Abstract
Isolation of DNA using magnetic particles is a field of high importance in biotechnology and molecular biology research. This protocol describes the evaluation of DNA-magnetic particles binding via dynamic light scattering (DLS) and electrophoretic light scattering (ELS). Analysis by DLS provides valuable information on the physicochemical properties of particles including particle size, polydispersity, and zeta potential. The latter describes the surface charge of the particle which plays major role in electrostatic binding of materials such as DNA. Here, a comparative analysis exploits three chemical modifications of nanoparticles and microparticles and their effects on DNA binding and elution. Chemical modifications by branched polyethylenimine, tetraethyl orthosilicate and (3-aminopropyl)triethoxysilane are investigated. Since DNA exhibits a negative charge, it is expected that zeta potential of particle surface will decrease upon binding of DNA. Forming of clusters should also affect particle size. In order to investigate the efficiency of these particles in isolation and elution of DNA, the particles are mixed with DNA in low pH (~6), high ionic strength and dehydration environment. Particles are washed on magnet and then DNA is eluted by Tris-HCl buffer (pH = 8). DNA copy number is estimated using quantitative polymerase chain reaction (PCR). Zeta potential, particle size, polydispersity and quantitative PCR data are evaluated and compared. DLS is an insightful and supporting method of analysis that adds a new perspective to the process of screening of particles for DNA isolation.
Keywords: Chemistry, Issue 129, DNA isolation, magnetic particle, chemical modification, dynamic light scattering, quantitative PCR
Introduction
DNA isolation is one of the most essential steps in molecular biology. The development of nucleic acid extraction methods has great impact on the emerging fields of genomics, metagenomics, epigenetics, and transcriptomics. There is a wide range of biotechnological applications for DNA isolation including medical (forensic/diagnostic tools and prognostic biomarkers), and environmental applications (metagenomic biodiversity, pathogen prevalence, and surveillance). There has been increasing demand to purify and isolate DNA from different materials and in different scales such as blood, urine, soil, wood, and other kinds of samples.1,2,3,4
Nano- and micro-sized particles are suitable for DNA isolation due to their high surface area and particularly when they can be immobilized by a magnetic field. Physicochemical properties of particles, such as size or charge, can greatly influence their ability to bind target biomolecules.5 To further enhance binding of biomolecules and to stabilize particles, different chemical modifications (surface coatings) can be utilized. The many different strategies for binding are classified according to covalent and non-covalent interactions.6 The size of particles directly affects their magnetization properties, whereas particle composition can be tailored by incorporation of metallic, alloy or other materials that can influence its density, porosity, and surface.7 There is no reliable way to measure surface charge of small particles. Instead, electric potential at the slipping plane (some distance away from nanoparticle surface) can be measured.8 This value is called zeta potential and it is a potent tool that is usually used for evaluation of nano- and microparticle stability via DLS.9 Since its value is highly dependent not only on the pH and ionic strength of the dispersive environment, but also on the surface characteristics of the particles, it can also prove the changes in this surface caused by the interaction between the particles and molecule of interest.10
On the other hand, DNA structure in dehydrated conditions (A-DNA form) exhibits compacted conformations that facilitate its precipitation (aggregation) when compared to commonly occurring B-DNA form. Electrostatic (ionic and H-bond) are the major forces controlling the binding of DNA to other materials due to their sterically accessible phosphate and nitrogen bases (particularly guanine).7,10
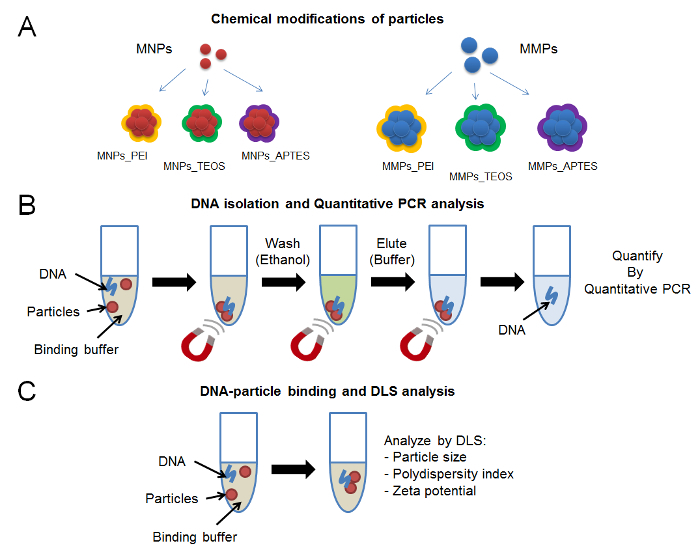
In this work, three representative chemical modifications of magnetic nanoparticles and microparticles are analyzed (Figure 1A). The method of synthesis and chemical modification of nanoparticles and microparticles is described. A binding solution, that accords to theoretical principles of DNA precipitation (pH, ionic strength, and dehydration), is used to evaluate DNA binding and elution. Quantitative PCR is used to evaluate the elution efficiency of DNA from the representative nanoparticles and microparticles (Figure 1B). Particle size, polydispersity index, and zeta potential are important parameters that are used to visualize the physicochemical changes that occur on particle surface (Figure 1C). It is important to emphasize on the chemical characterization of magnetic particle surface. While this step was beyond the scope of this protocol, several modern techniques can be applied to investigate the efficiency of chemical modifications.11,12,13,14 Fourier transform infrared spectroscopy (FTIR) can be used to evaluate the infrared spectrum of particle surface and compare it to the spectrum of free chemical modifiers. X-ray photoelectron spectroscopy (XPS) is another technique that can be used to identify the elemental composition of material surface. Other electrochemical, microscopic and spectroscopic methods can be used to shed the light on the quality of particle synthesis. This work highlights a new perspective to analyzing DNA-magnetic particles interactions via DLS.
Protocol
1. Magnetic Nanoparticles Synthesis
- Synthesis of magnetic nanoparticles stabilized with citrate (MNPs)
- Add 20 mmol of FeCl3 ∙ 6H2O (5.406 g) and 10 mmol of FeCl2 ∙ 4H2O (1.988 g) in 20 mL of deoxygenated double distilled water (ddd water). Stir obtained solution vigorously in beaker under N2 atmosphere using a mechanical stirrer until a transparent solution is obtained.
- Under rapid mechanical stirring, add 150 mL of 1 M NH4OH, drop by drop using dropping funnel (take 1 h or more if needed). Transfer the solution into a screwable vessel after all NH4OH is added. Screw the vessel and heat solution at 75 °C for 30 min. CAUTION: NH4OH is classified as corrosive and dangerous for environment. NOTE: Prepare NH4OH solution from high grade 28% NH4OH in water using deoxygenated water.
- Collect the black precipitate using magnet and wash it using 100 mL of ddd water. Repeat 3 times. Add 50 mL of 0.5 M tri-sodium citrate dihydrate to precipitate. Screw the vessel. NOTE: Use deoxygenated solution of tri-sodium citrate dihydrate.
- Re-disperse the precipitate via sonication (35 kHz, amplitude of 100%) and heat vessel at 80 °C for 1 h. NOTE: A sonicator of sufficient power is needed.
- Let the solution cool to room temperature. Collect precipitate using a magnet and remove supernatant. Re-disperse black mud in 50 mL of ddd water using sonicator (35 kHz, amplitude of 100%).
- Transfer aliquots (~25 mL) into 50 mL centrifuge tubes and add 20 mL of acetone. Centrifuge at 2400 x g for 10 min and remove supernatants.
- Re-disperse precipitate and dialyze solution against water using low molecular weight cut-off dialysis device (500 - 1,000 Da) of suitable volume for 24 h. NOTE: Prepare the dialysis device according to manufacturer's requirements.
- Synthesis of magnetic nanoparticles modified with branched polyethylenimine (MNPs_PEI)
- Add 2 mL of obtained MNPs solution (~0.1 g/mL) into a glass beaker. Add 38 mL of ddd water and sonicate (35 kHz, amplitude of 100%) the suspension (10 min).
- Under rapid mechanical stirring, add 10 mL of 10% branched polyethylenimine (average 25 kDa). Stir solution for subsequent 3 h.
- Collect precipitate using magnet and remove supernatant. Re-disperse precipitate in 20 mL ddd water. Repeat 5 times then sonicate the final suspension (35 kHz, amplitude of 100%).
- Synthesis of magnetic nanoparticles particles modified with silica (MNPs_TEOS)
- Add 10 mL of obtained MNPs solution (~0.1 g/mL) into a glass beaker. Add 60 mL of water and 130 mL of ethanol. Sonicate solution and add 6 mL of 28% NH4OH.
- Under rapid mechanical stirring, add 1 mL of tetraethyl orthosilicate. Stir solution for subsequent 12 h.
- Collect black precipitate using magnet and wash it using 50 mL of ethanol (3 times) and 50 mL of water (3 times). Re-disperse particles in 10 mL of ddd water and sonicate the suspension (35 kHz, amplitude of 100%).
- Synthesis of magnetic nanoparticles modified with amino groups (MNPs_APTES)
- Add 5 mL of obtained MNPs_TEOS solution (~0.1 g/mL) into a glass beaker. Add 45 mL of water and 50 mL of ethanol. Sonicate solution (35 kHz, amplitude of 100%).
- Under rapid mechanical stirring, add 0.5 mL of (3-aminopropyl)triethoxysilane. Stir solution for subsequent 3 h.
- Collect black precipitate using magnet and wash it using 50 mL of ethanol (3 times) and 50 mL of water (3 times). Re-disperse particles in 5 mL of ddd water and sonicate the suspension (35 kHz, amplitude of 100%).
2. Magnetic Microparticles Synthesis
- Synthesis of magnetic microparticles stabilized with citrate (MMPs)
- Add 25 mL of 2 M KNO3, 12.5 mL of 1 M KOH and 205.875 mL of ddd water into a screwable vessel. Add 6.675 mL of 1 M FeSO4 during magnetic stirring. Immediately transfer vessel to a preheated water bath (90 °C) for 2 h. NOTE: Prepare KNO3 and KOH solutions in ddd water.
- Let the solution cool to room temperature. Collect black precipitate using magnet and wash it using 50 mL of ddd water (5 times). Add 50 mL of 0.5 M tri-sodium citrate dihydrate to precipitate. Screw the vessel.
- Redisperse the precipitate via sonication (35 kHz, amplitude of 100%) and heat vessel at 80 °C for 1 h. Collect black precipitate using magnet and wash it using 50 mL of ddd water (10 times).
- Modify MMPs with branched polyethylenimine (MMPs_PEI), silica (MMPs_TEOS) and amino groups (MMPs_APTES) similar to MNPs protocol. Refer to sections 2.2, 2.3 and 2.4, respectively for more details.
- Synthesis of magnetic microparticles modified with branched polyethylenimine (MMPs_PEI)
- Add 5 mL of MMPs solution (~0.1 g/mL) into a glass beaker. Add 38 mL of ddd water and sonicate the solution (35 kHz, amplitude of 100%, 10 min). Under rapid mechanical stirring add 10 mL of 10% branched polyethylenimine (average 25 kDa). Stir solution for subsequent 3 h.
- Collect precipitate using a magnet and remove supernatant. Redisperse precipitate in 20 mL of double distilled water. Repeat 5 times then sonicate the final solution (35 kHz, amplitude of 100%).
- Synthesis of magnetic microparticles modified with silica (MMPs_TEOS)
- Add 5 mL of MMPs solution (~0.1 g/mL) into a glass beaker. Add 60 mL of water and 130 mL of ethanol. Sonicate suspension (35 kHz, amplitude of 100%) and add 6 mL of 28% NH4OH.
- Under rapid mechanical stirring, add 1 mL of tetraethyl orthosilicate. Stir solution for subsequent 12 h.
- Collect black precipitate using a magnet and wash it using 50 mL of ethanol (3 times) and 50 mL of water (3 times). Redisperse particles in 5 mL of ddd water and sonicate the suspension (35 kHz, amplitude of 100%).
- Synthesis of magnetic microparticles modified with amino groups (MMPs_APTES)
- Add 3 mL of MMPs_TEOS solution (~0.1 g/mL) into a glass beaker. Add 45 mL of water and 50 mL of ethanol. Sonicate the suspension (35 kHz, amplitude of 100%).
- Under rapid mechanical stirring, add 0.5 mL of (3-Aminopropyl) triethoxysilane. Stir solution for subsequent 3 h.
- Collect black precipitate using magnet and wash it using 50 mL of ethanol (3 times) and 50 mL of water (3 time). Redisperse particles in 3 mL of ddd water and sonicate the suspension (35 kHz, amplitude of 100%).
3. Magnetic Particle Size Analysis Using DLS
- Particle size in water
- Use a DLS instrument that applies a back scattering detector angle (173°) and a beam wavelength of 633 nm. NOTE: Back scattering DLS is suitable for concentrated samples where multiple scattering can be avoided. Side scattering (detector angle 90°) and forward scattering (detector angle 15°) DLS are suitable for smaller particles and mixed particles, respectively.
- Prior to measurement in water, sonicate particles for 3 min. NOTE: Use diluted solution of the particles. For this purpose, mix 9.6 µL of particles with 96 µL of water.
- Carefully pipette 50 µL of particle solution into polystyrene latex cuvettes and avoid formation of bubbles. NOTE: For each sample, use disposable cuvette (See table of materials).
- Note the clarity of the cuvette and the direction of light beam. Insert the cell in the sample holder and close the chamber.
- Use the following parameters for measurement: temperature: 25 °C; refractive index of dispersive phase (Iron): 2.344; refractive index of dispersive environment (water): 1.333 NOTE: Perform measurement of the refractive index using a refractometer for more accurate results, particularly when developing novel or hybrid materials.
4. Preparing DNA Stock, Magnetic Particles and Binding Buffer
- DNA stock
- Use 10 mM Tris-HCL buffer (pH = 8) for dilution and storage of DNA. Use a standard DNA sample that can be amplified specifically with a direct PCR protocol in the laboratory (Refer to section 5.2 for reaction settings and conditions). To amplify 498 base pair (bp) of the xis gene from bacteriophage λ control, use the following primers. Forward primer: 5'-CCTGCTCTGCCGCTTCACGC-3'; Reverse primer: 5'-TCCGGATAAAAACGTCGATGACATTTGC-3'
- Purify PCR product using commercial DNA purification kit. NOTE: Commercial kits using silica column-based procedure yield > 100 ng/µL purified DNA.
- Measure concentration of DNA using spectrophotometric method (absorbance at 260 nm). Dilute the DNA stock of 1011 copies/µL as instructed above to working solutions of 1010, 109, 108, and 107 copies/µL. NOTE: The number of copies of PCR product can be calculated according to the following equation, assuming average molecular weight is 650 Da for each bp: DNA copies/µL = (quantity in ng/µL * 6.022x1023 copies/mol) / (498 bp * 650 g/mol of bp * 1x109 ng/g)
- Magnetic particles
- Allow particles to sediment without the magnet and remove supernatant. Mix 1: 20 v/v ratio of magnetic particles with ddd water (particles: water). NOTE: Volume-based approach is used to describe quantities of magnetic particles since equal masses of MNPs and MMPs were readily difficult to compare.
- Binding buffer
- For rapid preparation of 0.75 M sodium acetate-HCL solution (pH = 6), mix 1 mL of 3 M sodium acetate with 3 mL of 1 mM HCL. NOTE: All solutions used in binding can be stored at 4 °C. Note that low temperature significantly increases binding particularly in the case of ethanol solution. Carefully control the temperature of solutions for better reproducibility and accuracy of screening.
5. Testing Efficiency of DNA Isolation Using Quantitative PCR
- DNA isolation by modified MMPs or MNPs
- Use a 96-well plate and a magnet customized for 96-well plate for small volume experiments.
- Use the following binding solution mixture: 8 µL modified MMPs or MNPs, 10 µL sodium acetate-HCl (0.75 M, pH = 6), 60 µL of 85% ethanol, 10 µL DNA (1010 copies solution).
- Mix well by pipetting and place the plate on the magnet for 3 min. While the plate is on the magnet, remove the solution and add 100 µL of 85% ethanol for washing.
- Remove ethanol. NOTE: Allow the ethanol to dry for few seconds but do not let the particles over-dry or show cracks. If the particles over-dry they will be difficult to dissolve with elution solution.
- Remove the plate from the magnet and add 40 µL of elution solution. Mix well by pipetting. Place the plate back on the magnet and collect eluted DNA (~37 µL). Store eluent at -20 °C until further analysis.
- Quantitative PCR
- Use the primers that were used to prepare stock DNA in step 4.1.1 to quantify copies of the xis gene, according to the protocol by Haddad et al.10
- Prepare master mix solution according to the amount of samples to be tested, then distribute the mixture into PCR tubes or a white background 96-well plate.
- Use the following quantities for a complete volume of 20 µL PCR mixture: 10 µL polymerase / intercalating dye mixture (see table of materials), 2 µL DNA sample, 2 µL forward primer (10 µM), 2 µL reverse primer (10 µM)
- Set up the thermocycler according to the following program: 95 °C for 120 s, 40 cycles (95 °C for 15 s, 64 °C for 15 s, 68 °C for 45 s), 68 °C for 5 min.
- Using triplicates for standards and samples, calculate the cycle threshold and construct a standard curve to estimate the number of copies of DNA retrieved in each sample. The instrument used here performs this automatically.
- Here, the number of copies of DNA is estimated for total elution volume (~40 µL). Percentage of DNA retrieved corresponding to original amount added to binding solution (totally 1011 copies) can vary beyond the 0 to 100 interval.
6. DNA-magnetic Particles Binding Analysis Using DLS
- Binding of DNA
- For large volume experiments (in 1.5 mL tube), use the following binding solution mixture: 96 µL modified MMPs or MNPs, 120 µL sodium acetate-HCl (0.75 M, pH = 6), 720 µL of 85% ethanol, 120 µL DNA (1010 copies solution) or 120 µL Tris-HCL buffer (10 mM, pH = 8) for control.
- Zeta potential analysis
- Prepare two binding solutions, one with DNA and another with Tris-HCL buffer for control as instructed in step 6.1.1. To avoid formation of bubbles, carefully pipette 800 µL of the binding mixture into a disposable cell. NOTE: Use disposable cuvette for measurement of surface zeta potential (See table of materials).
Use the following parameters for measurement on DLS instrument: Smoluchowski model with an F(ka) of 1.5, Temperature: 25 °C, refractive index of dispersive phase (Iron): 2.344, refractive index of dispersive environment (Water): 1.333. NOTE: The rate of DNA-magnetic particle interaction directly affects the measurement of replicates. A single measurement by the instrument takes 60-70 s. Here, the instrument was instructed to take 10 measurements with 1 min delay intervals. For simplicity, the results will be shown at time = 0, 2, 4, 6, 8, 10, 12, 14, 16 and 18 min from the first reading.
Representative Results
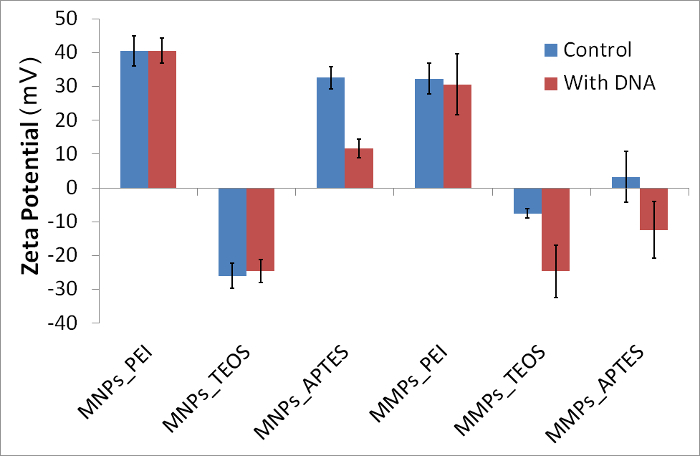
Using the protocol described here for chemical synthesis and modification of magnetic particles, six magnetic particles were synthesized and analyzed for DNA binding. A summary of the analysis is shown in Table 1. By comparing the particle size in water and in binding solution, it is clear that all particles aggregated in binding solution by 2 - 22 folds. Some particles further aggregated to more folds in the presence of DNA; however this was not directly correlated with DNA retrieval detected by quantitative PCR (Table 1). Comparison of zeta potential (10 measurements in ~2 min intervals) of particles in controls versus with DNA showed sharp drop in three particles; namely: MNPs_APTES, MMPs_TEOS and MMPs_APTES (Figure 2). Trends in zeta potential over time were also observed, showing variations in the first three readings before they are stabilized in most cases (Figure 3). Variation after exposure to DNA was mostly observed in MMPs (Figure 2). Lower standard deviation was observed by omitting the first three readings representing first 5 min after exposure of particles to DNA (Table 1).
Overall view of the analysis in Table 1 shows the different characteristics of DNA-particle binding as attributed to magnetic particle size and chemical modifications. Briefly, MNPs_PEI nanoparticles increased in size after binding DNA by four folds with no significant change in polydispersity index or zeta potential. However, DNA was not retrievable in this procedure. MNPs_TEOS nanoparticles increased in size and decreased in polydispersity index with unnoticeable change in zeta potential after DNA exposure, yet they were the best particles in their retrieval rates at 20.3%. MNPs_APTES nanoparticles decreased slightly in size and noticeably in zeta potential upon DNA exposure. MMPs_PEI microparticles did not show change in size upon DNA exposure, however they increased in polydispersity index and decreased slightly in zeta potential. MMPs_TEOS microparticles showed alternative size characteristics to their nanoparticle counterparts i.e. MNPs_TEOS. Surprisingly, they decreased noticeably in zeta potential but not beyond the range displayed by MNPs_TEOS. MMPs_APTES increased in particle size upon DNA exposure and also decreased in zeta potential. It is important to note that both MNPs_APTES and MMPs_APTES retained similar sizes after chemical modification despite the fact that they were made from different sized precursors. They have also displayed similar binding properties. MNPs_TEOS showed no significant change in zeta potential by their values were in the lowest extreme detected in this study.
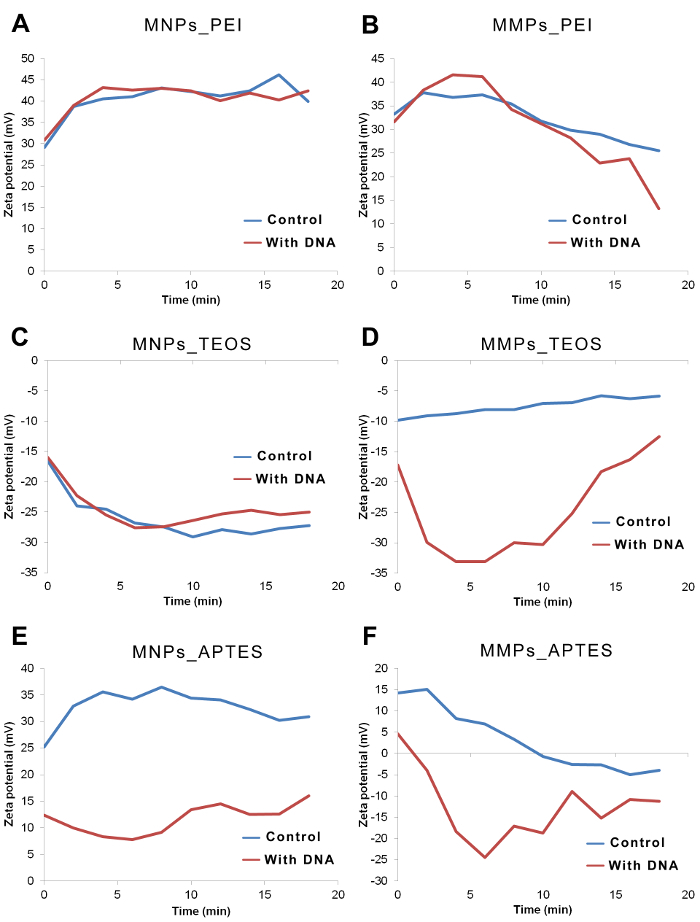
The follow up over time for zeta potential showed stabilizing values after 5 - 10 min, particularly in the case of MNPs (Figure 3A, 3C and 3E). A continuous trend in zeta potential was found in some MMPs readings (Figure 3B, 3D and 3F). In some cases, variations in zeta potential corresponded to variations in polydispersity index and particle size but not with DNA retrieval by elution.
Figure 1: Protocol scheme.(A) Chemical modifications of magnetic particles including branched polyethyleneimine, silica and amino groups. (B) DNA isolation steps using binding buffer and magnetic particles immobilized on magnet, followed by quantitative PCR. (C) DNA-particle binding analysis using DLS. Please click here to view a larger version of this figure.
Figure 2: Zeta potential of representative magnetic particles in presence and absence of DNA. Exposure to DNA caused visible decrease of zeta potential in MNPs_APTES, MMPs_TEOS and MMPs_APTES. Error bars represent standard deviation (N = 10). Please click here to view a larger version of this figure.
Figure 3: Zeta potential of representative magnetic particles over time in presence and absence of DNA. Zeta potential of particles in binding solution was measured at time = 0, 2, 4, 6, 8, 10, 12, 14, 16 and 18 min. Gap between the zeta potential curves of control vs. with DNA are obvious in cases of MMPs_TEOS, MNPs_APTES and MMPs_APTES. (A) Zeta potential of MNPs_PEI is highly positive indicating no interactions. (B) Zeta potential of MMPs_PEI decreased over time suggesting a very slow interaction between particles and DNA. (C) Zeta potential of MNPs_TEOS was very negative in absence and presence of DNA. (D) Zeta potential of MMPs_TEOS shows lower values in presence of DNA when compared to controls. (E) Zeta potential of MNPs_APTES shows lower values in presence of DNA when compared to controls. (F) Zeta potential of MMPs_APTES shows lower values in presence of DNA when compared to controls. Please click here to view a larger version of this figure.
| Characteristics | MNPs_PEI | MNPs_TEOS | MNPs_APTES | MMPs_PEI | MMPs_TEOS | MMPs_APTES |
| Particle Size (nm) | ||||||
| in water | 141.8 | 91.3 | 1106.4 | 825.0 | 1281.3 | 1281.3 |
| in binding solution | 531.2 | 1990.1 | 3091.0 | 2669.0 | 2669.0 | 1990.1 |
| in binding solution with DNA | 1990.1 | 3091.0 | 2669.0 | 2669.0 | 1990.1 | 4145.4 |
| observed change | increased | increased | increased | |||
| Polydispersity index | ||||||
| in water | 0.237 | 0.240 | 0.219 | 0.410 | 0.281 | 0.206 |
| in binding solution | 0.225 | 0.773 | 0.090 | 0.107 | 0.374 | 0.397 |
| in binding solution with DNA | 0.240 | 0.298 | 0.282 | 0.301 | 0.387 | 0.319 |
| observed change | decreased | increased | increased | |||
| Zeta potential (mV)* | ||||||
| in binding solution | 42.29 | -27.83 | 33.23 | 30.80 | -6.86 | -0.66 |
| (±SD) | 1.15 | 1.17 | 2.89 | 8.99 | 7.99 | 5.42 |
| in binding solution with DNA | 41.81 | -25.97 | 12.28 | 27.81 | -23.67 | -15.22 |
| (±SD) | 2.02 | 0.80 | 2.21 | 4.33 | 0.95 | 4.28 |
| observed change | decreased | decreased | decreased | |||
| Quantitative PCR retrieval** | ||||||
| DNA Copy number retrieved | 1.22E+03 | 1.01E+09 | 3.18E+06 | 1.44E+06 | 3.76E+08 | 6.88E+06 |
| (±SD) | 8.05E+02 | 7.55E+07 | 2.83E+06 | 1.98E+06 | 1.08E+08 | 3.33E+06 |
| DNA retrieval percentage | 0.0 | 20.3 | 0.1 | 0.0 | 7.5 | 0.1 |
| (±SD) | 0.0 | 1.5 | 0.1 | 0.0 | 2.2 | 0.1 |
| * Only stable measurements after 5 minutes of binding are shown (N=7). | ||||||
| ** R² = 0.940 for triplicates of standards. |
Table 1: Characterization of representative magnetic particles and their abilities to bind and retrieve DNA. Upon exposure to binding solution, all particles showed aggregation (increase in particle size). When DNA was present in binding solution, particle size increase was visible in MNPs_PEI, MNPs_TEOS and MMPs_APTES. Particles were polydisperse systems in binding solution, particularly MNPs_TEOS which became less polydispersed upon binding of DNA. MNPs_APTES showed increase in polydispersity index and decrease in zeta potential upon exposure to DNA. MMPs_TEOS and MMPs_APTES showed large decrease in zeta potential upon exposure to DNA. MNPs_TEOS and MMPs_TEOS showed the highest DNA retrieval rates of 20.3% and 7.5%, respectively, as shown by quantitative PCR. MNPs_APTES and MMPs_APTES showed 0.1% DNA retrieval rates.
Discussion
In this protocol, the theoretical principles that explain DNA binding to magnetic particles via zeta potential were under question. The protocol describes the synthesis and modification of magnetic nanoparticles and microparticles. Method for preparation of DNA control and binding solution are also described. Two strategies are shown here for screening of DNA-particle interactions: quantitative PCR and DLS approaches. DLS provides three indicators of physicochemical changes in particles: particle size, polydispersity index and zeta potential. Particle size changes directly indicate aggregation or disintegration. Polydispersity index value describes the distribution of particle species in the system ranging from monodispersed (index < 0.05) to highly polydispersed (index >0.7). Zeta potential can classify particles according to surface charge where highly positive particles show values greater than 30 mV and highly negative particles show values lower than -30 mV. As predicted, the zeta potential of polycharged magnetic particles in binding solution relatively decreased when the particles were exposed to DNA. The exception to this rule is MNPs_TEOS. These particles only differ in that they possess a unique highly negative surface charge (Figure 2). This suggests that ionic strength played an important role in holding the particles, DNA and positive ions together. On the other hand, the changes in particle size can indicate the role of precipitation/aggregation of DNA-particles during the binding process, particularly in presence of longer DNA strands.
During synthesis of magnetic particles and chemical modifications, there are several critical steps which require attention: firstly, the reproducibility of the method of synthesis depends greatly on accurate quantities of chemical precursors used in synthesis as well as good stirring and timely controlled addition of NH4OH. To acquire a homogenized population of particles (of similar size, shape, density and composition), it is important to follow sonication and stirring steps thoroughly when instructed. Secondly, in many realistic cases the chemical modifications do not predict the reality of what is on the surface of magnetic particles. A comprehensive characterization step is required to confirm the chemical composition of the particle surface, as described in the introduction section. Among the commonly used techniques, researchers rely on FTIR, XPS, electrochemistry, spectroscopy as well as DLS.
In addition, several critical steps are required during the DLS measurements and analysis: firstly, the choice of binding solution affects various steps including chemical compatibility with particles and DNA and also spectral properties. Choice of refractive indices of binding solution and particle material can be based on previous studies or evaluated by refractometry. Secondly, interactions between the particles and the electrodes in the cuvettes should be monitored carefully as oxidation/reduction process can affect measurement. Thirdly, as shown in Figure 3, the rate of binding can be a function of time. It is critical to observe the stability of interaction, and this is a major advantage for application of DLS in such studies. Here, the observer is monitoring particle aggregation and physiochemical changes in real-time. Fourthly, polydispersity of the system can set limitations to the scope of analysis. Polydispersity index value below 0.05 describes a monodispersed system while values above 0.7 describe a polydispersed system of various sizes of particles. The size of particles and distribution of various particle species can be studied using various types of DLS, including back-scattered, side-scattered and forward scattered, as mentioned in protocol section 3.1. Finally, sonication of particles prior to testing can obviously affect their physical properties (e.g. size and surface area) and if done excessively it can cause release of some chemical modifications.
In this protocol, information acquired via DLS is compared to data acquired by quantitative PCR, also known as real-time PCR. The latter is the gold standard for detection and quantitation of DNA.15 PCR technique is widely used in laboratories, hospitals and universities. However, when testing DNA exposure to novel and synthesized materials it is important to take the chemistry of PCR into consideration. Polymerase enzyme, the key element of polymerase chain reaction, is sensitive to certain chemical inhibitors and enhancers. Addition of internal controls, testing of the synthetic materials, and good laboratory practice can help reduce biases in PCR results. Mechanisms of action for inhibitors and enhancers vary between chemicals that directly bind to polymerase and others that chelate ions from the Mg2+-dependent polymerase in solution. Some insights on DNA isolation are difficult to get using PCR screening. DLS can show cases when DNA binds to particles but is rather washed or does not elute (DNA not detected by PCR). For example, both MNPs_APTES and MMPs_APTES showed significant decrease in zeta potential upon DNA exposure (Table 1) but the DNA detected in elution did not correlate with these results. It is possible to modify the binding, washing, elution solutions to control the release of DNA from particles. DLS is a relatively faster method for evaluation of DNA-particle interactions. The stability and rate of interaction can also be observed by this method (Figure 3), which can impact the time used in magnetic particles- based DNA isolation protocols.
The challenges for using DLS in analysis are straight forward. This technique requires a relatively large sample volume for analysis (~1 mL) when compared to PCR. Some parameters like refractive indices require careful evaluation when using new solutions in the samples. Furthermore, it is very common to observe oxidation/reduction reactions on the electrodes of cuvettes when testing novel material. Careful assessment and knowledge of the chemical composition of the sample can help evaluate the feasibility of re-using the same cuvette. It is worth mentioning that DLS analysis is limited to the specificity and sensitivity provided by PCR, yet it provides different kind of insight to the DNA-particle binding process.
Like all spectroscopic techniques, modifications and troubleshooting of DLS aim for development of more accurate and sensitive measurements. This can be achieved by providing better understanding of the nature of the material tested physically and chemically. In other words, optical properties and details of chemical composition are the key to gain accurate and sensitive data from DLS. Optical properties can be acquired by refractometry, while chemical composition of particles / particles' surface can be studied by various techniques such as FTIR, XPS, electrochemistry and other spectroscopic methods. In this protocol, binding solution can be improved by using various ionic strengths, other dehydrating agents (e.g. isopropanol), various pH and low temperatures. On the other hand, the chemical modifications of particle surface are left to the chemists' imagination.
The development of materials for isolation of DNA will continue to rise in the next decades with more focus on specificity, purity and limit of detection. The quality of extraction methods plays important role in studies of single cell scale, metagenomic scales and samples of unusual origins. At the present time, it is important to perform DLS analysis of nanoparticles and microparticles modified with various kinds of materials. This step is required before establishing a strong theoretical model that describes the behavior of DNA-particle interactions.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The financial support by Czech Science Foundation (project GA CR 17-12816S) and CEITEC 2020 (LQ1601) is greatly acknowledged.
References
- Kulinski MD, et al. Sample preparation module for bacterial lysis and isolation of DNA from human urine. Biomed Microdevices. 2009;11(3):671–678. doi: 10.1007/s10544-008-9277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonen AJM, et al. Comparison of Pathogen DNA Isolation Methods from Large Volumes of Whole Blood to Improve Molecular Diagnosis of Bloodstream Infections. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0072349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi N, Slater GF, Fulthorpe RR. Comparison of commercial DNA extraction kits for isolation and purification of bacterial and eukaryotic DNA from PAH-contaminated soils. Can J Microbiol. 2011;57(8):623–628. doi: 10.1139/w11-049. [DOI] [PubMed] [Google Scholar]
- Rachmayanti Y, Leinemann L, Gailing O, Finkeldey R. DNA from processed and unprocessed wood: Factors influencing the isolation success. Forensic Sci Int Genet. 2009;3(3):185–192. doi: 10.1016/j.fsigen.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Munir MT, Umar S, Shahzad KA, Shah MA. Potential of Magnetic Nanoparticles for Hepatitis B Virus Detection. J Nanosci Nanotechnol. 2016;16(12):12112–12123. [Google Scholar]
- Ulbrich K, et al. Targeted drug delivery with polymers and magnetic nanoparticles: covalent and noncovalent approaches, release control, and clinical studies. Chem Rev. 2016;116(9):5338–5431. doi: 10.1021/acs.chemrev.5b00589. [DOI] [PubMed] [Google Scholar]
- Pershina AG, Sazonov AE, Filimonov VD. Magnetic nanoparticles-DNA interactions: design and applications of nanobiohybrid systems. Rus Chem Rev. 2014;83(4):299. [Google Scholar]
- Xu RL. Progress in nanoparticles characterization: Sizing and zeta potential measurement. Particuol. 2008;6(2):112–115. [Google Scholar]
- Krickl S, Touraud D, Kunz W. Investigation of ethanolamine stabilized natural rubber latex from Taraxacum kok-saghyz and from Hevea brasiliensis using zeta-potential and dynamic light scattering measurements. Ind Crops Prod. 2017;103:169–174. [Google Scholar]
- Haddad Y, et al. The Isolation of DNA by Polycharged Magnetic Particles: An Analysis of the Interaction by Zeta Potential and Particle Size. Int J Mol Sci. 2016;17(4) doi: 10.3390/ijms17040550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio-Neto ET, et al. Submicron magnetic core conducting polypyrrole polymer shell: Preparation and characterization. Mater Sci Eng C Mater Biol Appl. 2016;61:688–694. doi: 10.1016/j.msec.2015.12.052. [DOI] [PubMed] [Google Scholar]
- Baharvand H. Encapsulation of ferromagnetic iron oxide particles by polyester resin. e-Polym. 2008;8(1):1–9. [Google Scholar]
- Ghorbani Z, Baharvand H, Nezhati MN, Panahi HA. Magnetic polymer particles modified with beta-cyclodextrin. J Polym Res. 2013;20(7) [Google Scholar]
- Heger Z, et al. Paramagnetic Nanoparticles as a Platform for FRET-Based Sarcosine Picomolar Detection. Sci Rep. 2015;5 doi: 10.1038/srep08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro E, Serrano-Heras G, Castaño MJ, Solera J. Real-time PCR detection chemistry. Clin Chim Acta. 2015;439:231–250. doi: 10.1016/j.cca.2014.10.017. [DOI] [PubMed] [Google Scholar]


