Abstract
An accumulating number of studies have reported that the expression levels of microRNAs (miRNAs/miRs) are dysregulated in a variety of human cancer types, including renal cell carcinoma (RCC). miRNAs play essential functions in tumorigenesis and the progression of tumors by serving as oncogenes or tumor suppressors. Recently, the expression and functions of miR-138 have been studied in a number of human cancer types; however, its role in RCC remains poorly understood. In the present study, the results revealed that miR-138 was significantly downregulated in RCC cell lines and tissues, and that low expression levels of miR-138 were correlated with histological grade, tumor stage and lymph node metastasis. In functional studies, restoration of miR-138 expression inhibited cell proliferation and invasion of ACHN and A498 cells. In addition, SOX9 was validated as a direct target gene of miR-138 in RCC. SOX9 knockdown inhibited cell proliferation and invasion of RCC, with a similar effect to that induced by miR-138, rendering SOX9 a functional target of miR-138 in the disease. These findings indicate that miR-138 may present a novel target for therapeutic strategies in RCC.
Keywords: microRNA-138, renal cell carcinoma, proliferation, invasion, SOX9
Introduction
Renal cell carcinoma (RCC) is the most common malignant cancer of the kidney in adults, accounting for almost 4% of all tumors (1). It is estimated that there are ~30,000 new cases and 12,000 mortalities due to RCC every year in the USA (2). RCC may be histologically classified into three subtypes, including clear cell RCC, papillary RCC and chromophobe RCC. Clear cell RCC is the most common of these subtypes, accounting for 75 to 80% of RCC cases, and also is the most aggressive (3–5). The standard curative treatment for patients with localized RCC is surgical excision with total nephrectomy. However, in ~30% of cases the tumors have already metastasized at the time of initial presentation (6,7). In addition, the benefit of chemotherapeutic and radiological approaches for RCC is limited, which results in a poor prognosis and a low five-year survival rate (8). It is therefore an urgent requirement to investigate novel molecular biomarkers which may be used for early detection and targeted therapy, based on the developing knowledge of the tumorigenesis and progression mechanisms of RCC.
MicroRNAs (miRNAs/miRs), small non-coding RNA molecules of ~19–25 nucleotides (nt) in length, exist in a number of organisms and are cleaved from 70 to 100-nt-long hairpin pre-miRNA precursors by the enzyme Drosha (9,10). miRNAs suppress gene expression at the post-transcriptional level through imperfect complementary sequence pairing to the 3′ untranslated regions (3′UTRs) of their target genes and inducing mRNA degradation or translational repression (11,12). It is estimated that >30% of human genes are regulated by miRNAs, and that they are involved in a variety of biological functions, including proliferation, differentiation, cell cycle, apoptosis, angiogenesis, invasion and metastasis (13,14). An accumulating number of studies have reported that miRNA expression levels are dysregulated in a variety of human cancer types (15–17). miRNAs play key functions in the initiation and progression of tumors by serving as oncogenes or tumor suppressors, primarily depending on the roles of their target genes (18–20). Therefore, miRNAs may be therapeutic targets for novel treatment strategies against RCC.
In this study, miR-138 expression was observed to be downregulated in RCC cell lines and tissues, and the expression of miR-138 was correlated with histological grade, tumor stage and lymph node metastasis. miR-138 was identified as a tumor suppressor in RCC, by inhibiting RCC cell proliferation and invasion. Furthermore, SOX9 was demonstrated to be a direct and functional target gene of miR-138 in RCC. These findings from the present study indicate that miR-138 may be a potential therapeutic target for RCC.
Materials and methods
Tissue samples and cell lines
Sixty-two pairs of RCC tissues and matched normal adjacent tissues (NATs) were collected from patients with RCC who had undergone surgery at Shandong Provincial Hospital Affiliated to Shandong University, China. The matched NATs were kidney tissues located 2 cm away from the visible tumor lesions. All tissue samples were frozen in liquid nitrogen immediately after resection and stored at −80°C until RNA extraction. This study was approved by the ethics committee of Shandong Provincial Hospital Affiliated to Shandong University.
The human RCC cell lines 786-O, ACHN, A498, OS-RC-2 and Caki-1 were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA), and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.) and 100 mg/l streptomycin (Gibco; Thermo Fisher Scientific, Inc.). The non-malignant SV-40 immortalized renal cell line HK-2 was obtained from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China), and maintained in keratinocyte serum-free medium (Invitrogen; Thermo Fisher Scientific, Inc.). All cells were maintained at 37°C in a humidified incubator (5% CO2).
Cell transfection
The following oligonucleotides were used in the present study: miR-138 mimics, scrambled control (NC), SOX9 siRNA (si-SOX9) and control siRNA (si-NC). Cells were transfected with these oligonucleotides using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA was extracted from cell lines or tissue samples using TRIzol (Invitrogen), following to the manufacturer's protocol. For miR-138 expression, RT-qPCR was performed in a CFX96 Real-Time PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using a One Step SYBR® PrimeScript™ miRNA RT-PCR kit (Takara Bio, Inc., Otsu, Japan). For analysis of SOX9 mRNA, cDNA was synthesized using the Moloney Murine Leukemia Virus Reverse Transcription system (Promega Corporation, Madison, WI, USA), and qPCR was performed with a SYBR-Green I mix (Takara Bio), according to the manufacturer's protocol. The relative expression of miR-138 and SOX9 mRNA was determined using the 2−∆∆Ct analysis method, where U6 was used as an internal control for miR-138 and GADPH for SOX9 mRNA.
MTT assay
The capacity for cellular proliferation was determined with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich; EMD Millipore, Billerica, MA, USA) assay. Cells were plated at 3,000 cells per well in 96-well culture plates. Following transfection for 24, 48, 72 and 96 h, 20 µl MTT solution (5 mg/ml) was added to the wells and incubated for an additional 4 h. The supernatant was then removed and 200 µl dimethylsulphoxide was added to each well to dissolve the precipitate. Finally, absorbance was measured at a wavelength of 490 nm using a microplate reader (model 680; Bio-Rad Laboratories, Inc.).
Transwell invasion assay
The capacity for cellular invasion was evaluated using Transwell chambers (8.0 mm pore size; BD Biosciences, San Jose, CA, USA). The chambers were coated with Matrigel (BD Biosciences) and incubated for another 5 h. Transfected cells were collected 48 h after transfection, and re-suspended in FBS-free DMEM. A total of 1×104 cells were plated into the upper chamber, while culture medium containing 20% FBS in the lower chamber served as a chemoattractant. Following incubation for 48 h, the cells that did not invade through the pores were carefully removed with a cotton swab. The invaded cells were fixed with 95% ethanol, stained with 0.1% crystal violet and washed with phosphate-buffered saline (Gibco; Thermo Fisher Scientific, Inc.). The number of invaded cells was counted in five randomly selected fields using an Eclipse TS 100 microscope (Nikon Corporation, Tokyo, Japan).
Western blot analysis
At 72 h after transfection, total protein was extracted using radioimmunoprecipitation assay lysis buffer (50 mM Tris Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS/1% aprotinin, 50 mM NaF and 0.1 mM Na3VO4) supplemented with a protease inhibitor cocktail (Promega Corporation). After denaturing at 100°C for 5 min, equal amounts of proteins were separated using 10% SDS-PAGE, and then transferred onto polyvinylidene difluoride membranes (EMD Millipore). Subsequently, the membranes were blocked with Tris-buffered saline containing 5% skimmed milk, and probed with primary antibodies, including mouse anti-human monoclonal SOX9 antibody (1:1,000 dilution; sc-166505; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse anti-human monoclonal β-actin antibody (1:1,000 dilution; sc-130301; Santa Cruz Biotechnology, Inc.), at 4°C for 12 h. Finally, protein signals were detected using goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Inc.). Bands were visualized using enhanced chemiluminescence solution (Pierce; Thermo Fisher Scientific, Inc.). The relative expression of SOX9 was determined after normalizing to β-actin.
Luciferase reporter assay
For the luciferase reporter assay, PGL3-SOX9-3′UTR wild-type (Wt) and PGL3-SOX9-3′UTR mutant (Mut) luciferase reporter vectors were synthesized and purified by GenePharma (Shanghai, China). In brief, cells were seeded into 24-well plates at a confluence of 60–70%. Following incubation overnight, cells were transfected with miR-138 mimics or NC together with luciferase reporter vectors using Lipofectamine® 2000 transfection reagent. At 48 h after transfection, luciferase activity was quantified using the Dual-Luciferase Reporter Assay system (Promega Corporation). The results were normalized to Renilla luciferase activity.
Statistical analysis
Data are expressed as the means ± standard deviation from at least three separate experiments, and compared using SPSS version 18.0 software (SPSS, Inc., Chicago, IL, USA). Values of P<0.05 were considered to indicate a statistically significant difference.
Results
miR-138 is downregulated in RCC and is associated with clinicopathological features
First, miR-138 expression was measured in five RCC cell lines (786-O, ACHN, A498, OS-RC-2 and Caki-1) and the non-malignant SV-40 immortalized renal cell line (HK-2). Expression levels of miR-138 were observed to be significantly lower in RCC cell lines than in HK-2 (Fig. 1A, P<0.05). miR-138 expression was also evaluated in RCC tissues and matched NATs. As shown in Fig. 1B, miR-138 was downregulated in RCC tissues compared with matched NATs (P<0.05).
Figure 1.
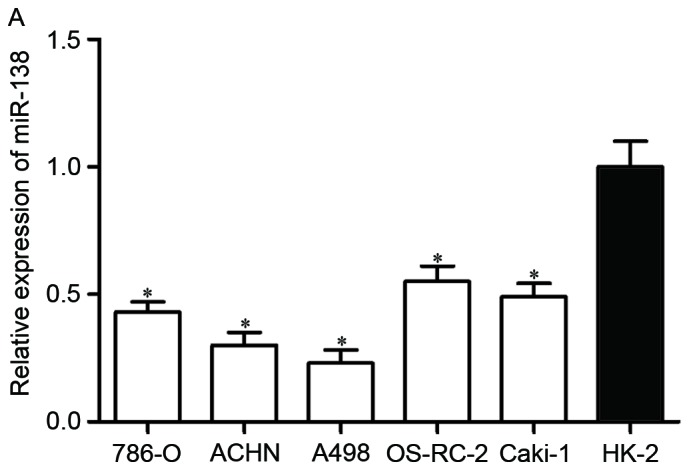
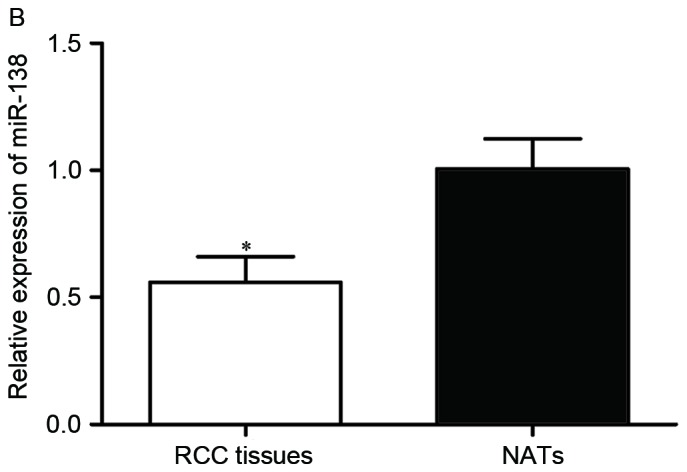
Expression of miR-138 in RCC. (A) Relative expression levels of miR-138 in RCC cell lines. (B) Relative expression levels of miR-138 in RCC tissues and matched NATs. *P<0.05. miR, microRNA; RCC, renal cell carcinoma; NATs, normal adjacent tissues.
Next, the association between miR-138 expression levels and clinicopathological factors of RCC patients was assessed. As shown in Table I, low expression levels of miR-138 were significantly correlated with histological grade, tumor stage and lymph node metastasis (P<0.05). However, there were no notable differences between miR-138 expression and other clinicopathological features in this study (P>0.05). These findings suggest that miR-138 was downregulated in RCC, and that low expression levels of miR-138 may serve a significant role in RCC carcinogenesis and progression.
Table I.
Correlation between miR-138 expression and clinicopathological features.
| miR-138 expression | ||||
|---|---|---|---|---|
| Parameter | Cases | Low | High | P-value |
| Gender | 0.933 | |||
| Male | 28 | 17 | 11 | |
| Female | 34 | 21 | 13 | |
| Age | 0.275 | |||
| <60 years | 36 | 20 | 16 | |
| ≥60 years | 26 | 18 | 8 | |
| Tumor size | 0.213 | |||
| <4 cm | 30 | 16 | 14 | |
| ≥4 cm | 32 | 22 | 10 | |
| Histological grade | 0.001 | |||
| I–II | 32 | 13 | 19 | |
| III–IV | 30 | 25 | 5 | |
| Tumor stage | 0.009 | |||
| T1-T2 | 26 | 11 | 15 | |
| T3-T4 | 36 | 27 | 9 | |
| Lymph node metastasis | 0.016 | |||
| Negative | 32 | 15 | 17 | |
| Positive | 30 | 23 | 7 | |
miR, microRNA.
miR-138 inhibits proliferation and invasion of RCC cells
The significantly low expression of miR-138 in RCC cell lines and tissues prompted the investigation of the contribution of miR-138 in RCC carcinogenesis and progression. To determine this, its expression was increased in RCC cells. miR-138 mimics or NC were transfected into ACHN and A498 cells. Upregulation of miR-138 in ACHN and A498 cells was confirmed by RT-qPCR (Fig. 2A, P<0.05).
Figure 2.
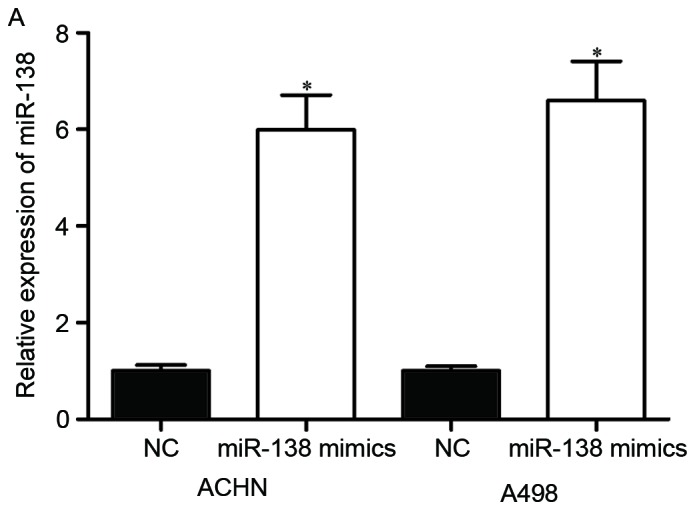
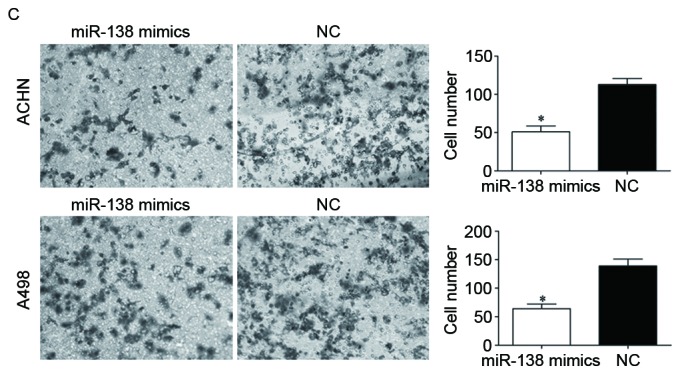
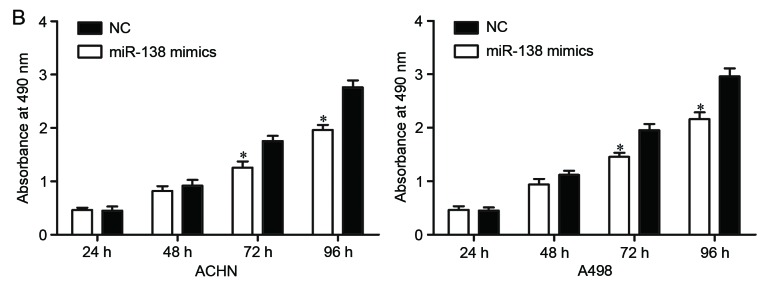
Effects of miR-138 on RCC cell proliferation and invasion. (A) miR-138 expression levels in ACHN and A498 cells transfected with miR-138 mimics or NC were detected by reverse transcription-quantitative polymerase chain reaction. (B) MTT assay was used to investigate the effect of miR-138 on RCC cell proliferation. (C) Transwell invasion assay was performed to assess the effect of miR-138 on RCC cell invasion (×200 magnification). *P<0.05. miR, microRNA; RCC, renal cell carcinoma; NC, negative control.
The effect of miR-138 on RCC cell proliferation was evaluated by MTT assay. As shown in Fig. 2B, ectopic miR-138 expression significantly inhibited the proliferation of ACHN and A498 cells compared with NC (Fig. 2B, P<0.05).
Furthermore, the effect of miR-138 on RCC cell invasion was assessed by Transwell invasion assay. The results revealed that overexpression of miR-138 decreased RCC cell invasion ability (Fig. 2C, P<0.05). This indicated that miR-138 acted as a tumor suppressor in RCC by inhibiting cell proliferation and invasion.
SOX9 is a target gene of miR-138
To elucidate the underlying molecular mechanisms by which miR-138 exerts its suppressive functions, the direct target genes of miR-138 were explored. Using TargetScan (http://www.targetscan.org/vert_60/), SOX9 was observed to be a potential target of miR-138 based on putative target sequences at position 1082–1089 of the SOX9 3′UTR (Fig. 3A).
Figure 3.

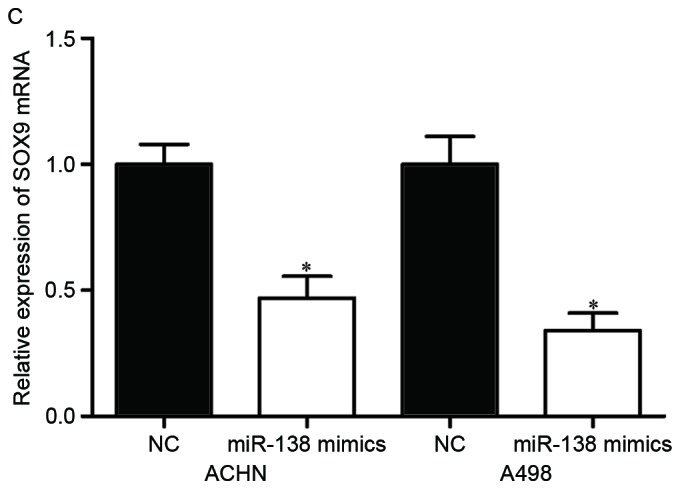
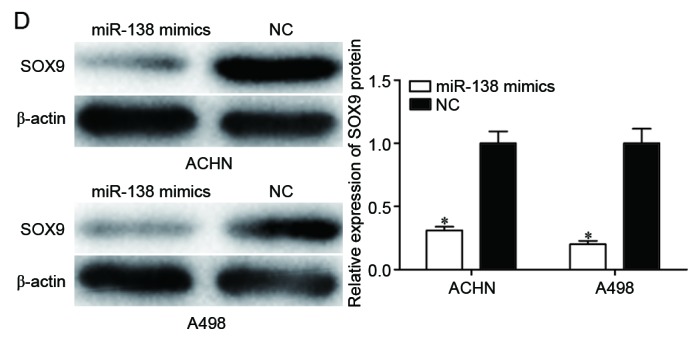
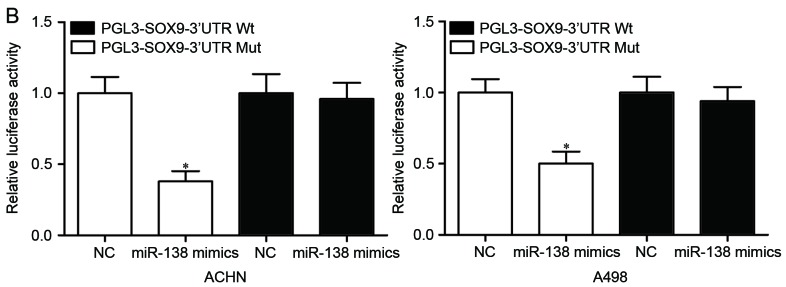
SOX9 is a direct target gene of miR-138. (A) Putative miR-138 targeting site in the SOX9 3′UTR and the generated mutant SOX9 3′UTR. (B) Luciferase activity of PGL3-SOX9-3′UTR Wt and PGL3-SOX9-3′UTR Mut co-transfected with miR-138 mimics or NC were measured. (C) Reverse transcription-quantitative polymerase chain reaction analysis revealed that miR-138 decreased SOX9 mRNA in ACHN and A498 cells. (D) Western blot analysis revealed that miR-138 downregulated SOX9 protein in ACHN and A498 cells. *P<0.05. miR, microRNA; 3′UTR, 3′ untranslated region; Wt, wild type; Mut, mutant; NC, negative control.
To investigate whether SOX9 was a direct target of miR-138, luciferase reporter assay was performed. PGL3-SOX9-3′UTR Wt or PGL3-SOX9-3′UTR Mut luciferase reporter vectors, together with miR-138 mimics or NC, were co-transfected into ACHN and A498 cells. The results revealed that luciferase activity was notably suppressed by miR-138 mimics in PGL3-SOX9-3′UTR Wt (Fig. 3B, P<0.05), whereas the inhibitory effect of miR-138 mimics was abolished in PGL3-SOX9-3′UTR Mut.
To examine the inhibitory effect of miR-138 on SOX9 expression, RT-qPCR and western blot analysis were performed in ACHN and A498 cells transfected with miR-138 mimics or NC. The results revealed that the mRNA (Fig. 3C, P<0.05) and protein (Fig. 3D, P<0.05) levels of SOX9 in ACHN and A498 cells transfected with miR-138 mimics were significantly decreased compared with those in the NC groups. Taken together, these results strongly indicated that SOX9 was a direct target of miR-138 in RCC.
SOX9 knockdown inhibits RCC cell proliferation and invasion
To explore whether SOX9 affected RCC cell proliferation and invasion, si-SOX9 or si-NC was introduced into ACHN and A498 cells. Following transfection, the expression of SOX9 mRNA was significantly downregulated in ACHN and A498 cells transfected with si-SOX9 (Fig. 4A, P<0.05).
Figure 4.
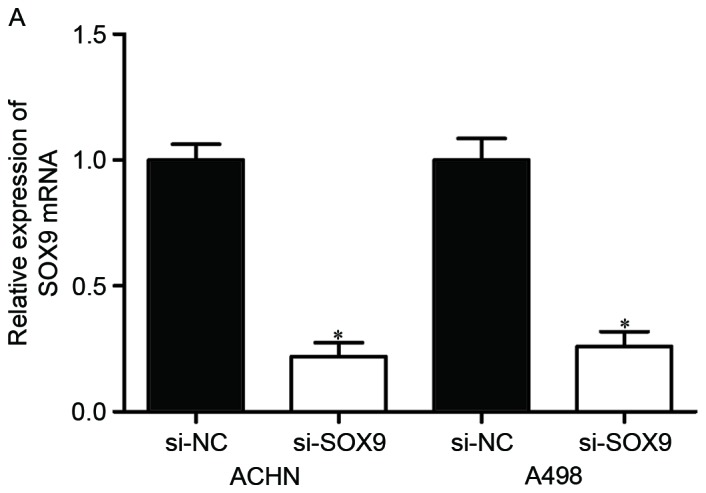
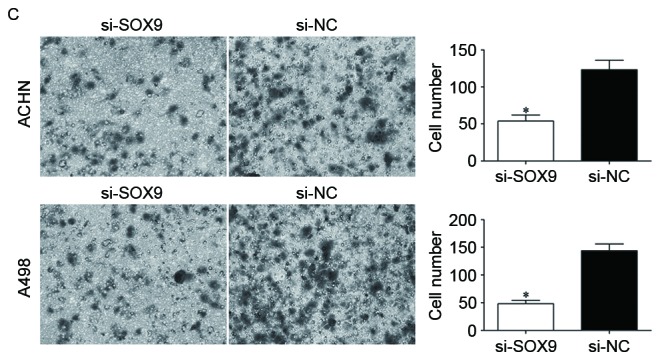
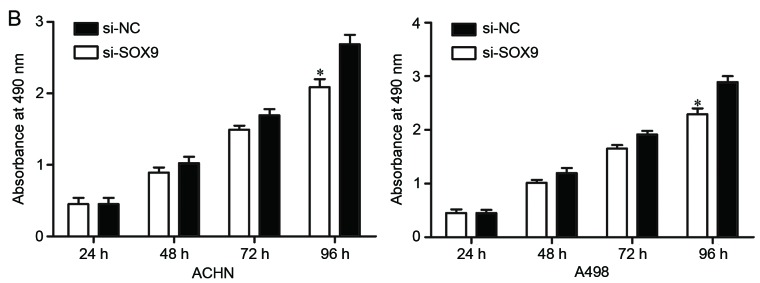
Effects of SOX9 knockdown on RCC cell proliferation and invasion. (A) si-SOX9 downregulated SOX9 mRNA in ACHN and A498 cells. The expression levels of SOX9 mRNA were measured by reverse transcription-quantitative polymerase chain reaction. (B) MTT assay was used to investigate the effect of SOX9 knockdown on RCC cell proliferation. (C) Transwell invasion assay was performed to assess the effect of SOX9 knockdown on RCC cell invasion (×200 magnification). *P<0.05. RCC, renal cell carcinoma; NC, negative control.
To determine the effect of SOX9 knockdown on RCC cell proliferation, MTT assay was performed in ACHN and A498 cells transfected with si-SOX9 or si-NC. As shown in Fig. 4B, cell proliferation was significantly suppressed by si-SOX9 in ACHN and A498 cells (P<0.05).
Transwell invasion assay was adopted to evaluate the effect of SOX9 knockdown on RCC cell invasion. As shown in Fig. 4C, the number of invading cells was significantly decreased in ACHN and A498 cells transfected with si-SOX9 compared with the si-NC groups. Collectively, knockdown of SOX9 notably inhibited RCC cell proliferation and invasion, with a similar effect to that induced by miR-138, rendering SOX9 a functional target of miR-138 in RCC.
Discussion
In the present study, it was revealed that miR-138 was significantly downregulated in RCC cell lines and tissue samples compared with the non-malignant SV-40 immortalized renal cell line and matched NATs, respectively. Decreased expression levels of miR-138 were notably correlated with histological grade, tumor stage and lymph node metastasis in RCC patients. In addition, enforced miR-138 expression reduced cell proliferation and invasion of RCC. Next, SOX9 was validated as a direct target gene of miR-138 in RCC. The effects of SOX9 knockdown on RCC cell proliferation and invasion were similar to those induced by miR-138 overexpression, rendering SOX9 a functional target of miR-138 in RCC. From these data, it was concluded that miR-138 may serve as a potential treatment target to inhibit the growth and metastasis of RCC.
Downregulation of miR-138 has been identified in several cancer types, including breast cancer (21), bladder cancer (22), larynx carcinoma (23), colorectal cancer (24,25), non-small cell lung cancer (26,27), oral squamous cell carcinoma (28), gallbladder carcinoma (29), pancreatic cancer (30), hepatocellular carcinoma (31) and glioblastoma (32). More significantly, miR-138 expression levels were observed to be correlated with clinicopathological features of cancer patients. For example, in breast cancer, miR-138 expression was correlated with lymph node metastasis and invasion (21). In non-small cell lung cancer, low expression levels of miR-138 were significantly associated with advanced tumor-node-metastasis (TNM) stage and positive lymph node metastasis. In addition, non-small cell lung cancer patients with low miR-138 expression levels had a shorter overall survival time than those with high miR-138 expression levels (27). In hepatocellular carcinoma, miR-138 downregulation was significantly correlated with advanced TNM stage and the presence of portal vein invasion and lymph node metastasis. In addition, multivariate survival analysis indicated that low expression of miR-138 was an independent prognostic factor for patients with hepatocellular carcinoma (31). In glioblastoma, high expression levels of miR-138 were associated with a long overall and progression-free survival time (32). These findings suggested that miR-138 may represent a promising prognostic biomarker of human cancer.
Previous studies have reported that miR-138 is involved in tumorigenesis and progression of several tumor types. Zhang et al reported that miR-138 overexpression inhibited breast cancer metastasis and epithelial-mesenchymal transition through negative regulation of vimentin (21). Sun et al noted that miR-138 targeted zinc finger E-box-binding homeobox 2 to suppress bladder cancer invasion and metastases (22). Long et al demonstrated that inhibition of miR-138 in colorectal cancer cells resulted in a significant reduction of migration and invasion capacity by directly targeting TWIST2 (25). In non-small cell lung cancer, miR-138 decreased cell proliferation, migration in vitro and tumor growth in vivo, and increased cisplatin sensitivity through targeting multiple genes, including G protein-coupled receptor kinase interacting ArfGAP 1 (GIT1), semaphorin 4C, cyclin D3, Glucose regulated protein 124 (GRP124), enhancer of zeste homolog 2 and pyruvate dehydrogenase kinase 1 (26,33–35). In oral squamous cell carcinoma, miR-138 inhibited cell proliferation via blockade of yes-associated protein 1 (28). Ma et al revealed that miR-138 inhibited gallbladder carcinoma cell proliferation by targeting BCL2-associated athanogene 1 (29). In pancreatic cancer, miR-138 overexpression reduced pancreatic cancer cell growth through directly targeting forkhead box C1 (30). In hepatocellular carcinoma, restoration of miR-138 expression suppressed cell viability and colony formation, and decreased tumor cell growth in xenograft nude mice by directly targeting cyclin D3 (36). Thus, re-expression of miR-138 in these human cancer types may be a new potential therapeutic approach.
miRNAs serve crucial gene regulatory functions by affecting the expression of multiple genes. To better understand the roles of miR-138 in RCC carcinogenesis and progression, miR-138 target genes were searched. In the present study, SOX9 was identified as a novel direct target gene of miR-138. First, TargetScan analysis revealed that SOX9 was a potential target of miR-138 based on putative target sequences at position 1082–1089 of the SOX9 3′UTR. Second, luciferase reporter assay revealed that luciferase activity was suppressed by miR-138 in PGL3-SOX9-3′UTR Wt, whereas the inhibitory effect of miR-138 was abolished in PGL3-SOX9-3′UTR Mut, indicating that SOX9 could be targeted by miR-138. Third, RT-qPCR and western blot analysis demonstrated an inhibitory effect of miR-138 on SOX9 expression at the mRNA and protein levels. Finally, knockdown of SOX9 notably inhibited RCC cell proliferation and invasion, with an effect similar to that induced by miR-138, rendering SOX9 a functional target of miR-138 in RCC. These results demonstrated that SOX9 was a direct and functional target of miR-138 in RCC. miR-138 repressed RCC cell proliferation and invasion through negative regulation of SOX9 expression.
In conclusion, miR-138 was significantly downregulated in RCC, and low expression levels of miR-138 were associated with histological grade, tumor stage and lymph node metastasis. In addition, miR-138 acted as a tumor suppressor in RCC by inhibiting RCC cell proliferation and invasion. SOX9 was also identified as a direct target gene of miR-138. The miR-138/SOX9-based molecular network may serve a critical role in RCC carcinogenesis and progression, and serve as a novel therapeutic target for patients with RCC.
References
- 1.Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev. 2008;34:193–205. doi: 10.1016/j.ctrv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Peng J, Mo R, Ma J, Fan J. let-7b and let-7c are determinants of intrinsic chemoresistance in renal cell carcinoma. World J Surg Oncol. 2015;13:175. doi: 10.1186/s12957-015-0596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nerich V, Hugues M, Paillard MJ, Borowski L, Nai T, Stein U, Nguyen Tan Hon T, Montcuquet P, Maurina T, Mouillet G, et al. Clinical impact of targeted therapies in patients with metastatic clear-cell renal cell carcinoma. Onco Targets Ther. 2014;7:365–374. doi: 10.2147/OTT.S56370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Williamson SR, Wang M, Davidson DD, Zhang S, Baldridge LA, Du X, Cheng L. Molecular subtyping of metastatic renal cell carcinoma: Implications for targeted therapy. Mol Cancer. 2014;13:39. doi: 10.1186/1476-4598-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youssef YM, White NM, Grigull J, Krizova A, Samy C, Mejia-Guerrero S, Evans A, Yousef GM. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol. 2011;59:721–730. doi: 10.1016/j.eururo.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Motzer RJ, Russo P. Systemic therapy for renal cell carcinoma. J Urol. 2000;163:408–417. doi: 10.1016/S0022-5347(05)67889-5. [DOI] [PubMed] [Google Scholar]
- 7.Kurozumi A, Kato M, Goto Y, Matsushita R, Nishikawa R, Okato A, Fukumoto I, Ichikawa T, Seki N. Regulation of the collagen cross-linking enzymes LOXL2 and PLOD2 by tumor-suppressive microRNA-26a/b in renal cell carcinoma. Int J Oncol. 2016;48:1837–1846. doi: 10.3892/ijo.2016.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): A cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 12.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannakakis A, Coukos G, Hatzigeorgiou A, Sandaltzopoulos R, Zhang L. miRNA genetic alterations in human cancers. Expert Opin Biol Ther. 2007;7:1375–1386. doi: 10.1517/14712598.7.9.1375. [DOI] [PubMed] [Google Scholar]
- 15.Nelson KM, Weiss GJ. MicroRNAs and cancer: Past, present and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 16.Goto Y, Kurozumi A, Enokida H, Ichikawa T, Seki N. Functional significance of aberrantly expressed microRNAs in prostate cancer. Int J Urol. 2015;22:242–252. doi: 10.1111/iju.12700. [DOI] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 18.Dias F, Teixeira AL, Santos JI, Gomes M, Nogueira A, Assis J, Medeiros R. Renal cell carcinoma development and miRNAs: A possible link to the EGFR pathway. Pharmacogenomics. 2013;14:1793–1803. doi: 10.2217/pgs.13.184. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Yu X, Shen J, Jiang Y. MicroRNA dysregulation in uveal melanoma: A new player enters the game. Oncotarget. 2015;6:4562–4568. doi: 10.18632/oncotarget.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv C, Bai Z, Liu Z, Luo P, Zhang J. MicroRNA-495 suppresses human renal cell carcinoma malignancy by targeting SATB1. Am J Transl Res. 2015;7:1992–1999. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Liu D, Feng Z, Mao J, Zhang C, Lu Y, Li J, Zhang Q, Li Q, Li L. MicroRNA-138 modulates metastasis and EMT in breast cancer cells by targeting vimentin. Biomed Pharmacother. 2016;77:135–141. doi: 10.1016/j.biopha.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 22.Sun DK, Wang JM, Zhang P, Wang YQ. MicroRNA-138 regulates metastatic potential of bladder cancer through ZEB2. Cell Physiol Biochem. 2015;37:2366–2374. doi: 10.1159/000438590. [DOI] [PubMed] [Google Scholar]
- 23.Gao S, Wang J, Xie J, Zhang T, Dong P. Role of miR-138 in the regulation of larynx carcinoma cell metastases. Tumour Biol. 2015 Oct 24; doi: 10.1007/s13277-015-4244-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 24.Cristobal I, Torrejon B, González-Alonso P, Manso R, Rojo F, García-Foncillas J. Downregulation of miR-138 as a contributing mechanism to Lcn-2 overexpression in colorectal cancer with liver metastasis. World J Surg. 2016;40:1021–1022. doi: 10.1007/s00268-015-3241-z. [DOI] [PubMed] [Google Scholar]
- 25.Long L, Huang G, Zhu H, Guo Y, Liu Y, Huo J. Down-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2. J Transl Med. 2013;11:275. doi: 10.1186/1479-5876-11-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Zhang H, Zhao M, Lv Z, Zhang X, Qin X, Wang H, Wang S, Su J, Lv X, et al. MiR-138 inhibits tumor growth through repression of EZH2 in non-small cell lung cancer. Cell Physiol Biochem. 2013;31:56–65. doi: 10.1159/000343349. [DOI] [PubMed] [Google Scholar]
- 27.Han L, Zhang G, Zhang N, Li H, Liu Y, Fu A, Zheng Y. Prognostic potential of microRNA-138 and its target mRNA PDK1 in sera for patients with non-small cell lung cancer. Med Oncol. 2014;31:129. doi: 10.1007/s12032-014-0129-y. [DOI] [PubMed] [Google Scholar]
- 28.Xu R, Zeng G, Gao J, Ren Y, Zhang Z, Zhang Q, Zhao J, Tao H, Li D. miR-138 suppresses the proliferation of oral squamous cell carcinoma cells by targeting Yes-associated protein 1. Oncol Rep. 2015;34:2171–2178. doi: 10.3892/or.2015.4144. [DOI] [PubMed] [Google Scholar]
- 29.Ma F, Zhang M, Gong W, Weng M, Quan Z. MiR-138 suppresses cell proliferation by targeting bag-1 in gallbladder carcinoma. PLoS One. 2015;10:e0126499. doi: 10.1371/journal.pone.0126499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu C, Wang M, Li Z, Xiao J, Peng F, Guo X, Deng Y, Jiang J, Sun C. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell Oncol (Dordr) 2015;38:173–181. doi: 10.1007/s13402-014-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang B, Li H, Huang L, Luo C, Zhang Y. Clinical significance of microRNA 138 and cyclin D3 in hepatocellular carcinoma. J Surg Res. 2015;193:718–723. doi: 10.1016/j.jss.2014.03.076. [DOI] [PubMed] [Google Scholar]
- 32.Qiu S, Huang D, Yin D, Li F, Li X, Kung HF, Peng Y. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim Biophys Acta. 2013;1832:1697–1707. doi: 10.1016/j.bbadis.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Ye XW, Yu H, Jin YK, Jing XT, Xu M, Wan ZF, Zhang XY. miR-138 inhibits proliferation by targeting 3-phosphoinositide-dependent protein kinase-1 in non-small cell lung cancer cells. Clin Respir J. 2015;9:27–33. doi: 10.1111/crj.12100. [DOI] [PubMed] [Google Scholar]
- 34.Han LP, Fu T, Lin Y, Miao JL, Jiang QF. MicroRNA-138 negatively regulates non-small cell lung cancer cells through the interaction with cyclin D3. Tumour Biol. 2015;37:291–298. doi: 10.1007/s13277-015-3757-8. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Wang Q, Wen R, Liang J, Zhong X, Yang W, Su D, Tang J. miR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med. 2015;19:2793–2805. doi: 10.1111/jcmm.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis. 2012;33:1113–1120. doi: 10.1093/carcin/bgs113. [DOI] [PMC free article] [PubMed] [Google Scholar]