Scholar]
- Burreson EM, Reece KS, Dungan CF.. 2005. Molecular, morphological, and experimental evidence support the synonymy of Perkinsus chesapeaki and Perkinsus andrewsi. J Eukaryot Microbiol. 52(3):258–270.http://dx.doi.org/10.1111/j.1550-7408.2005.05-00035.x [DOI] [PubMed] [Google Scholar]
- Bussell JD, Behrens C, Ecke W, Eubel H.. 2013. Arabidopsis peroxisome proteomics. Front Plant Sci. 4:101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield ER, Howe CJ, Nisbet RER.. 2013. An analysis of dinoflagellate metabolism using EST data. Protist 164(2):218–236.http://dx.doi.org/10.1016/j.protis.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1987. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann N Y Acad Sci. 503:55–71.http://dx.doi.org/10.1111/j.1749-6632.1987.tb40597.x [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE.. 2004. Protalveolate phylogeny and systematics and the origins of Sporozoa and dinoflagellates (phylum Myzozoa nom. nov.). Eur J Protistol. 40(3):185–212.http://dx.doi.org/10.1016/j.ejop.2004.01.002 [Google Scholar]
- Chevreux B, et al. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14(6):1147–1159.http://dx.doi.org/10.1101/gr.1917404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao M-C.. 1996. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet. 30:557–578.http://dx.doi.org/10.1146/annurev.genet.30.1.557 [DOI] [PubMed] [Google Scholar]
- Ding M, Clayton C, Soldati D.. 2000. Toxoplasma gondii catalase: are there peroxisomes in Toxoplasma? J Cell Sci. 113:2409–2419. [DOI] [PubMed] [Google Scholar]
- Ding M, Kwok LY, Schlüter D, Clayton C, Soldati D.. 2004. The antioxidant systems in Toxoplasma gondii and the role of cytosolic catalase in defence against oxidative injury. Mol Microbiol. 51(1):47–61. [DOI] [PubMed] [Google Scholar]
- Duhita N, et al. 2010. The origin of peroxisomes: the possibility of an actinobacterial symbiosis. Gene 450(1–2):18–24.http://dx.doi.org/10.1016/j.gene.2009.09.014 [DOI] [PubMed] [Google Scholar]
- de Duve C. 1982. Peroxisomes and related particles in historical perspective. Ann N Y Acad Sci. 386:1–4.http://dx.doi.org/10.1111/j.1749-6632.1982.tb21402.x [DOI] [PubMed] [Google Scholar]
- de Duve C. 2007. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 8(5):395–403.http://dx.doi.org/10.1038/nrg2071 [DOI] [PubMed] [Google Scholar]
- de Duve C, Baudhuin P.. 1966. Peroxisomes (microbodies and related particles). Physiol Rev. 46(2):323–357. [DOI] [PubMed] [Google Scholar]
- de Figueiredo LF, Schuster S, Kaleta C, Fell DA.. 2009. Can sugars be produced from fatty acids? A test case for pathway analysis tools. Bioinformatics 25(1):152–158.http://dx.doi.org/10.1093/bioinformatics/btn621 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Graham IA.. 2001. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6(2):72–77.http://dx.doi.org/10.1016/S1360-1385(00)01835-5 [DOI] [PubMed] [Google Scholar]
- El Magraoui F, et al. 2014. The cytosolic domain of Pex22p stimulates the Pex4p-dependent ubiquitination of the PTS1-receptor. PLoS ONE. 9(8):e105894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R, Kunau W-H.. 1992. A genetic approach to the biogenesis of peroxisomes in the yeast Saccharomyces cerevisiae. Cell Biochem Funct. 10(3):167–174.http://dx.doi.org/10.1002/cbf.290100306 [DOI] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Mertens D, Kunau WH.. 1989. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 86(14):5419–5423.http://dx.doi.org/10.1073/pnas.86.14.5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Rachubinski RA.. 2007. Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol. 23:321–344.http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123456 [DOI] [PubMed] [Google Scholar]
- Gabaldón T, et al. 2006. Origin and evolution of the peroxisomal proteome. Biol Direct. 1:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T. 2010. Peroxisome diversity and evolution. Philos Trans R Soc B Biol Sci. 365(1541):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Capella-Gutiérrez S.. 2010. Lack of phylogenetic support for a supposed actinobacterial origin of peroxisomes. Gene 465(1–2):61–65. [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Ginger ML, Michels PAM.. 2016. Peroxisomes in parasitic protists. Mol Biochem Parasitol. 209(1-2):35–45. [DOI] [PubMed] [Google Scholar]
- Gajria B, et al. 2007. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36(Database):553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawryluk RMR, Chisholm KA, Pinto DM, Gray MW.. 2014. Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants. J Proteomics. 1:400–416. [DOI] [PubMed] [Google Scholar]
- Hayashi M, et al. 2000. Functional transformation of plant peroxisomes. Cell Biochem Biophys. 32(1–3):295–304.http://dx.doi.org/10.1385/CBB:32:1-3:295 [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF.. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122(1):85–95. [DOI] [PubMed] [Google Scholar]
- Hogg JF, Kornberg HL.. 1963. The metabolism of C2-compounds in micro-organisms. 9. Role of the glyoxylate cycle in protozoal glyconeogenesis. Biochem J. 86(3):462–468.http://dx.doi.org/10.1042/bj0860462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, Horák A, Oborník M, Lukes J, Keeling PJ.. 2010. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 107(24):10949–10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasch AJ, Joiner KA.. 2000. Targeting and subcellular localization of Toxoplasma gondii catalase. Identification of peroxisomes in an apicomplexan parasite. J Biol Chem. 275(2):1112–1118. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30.http://dx.doi.org/10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienle N, Kloepper D, Fasshauer D.. 2016. Shedding light on the expansion and diversification of the Cdc48 protein family during the rise of the eukaryotic cell. BMC Evol Biol. 16(1):215..http://dx.doi.org/10.1186/s12862-016-0790-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg HL, Krebs HA.. 1957. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179(4568):988–991.http://dx.doi.org/10.1038/179988a0 [DOI] [PubMed] [Google Scholar]
- Kunze M, Hartig A.. 2013. Permeability of the peroxisomal membrane: lessons from the glyoxylate cycle. Front Physiol. 4:204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze M, Pracharoenwattana I, Smith SM, Hartig A.. 2006. A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim Biophys Acta. 1763(12):1441–1452. [DOI] [PubMed] [Google Scholar]
- Lanyon-Hogg T, Warriner SL, Baker A.. 2010. Getting a camel through the eye of a needle: the import of folded proteins by peroxisomes. Biol Cell. 102(4):245–263. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y.. 1985. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1:489–530.http://dx.doi.org/10.1146/annurev.cb.01.110185.002421 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25(7):1307–1320.http://dx.doi.org/10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Lige B, Jayabalasingham B, Zhang H, Pypaert M, Coppens I.. 2008. Role of an ancestral d-bifunctional protein containing two sterol-carrier protein-2 domains in lipid uptake and trafficking in Toxoplasma. Mol Biol Cell. 20(2):658–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi IJ, Semenkovich CF.. 2014. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19(3):380–392.http://dx.doi.org/10.1016/j.cmet.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham P, Collinge AJ.. 1987. Woronin bodies of filamentous fungi. FEMS Microbiol Lett. 46(1):1–11.http://dx.doi.org/10.1111/j.1574-6968.1987.tb02448.x [Google Scholar]
- Maurino VG, Peterhansel C.. 2010. Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol. 13(3):249–256. [DOI] [PubMed] [Google Scholar]
- McCammon MT, Veenhuis M, Trapp SB, Goodman JM.. 1990. Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J Bacteriol. 172(10):5816–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RB, et al. 2008. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451(7181):959–963.http://dx.doi.org/10.1038/nature06635 [DOI] [PubMed] [Google Scholar]
- Morales J, et al. 2016. Differential remodelling of peroxisome function underpins the environmental and metabolic adaptability of diplonemids and kinetoplastids. Proc R Soc B. 283(1830):1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DA. 2009. Evolution of the Apicomplexa: where are we now? Trends Parasitol. 25(8):375–382. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P.. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 24(1):34–35.http://dx.doi.org/10.1016/S0968-0004(98)01336-X [DOI] [PubMed] [Google Scholar]
- Oborník M, et al. 2012. Morphology, ultrastructure and life cycle of Vitrella brassicaformis n. sp., n. gen., a novel chromerid from the Great Barrier Reef. Protist 163(2):306–323. [DOI] [PubMed] [Google Scholar]
- Oborník M, Lukeš J.. 2015. The organellar genomes of Chromera and Vitrella, the phototrophic relatives of apicomplexan parasites. Annu Rev Microbiol. 69:129–144. [DOI] [PubMed] [Google Scholar]
- Ono K, et al. 2003. Presence of glyoxylate cycle enzymes in the mitochondria of Euglena gracilis. J Eukaryot Microbiol. 50(2):92–96.http://dx.doi.org/10.1111/j.1550-7408.2003.tb00239.x [DOI] [PubMed] [Google Scholar]
- Opperdoes FR, Borst P.. 1977. Localization of nine glycolytic enzymes in a microbody-like organelle in Trypanosoma brucei: the glycosome. FEBS Lett. 80(2):360–364.http://dx.doi.org/10.1016/0014-5793(77)80476-6 [DOI] [PubMed] [Google Scholar]
- Petersen J, et al. 2014. Chromera velia, endosymbioses and the Rhodoplex hypothesis – plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Genome Biol Evol. 6(3):666–684.http://dx.doi.org/10.1093/gbe/evu043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H. 1993. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res. 21(22):5264–5272.http://dx.doi.org/10.1093/nar/21.22.5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieuchot L, Jedd G.. 2012. Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu Rev Microbiol. 66:237–263.http://dx.doi.org/10.1146/annurev-micro-092611-150126 [DOI] [PubMed] [Google Scholar]
- Reddy JK, Mannaerts GP.. 1994. Peroxisomal lipid metabolism. Annu Rev Nutr. 14:343–370.http://dx.doi.org/10.1146/annurev.nu.14.070194.002015 [DOI] [PubMed] [Google Scholar]
- Reid AJ, et al. 2012. Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: Coccidia differing in host range and transmission strategy. PLoS Pathog. 8(3):e1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodin J. 1954. Correlation of ultrastructural organization and function in normal and experimentally changed proximal tubule cells of the mouse kidney. [Doctoral thesis] Stockholm: Karolinska Institute. Akitbolaget Godvil.
- Rucktäschel R, Girzalsky W, Erdmann R.. 2011. Protein import machineries of peroxisomes. Biochim Biophys Acta. 1808(3):892–900. [DOI] [PubMed] [Google Scholar]
- Scheibe R. 2004. Malate valves to balance cellular energy supply. Physiol Plant. 120(1):21–26. [DOI] [PubMed] [Google Scholar]
- Schliebs W, Girzalsky W, Erdmann R.. 2010. Peroxisomal protein import and ERAD: variations on a common theme. Nat Rev Mol Cell Biol. 11(12):885–890.http://dx.doi.org/10.1038/nrm3008 [DOI] [PubMed] [Google Scholar]
- Schlüter A, et al. 2006. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol Biol Evol. 23(4):838–845. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Real-Chicharro A, Gabaldón T, Sánchez-Jiménez F, Pujol A.. 2010. PeroxisomeDB 2.0: an integrative view of the global peroxisomal metabolome. Nucleic Acids Res. 38(Suppl 1):800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnarrenberger C, Martin W.. 2002. Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants: a case study of endosymbiotic gene transfer. Eur J Biochem. 269(3):868–883.http://dx.doi.org/10.1046/j.0014-2956.2001.02722.x [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD.. 2008. The peroxisome: still a mysterious organelle. Histochem Cell Biol. 129(4):421–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I. 1997. Biochemistry of peroxisomes in health and disease. Mol Cell Biochem. 167(1–2):1–29. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Aitchison JD.. 2009. Regulation of peroxisome dynamics. Curr Opin Cell Biol. 21(1):119–126.http://dx.doi.org/10.1016/j.ceb.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Aitchison JD.. 2013. Peroxisomes take shape. Nat Rev Mol Cell Biol. 14(12):803–817.http://dx.doi.org/10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speijer D. 2011. Oxygen radicals shaping evolution: why fatty acid catabolism leads to peroxisomes and neurons do without it: FADH2/NADH flux ratios determining mitochondrial radical formation were crucial for the eukaryotic invention of peroxisomes and catabolic tissue differentiation. BioEssays 33(2):88–99.http://dx.doi.org/10.1002/bies.201000097 [DOI] [PubMed] [Google Scholar]
- Speijer D. 2015. Birth of the eukaryotes by a set of reactive innovations: new insights force us to relinquish gradual models. BioEssays 37(12):1268–1276.http://dx.doi.org/10.1002/bies.201500107 [DOI] [PubMed] [Google Scholar]
- Speijer D. 2017. Evolution of peroxisomes illustrates symbiogenesis. BioEssays 39(9):1700050..http://dx.doi.org/10.1002/bies.201700050 [DOI] [PubMed] [Google Scholar]
- Spinazzola A, et al. 2006. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 38(5):570–575.http://dx.doi.org/10.1038/ng1765 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Alachiotis N.. 2010. Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics 26(12):132–139.http://dx.doi.org/10.1093/bioinformatics/btq205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, Mattie S, Prudent J, McBride HM.. 2017. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542(7640):251–254. [DOI] [PubMed] [Google Scholar]
- Sunila I, Hamilton RM, Dungan CF.. 2001. Ultrastructural characteristics of the in vitro cell cycle of the protozoan pathogen of oysters, Perkinsus marinus. J Eukaryot Microbiol. 48(3):348–361.http://dx.doi.org/10.1111/j.1550-7408.2001.tb00324.x [DOI] [PubMed] [Google Scholar]
- Swapna LS, Parkinson J.. 2017. Genomics of apicomplexan parasites. Crit Rev Biochem Mol Biol. 52(3):254–273.http://dx.doi.org/10.1080/10409238.2017.1290043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.http://dx.doi.org/10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gilson TJ, Plewniak F, Jeanmougin F, Higgins DG.. 1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert NE, Essner E.. 1981. Microbodies: peroxisomes and glyoxysomes. J. Cell Biol. 91(3):271–283.http://dx.doi.org/10.1083/jcb.91.3.271s [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei IJ, Veenhuis M.. 2006. PTS1-independent sorting of peroxisomal matrix proteins by Pex5p. Biochim Biophys Acta. 1763(12):1794–1800. [DOI] [PubMed] [Google Scholar]
- van den Bosch H, Schutgens RBH, Wanders RJA, Tager JM.. 1992. Biochemistry of peroxisomes. Annu Rev Biochem. 61:157–197. [DOI] [PubMed] [Google Scholar]
- van der Leij I, et al. 1992. Isolation of peroxisome assembly mutants from Saccharomyces cerevisiae with different morphologies using a novel positive selection procedure. J Cell Biol. 119(1):153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer KA, et al. 2013. Hammondia hammondi, an avirulent relative of Toxoplasma gondii, has functional orthologs of known T. gondii virulence genes. Proc Natl Acad Sci U S A. 110(18):7446–7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth J, Daub J, Peregrin-Alvarez JM, Finney CAM, Parkinson J.. 2009. The origins of apicomplexan sequence innovation. Genome Res. 19(7):1202–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Hackett JD.. 2011. Dinoflagellate genome evolution. Annu Rev Microbiol. 65:369–387.http://dx.doi.org/10.1146/annurev-micro-090110-102841 [DOI] [PubMed] [Google Scholar]
- Yang M-F, et al. 2009. Proteomic analysis of oil mobilization in seed germination and postgermination development of Jatropha curcas. J Proteome Res. 8(3):1441–1451.http://dx.doi.org/10.1021/pr800799s [DOI] [PubMed] [Google Scholar]
- Žárský V, Tachezy J.. 2015. Evolutionary loss of peroxisomes – not limited to parasites. Biol Direct. 10:74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.http://dx.doi.org/10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.